Submitted:
17 September 2024
Posted:
18 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
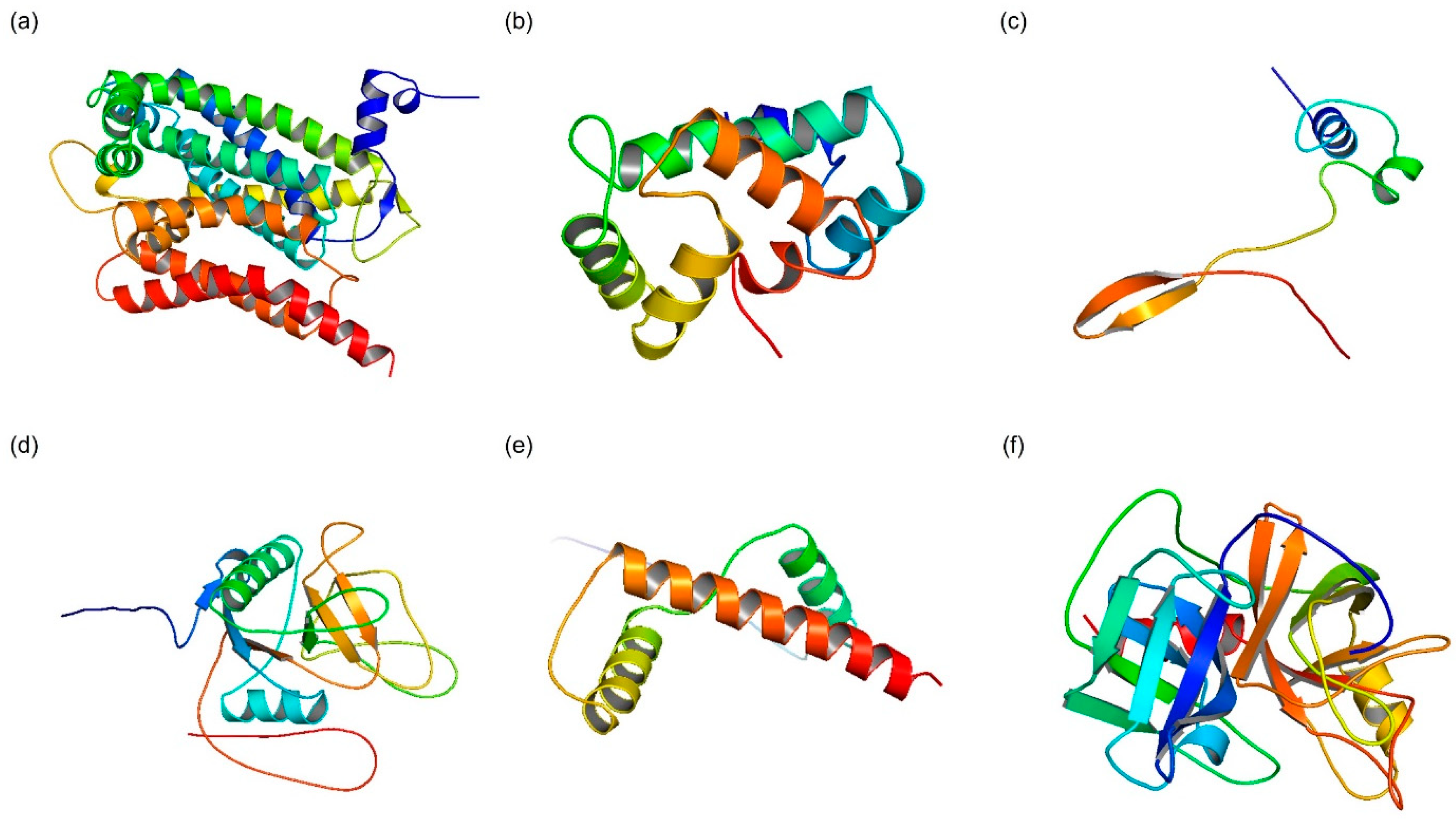
2.1.3. D Structure Modeling
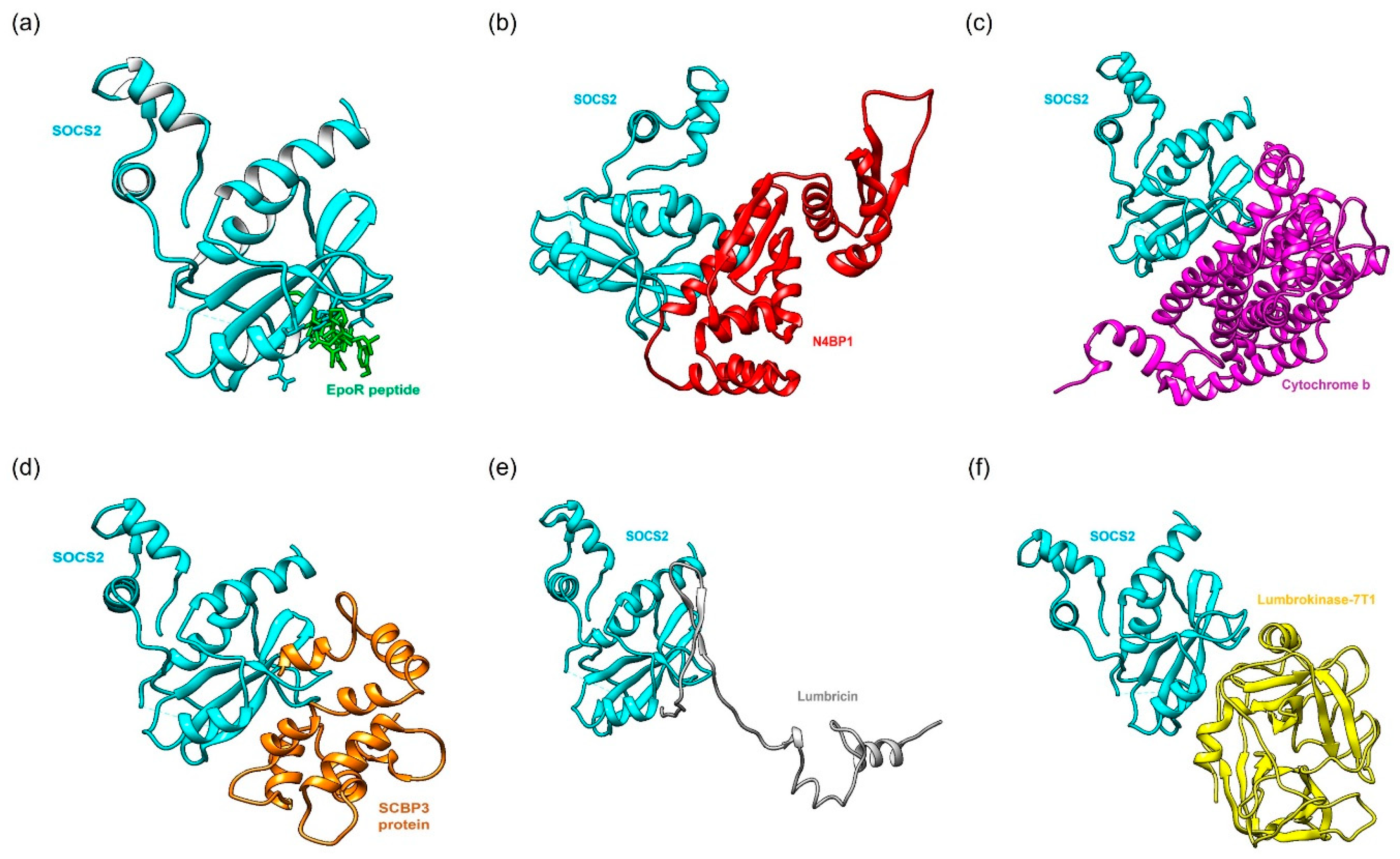
2.2. Protein-Protein Docking Simulation
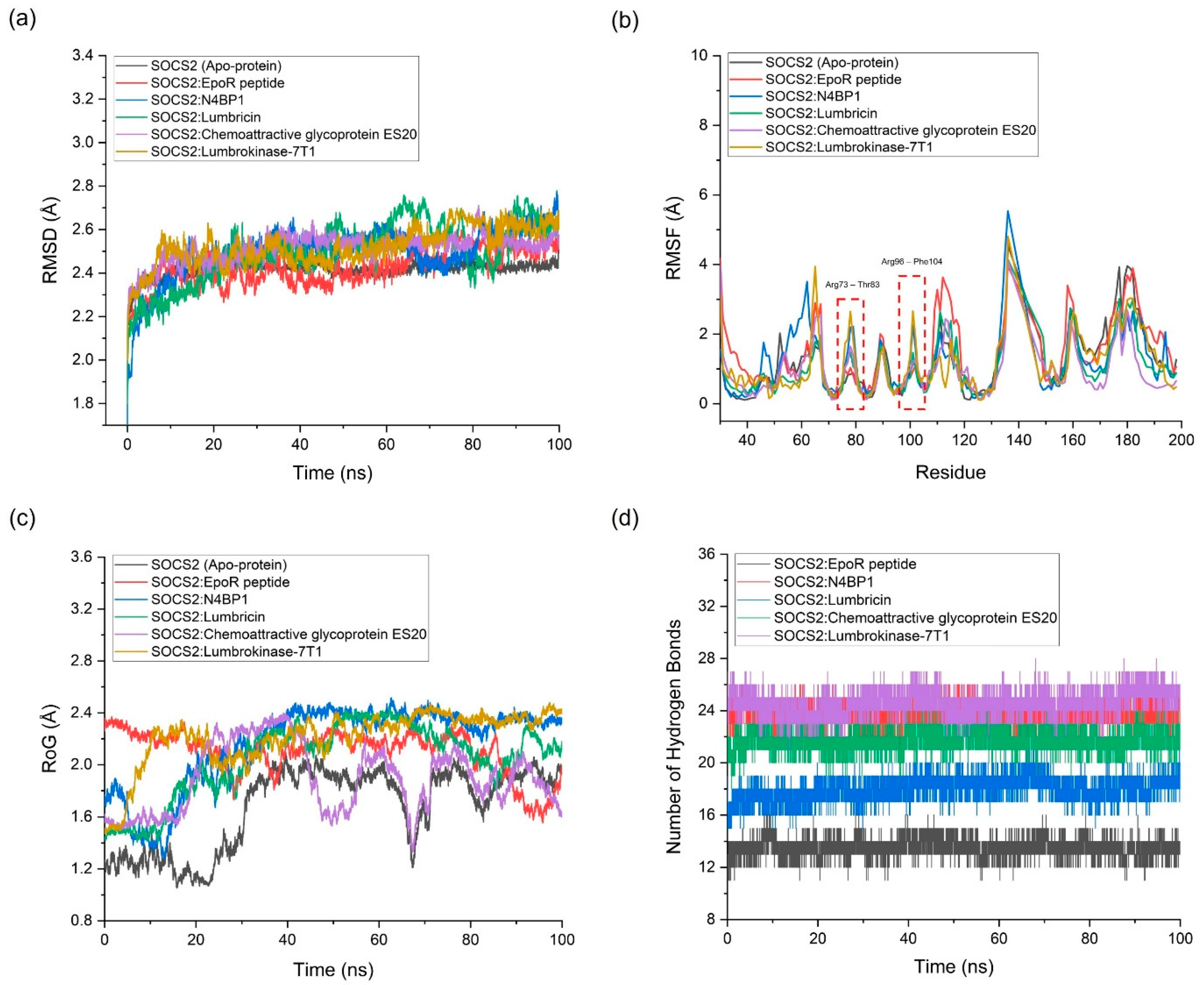
2.3. Molecular Dynamics (MD) Simulation
2.4. Molecular Mechanics/Poisson–Boltzmann Surface Area (MM/PBSA) Calculations
3. Discussion
4. Limitations, Clinical Implications, And Future Works
5. Materials and Methods
5.1. Materials
5.2. Computing Power
5.3. 3D Structure Modeling
5.4. Protein-Protein Docking Simulation
5.5. Molecular Dynamics (MD) Simulation
5.6. Molecular Mechanics/Poisson–Boltzmann Surface Area (MM/PBSA) Calculations
- ΔG_binding: the binding free energy associated with the formation of the protein-protein complex.
- ΔG_complex: the free energy of the fully solvated protein-protein complex.
- ΔG_proteinX1: the free energy of protein 1 in its solvated state when unbound.
- ΔG_proteinX2: the free energy of protein 2 in its solvated state when unbound.
5.7. Statistical Analysis
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Cardiovascular diseases (CVDs): World Health Organization; 2021 [cited 2024 01th April]. Available from: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds).
- Roth Gregory A, Mensah George A, Johnson Catherine O, Addolorato G, Ammirati E, Baddour Larry M, et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019. J Am Coll Cardiol. 2020;76(25):2982-3021. [CrossRef]
- Del Buono MG, Montone RA, Camilli M, Carbone S, Narula J, Lavie CJ, et al. Coronary Microvascular Dysfunction Across the Spectrum of Cardiovascular Diseases: JACC State-of-the-Art Review. J Am Coll Cardiol. 2021;78(13):1352-71. PubMed PMID: 34556322; PubMed Central PMCID: PMC8528638. [CrossRef]
- Gimbrone MA, García-Cardeña G. Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ Res. 2016;118(4):620-36. [CrossRef]
- Marc T. Cardiovascular Disease: An Introduction. 2018. p. 1-90.
- Ullah A, Kumar M, Sayyar M, Sapna F, John C, Memon S, et al. Revolutionizing Cardiac Care: A Comprehensive Narrative Review of Cardiac Rehabilitation and the Evolution of Cardiovascular Medicine. Cureus. 2023;15(10):1-11. Epub 20231004. PubMed PMID: 37927717; PubMed Central PMCID: PMC10624210. [CrossRef]
- Jansen-Chaparro S, López-Carmona MD, Cobos-Palacios L, Sanz-Cánovas J, Bernal-López MR, Gómez-Huelgas R. Statins and Peripheral Arterial Disease: A Narrative Review. Front Cardiovasc Med. 2021;8:1-18. Epub 20211122. PubMed PMID: 34881314; PubMed Central PMCID: PMC8645843. [CrossRef]
- Rousan TA, Thadani U. Stable Angina Medical Therapy Management Guidelines: A Critical Review of Guidelines from the European Society of Cardiology and National Institute for Health and Care Excellence. Eur Cardiol. 2019;14(1):18-22. PubMed PMID: 31131033; PubMed Central PMCID: PMC6523058. [CrossRef]
- Kario K, Hoshide S, Narita K, Okawara Y, Kanegae H, Aoki K, et al. Cardiovascular Prognosis in Drug-Resistant Hypertension Stratified by 24-Hour Ambulatory Blood Pressure: The JAMP Study. Hypertension. 2021;78(6):1781-90. [CrossRef]
- Wijkman MO, Malachias MVB, Claggett BL, Cheng S, Matsushita K, Shah AM, et al. Resistance to antihypertensive treatment and long-term risk: The Atherosclerosis Risk in Communities study. J Clin Hypertens (Greenwich). 2021;23(10):1887-96. Epub 20210921. PubMed PMID: 34547175; PubMed Central PMCID: PMC8678845. [CrossRef]
- Hu X, li J, Fu M, Zhao X, Wang W. The JAK/STAT signaling pathway: from bench to clinic. Signal Transduction and Targeted Therapy. 2021;6(1):402. [CrossRef]
- Morris R, Kershaw NJ, Babon JJ. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 2018;27(12):1984-2009. PubMed PMID: 30267440; PubMed Central PMCID: PMC6237706. [CrossRef]
- Liang C, Zhang L, Lian X, Zhu T, Zhang Y, Gu N. Circulating Exosomal SOCS2-AS1 Acts as a Novel Biomarker in Predicting the Diagnosis of Coronary Artery Disease. BioMed Research International. 2020;2020:1-10. [CrossRef]
- Zhang H, Dhalla NS. The Role of Pro-Inflammatory Cytokines in the Pathogenesis of Cardiovascular Disease. International Journal of Molecular Sciences. 2024;25(2):1082. PubMed PMID:. [CrossRef]
- Shook PL, Singh M, Singh K. Macrophages in the Inflammatory Phase following Myocardial Infarction: Role of Exogenous Ubiquitin. Biology (Basel). 2023;12(9). Epub 20230920. PubMed PMID: 37759657; PubMed Central PMCID: PMC10526096. [CrossRef]
- Klaeske K, Dix M, Adams V, Jawad K, Eifert S, Etz C, et al. Differential Regulation of Myocardial E3 Ligases and Deubiquitinases in Ischemic Heart Failure. Life (Basel). 2021;11(12). Epub 20211218. PubMed PMID: 34947961; PubMed Central PMCID: PMC8708923. [CrossRef]
- Rico-Bautista E, Flores-Morales A, Fernández-Pérez L. Suppressor of cytokine signaling (SOCS) 2, a protein with multiple functions. Cytokine Growth Factor Rev. 2006;17(6):431-9. Epub 20061027. PubMed PMID: 17070092. [CrossRef]
- Durham G, Williams J, Nasim T, Palmer T. Targeting SOCS Proteins to Control JAK-STAT Signalling in Disease. Trends in Pharmacological Sciences. 2019;40. [CrossRef]
- Mustafa R, Saiqa A, Domínguez J, Jamil M, Manzoor S, Wazir S, et al. Therapeutic Values of Earthworm Species Extract from Azad Kashmir as Anticoagulant, Antibacterial, and Antioxidant Agents. Can J Infect Dis Med Microbiol. 2022;2022(1):1-20. [CrossRef]
- Cooper EL, Balamurugan M, Huang CY, Tsao CR, Heredia J, Tommaseo-Ponzetta M, Paoletti MG. Earthworms dilong: ancient, inexpensive, noncontroversial models may help clarify approaches to integrated medicine emphasizing neuroimmune systems. Evid Based Complement Alternat Med. 2012;2012(1):1-11. Epub 20120725. PubMed PMID: 22888362; PubMed Central PMCID: PMC3410320. [CrossRef]
- Dong Y, Woo YM, Lee YH, Ahn MY, Lee DG, Lee SH, et al. Data on the potent fibrinolytic effects of the Lumbricus rubellus earthworm and the Perinereis linea lugworm. Data Brief. 2019;26(1):1-7. Epub 20190905. PubMed PMID: 31667249; PubMed Central PMCID: PMC6811935. [CrossRef]
- Lai CH, Han CK, Shibu MA, Pai PY, Ho TJ, Day CH, et al. Lumbrokinase from earthworm extract ameliorates second-hand smoke-induced cardiac fibrosis. Environ Toxicol. 2015;30(10):1216-25. Epub 20140405. PubMed PMID: 24706507. [CrossRef]
- Chen M, Wang L, Zheng C, Ma A, Hu K, Xiang A, et al. Novel ACE inhibitory peptides derived from bighead carp (Aristichthys nobilis) hydrolysates: Screening, inhibition mechanisms and the bioconjugation effect with graphene oxide. Food Biosci. 2023;52:1-14.
- Prakash M, Balamurugan M, Parthasarathi K, Gunasekaran G, Cooper E, Ranganathan L. Anti-ulceral and anti-oxidative properties of “earthworm paste” of Lampito mauritii (Kinberg) on Rattus Norvegicus. Eur Rev Med Pharmacol Sci. 2006;11:9-15.
- Dermawan D, Prabowo BA, Rakhmadina CA. In silico study of medicinal plants with cyclodextrin inclusion complex as the potential inhibitors against SARS-CoV-2 main protease (Mpro) and spike (S) receptor. Inform Med Unlocked. 2021;25:1-18.
- Challapa-Mamani MR, Tomás-Alvarado E, Espinoza-Baigorria A, León-Figueroa DA, Sah R, Rodriguez-Morales AJ, Barboza JJ. Molecular Docking and Molecular Dynamics Simulations in Related to Leishmania donovani: An Update and Literature Review. Trop Med Infect Dis. 2023;8(10):1-13. Epub 20230926. PubMed PMID: 37888585; PubMed Central PMCID: PMC10610989. [CrossRef]
- Sánchez-Gloria JL, Arellano-Buendía AS, Juárez-Rojas JG, García-Arroyo FE, Argüello-García R, Sánchez-Muñoz F, et al. Cellular Mechanisms Underlying the Cardioprotective Role of Allicin on Cardiovascular Diseases. Int J Mol Sci. 2022;23(16):1-21. Epub 20220813. PubMed PMID: 36012349; PubMed Central PMCID: PMC9409331. [CrossRef]
- Choudhury TZ, Garg V. Molecular genetic mechanisms of congenital heart disease. Curr Opin Genet Dev. 2022;75:1-13.
- Chen S, Fan G, Li J. Improving completeness and accuracy of 3D point clouds by using deep learning for applications of digital twins to civil structures. Adv Eng Inform. 2023;58:1-13. [CrossRef]
- Davis EM, Sun Y, Liu Y, Kolekar P, Shao Y, Szlachta K, et al. SequencErr: measuring and suppressing sequencer errors in next-generation sequencing data. Genome Biol. 2021;22(1):1-18. Epub 20210125. PubMed PMID: 33487172; PubMed Central PMCID: PMC7829059. [CrossRef]
- Faits T, Odom A, Castro-Nallar E, Crandall K, Johnson W. Metagenomic profiling pipelines improve taxonomic classification for 16S amplicon sequencing data2022.
- Zhang Y. I-TASSER: Fully automated protein structure prediction in CASP8. Proteins. 2009;77 Suppl 9:100-13. [CrossRef]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583-9. [CrossRef]
- Mohammadi S, Narimani Z, Ashouri M, Firouzi R, Karimi-Jafari MH. Ensemble learning from ensemble docking: revisiting the optimum ensemble size problem. Sci Rep. 2022;12(410):1-15. Epub 20220110. PubMed PMID: 35013496; PubMed Central PMCID: PMC8748946. [CrossRef]
- Saha A, Panja A, Shyamal M, Mandal. Influence of anionic co-ligands on the structural diversity and catecholase activity of copper(II) complexes with 2-methoxy-6-(8-iminoquinolinylmethyl)phenol. RSC Adv. 2014;4. [CrossRef]
- Kufareva I, Abagyan R. Methods of Protein Structure Comparison. Methods in molecular biology (Clifton, NJ). 2012;857:231-57. [CrossRef]
- Russell R, Alber F, Aloy P, Davis F, Korkin D, Pichaud M, et al. A structural perspective on protein-protein interactions. Curr Opin Struct Biol. 2004;14:313-24. [CrossRef]
- Dagliyan O, Proctor EA, D'Auria KM, Ding F, Dokholyan NV. Structural and dynamic determinants of protein-peptide recognition. Structure. 2011;19(12):1837-45. PubMed PMID: 22153506; PubMed Central PMCID: PMC3240807. [CrossRef]
- Guedes IA, Pereira FSS, Dardenne LE. Empirical Scoring Functions for Structure-Based Virtual Screening: Applications, Critical Aspects, and Challenges. Front Pharmacol. 2018;9:1-18. Epub 20180924. PubMed PMID: 30319422; PubMed Central PMCID: PMC6165880. [CrossRef]
- Desantis F, Miotto M, Di Rienzo L, Milanetti E, Ruocco G. Spatial organization of hydrophobic and charged residues affects protein thermal stability and binding affinity. Sci Rep. 2022;12(1):12087. Epub 20220715. PubMed PMID: 35840609; PubMed Central PMCID: PMC9287411. [CrossRef]
- Bitencourt-Ferreira G, Veit-Acosta M, De Azevedo Jr W. Van der Waals Potential in Protein Complexes. 20532019. p. 79-91.
- Cramer J, Krimmer S, Heine A, Klebe G. Paying the Price of Desolvation in Solvent-Exposed Protein Pockets: Impact of Distal Solubilizing Groups on Affinity and Binding Thermodynamics in a Series of Thermolysin Inhibitors. Journal of Medicinal Chemistry. 2017;60. [CrossRef]
- Zhou H-X, Pang X. Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation. Chemical Reviews. 2018;118. [CrossRef]
- Kastritis PL, Rodrigues JPGLM, Bonvin AMJJ. HADDOCK2P2I: A Biophysical Model for Predicting the Binding Affinity of Protein–Protein Interaction Inhibitors. J Chem Inf Model. 2014;54(3):826-36. [CrossRef]
- Kastritis PL, Bonvin AM. On the binding affinity of macromolecular interactions: daring to ask why proteins interact. J R Soc Interface. 2013;10(79):1-27. Epub 20121212. PubMed PMID: 23235262; PubMed Central PMCID: PMC3565702. [CrossRef]
- Dermawan D, Sumirtanurdin R, Dewantisari D. Simulasi dinamika molekular reseptor estrogen alfa dengan andrografolid sebagai anti kanker payudara. Indones J Pharm Sci Technol. 2019;6(2):65-76.
- Craveur P, Joseph AP, Esque J, Narwani TJ, Noël F, Shinada N, et al. Protein flexibility in the light of structural alphabets. Front Mol Biosci. 2015;2:1-20. Epub 20150527. PubMed PMID: 26075209; PubMed Central PMCID: PMC4445325. [CrossRef]
- Maspero E, Mari S, Valentini E, Musacchio A, Fish A, Pasqualato S, Polo S. Structure of the HECT:ubiquitin complex and its role in ubiquitin chain elongation. EMBO Rep. 2011;12(4):342-9. Epub 20110311. PubMed PMID: 21399620; PubMed Central PMCID: PMC3077247. [CrossRef]
- Sanusi ZK, Lobb KA. Insights into the Dynamics and Binding of Two Polyprotein Substrate Cleavage Points in the Context of the SARS-CoV-2 Main and Papain-like Proteases. Molecules. 2022;27(23):1-16. Epub 20221126. PubMed PMID: 36500348; PubMed Central PMCID: PMC9740519. [CrossRef]
- Hou T, Wang J, Li Y, Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model. 2011;51(1):69-82. Epub 20101130. PubMed PMID: 21117705; PubMed Central PMCID: PMC3029230. [CrossRef]
- Kryshtafovych A, Schwede T, Topf M, Fidelis K, Moult J. Critical assessment of methods of protein structure prediction (CASP)-Round XIII. Proteins. 2019;87(12):1011-20. Epub 20191023. PubMed PMID: 31589781; PubMed Central PMCID: PMC6927249. [CrossRef]
- Wang YH, Chen KM, Chiu PS, Lai SC, Su HH, Jan MS, et al. Lumbrokinase attenuates myocardial ischemia-reperfusion injury by inhibiting TLR4 signaling. J Mol Cell Cardiol. 2016;99:113-22. Epub 20160805. PubMed PMID: 27503317. [CrossRef]
- Lenselink EB, Louvel J, Forti AF, van Veldhoven JPD, de Vries H, Mulder-Krieger T, et al. Predicting Binding Affinities for GPCR Ligands Using Free-Energy Perturbation. ACS Omega. 2016;1(2):293-304. Epub 20160830. PubMed PMID: 30023478; PubMed Central PMCID: PMC6044636. [CrossRef]
- Salsbury FR, Jr. Molecular dynamics simulations of protein dynamics and their relevance to drug discovery. Curr Opin Pharmacol. 2010;10(6):738-44. PubMed PMID: 20971684; PubMed Central PMCID: PMC2981647. [CrossRef]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9(1):40. [CrossRef]
- Tian W, Chen C, Lei X, Zhao J, Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46(1):363-7. [CrossRef]
- Kung W-W, Ramachandran S, Makukhin N, Bruno E, Ciulli A. Structural insights into substrate recognition by the SOCS2 E3 ubiquitin ligase. Nature Communications. 2019;10(1):2534. [CrossRef]
- Laskowski RA, Jabłońska J, Pravda L, Vařeková RS, Thornton JM. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018;27(1):129-34. Epub 20171027. PubMed PMID: 28875543; PubMed Central PMCID: PMC5734310. [CrossRef]
- Johansson MU, Zoete V, Michielin O, Guex N. Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinformatics. 2012;13(173):1-10. Epub 20120723. PubMed PMID: 22823337; PubMed Central PMCID: PMC3436773. [CrossRef]
- Gitlin AD, Maltzman A, Kanno Y, Heger K, Reja R, Schubert AF, et al. N4BP1 coordinates ubiquitin-dependent crosstalk within the IκB kinase family to limit Toll-like receptor signaling and inflammation. Immunity. 2024;57(5):973-86.
- Ramachandran S, Makukhin N, Haubrich K, Nagala M, Forrester B, Lynch DM, et al. Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2. Nature Communications. 2023;14(1):6345. [CrossRef]
- Dominguez C, Boelens R, Bonvin AMJJ. HADDOCK: A Protein−Protein Docking Approach Based on Biochemical or Biophysical Information. Journal of the American Chemical Society. 2003;125(7):1731-7. [CrossRef]
- van Zundert GCP, Rodrigues J, Trellet M, Schmitz C, Kastritis PL, Karaca E, et al. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J Mol Biol. 2016;428(4):720-5. Epub 20150926. PubMed PMID: 26410586. [CrossRef]
- Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845-54. [CrossRef]
- Robertson MJ, Tirado-Rives J, Jorgensen WL. Improved Peptide and Protein Torsional Energetics with the OPLSAA Force Field. J Chem Theory Comput. 2015;11(7):3499-509. Epub 2015/07/21. PubMed PMID: 26190950; PubMed Central PMCID: PMC4504185. [CrossRef]
- Yuet P, Blankschtein D. Molecular Dynamics Simulation Study of Water Surfaces: Comparison of Flexible Water Models. The journal of physical chemistry B. 2010;114:13786-95. [CrossRef]
- Schrödinger. The PyMOL Molecular Graphics System. 2.4 ed2020.
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605-12. PubMed PMID: 15264254. [CrossRef]
- Tian S, Sun H, Pan P, Li D, Zhen X, Li Y, Hou T. Assessing an ensemble docking-based virtual screening strategy for kinase targets by considering protein flexibility. J Chem Inf Model. 2014;54(10):2664-79. Epub 20140929. PubMed PMID: 25233367. [CrossRef]
- Yuan Z, Chen X, Fan S, Chang L, Chu L, Zhang Y, et al. Binding Free Energy Calculation Based on the Fragment Molecular Orbital Method and Its Application in Designing Novel SHP-2 Allosteric Inhibitors. Int J Mol Sci [Internet]. 2024; 25(1):[1-24 pp.].
- Rifai EA, Ferrario V, Pleiss J, Geerke DP. Combined Linear Interaction Energy and Alchemical Solvation Free-Energy Approach for Protein-Binding Affinity Computation. J Chem Theory Comput. 2020;16(2):1300-10. [CrossRef]
- Valdés-Tresanco MS, Valdés-Tresanco ME, Valiente PA, Moreno E. gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. Journal of Chemical Theory and Computation. 2021;17(10):6281-91. [CrossRef]
- Miller BR, 3rd, McGee TD, Jr., Swails JM, Homeyer N, Gohlke H, Roitberg AE. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. Journal of chemical theory and computation. 2012;8(9):3314-21. [CrossRef]
- Panday SK, Alexov E. Protein-Protein Binding Free Energy Predictions with the MM/PBSA Approach Complemented with the Gaussian-Based Method for Entropy Estimation. ACS Omega. 2022;7(13):11057-67. Epub 2022/04/14. PubMed PMID: 35415339; PubMed Central PMCID: PMC8991903. [CrossRef]
- IBM. IBM SPSS Statistics for Windows. Version 25.0 ed. New York: IBM Corp; 2017.
- OriginLab. Origin(Pro). 2022 ed. Northampton, MA, USA: OriginLab Corporation; 2022.
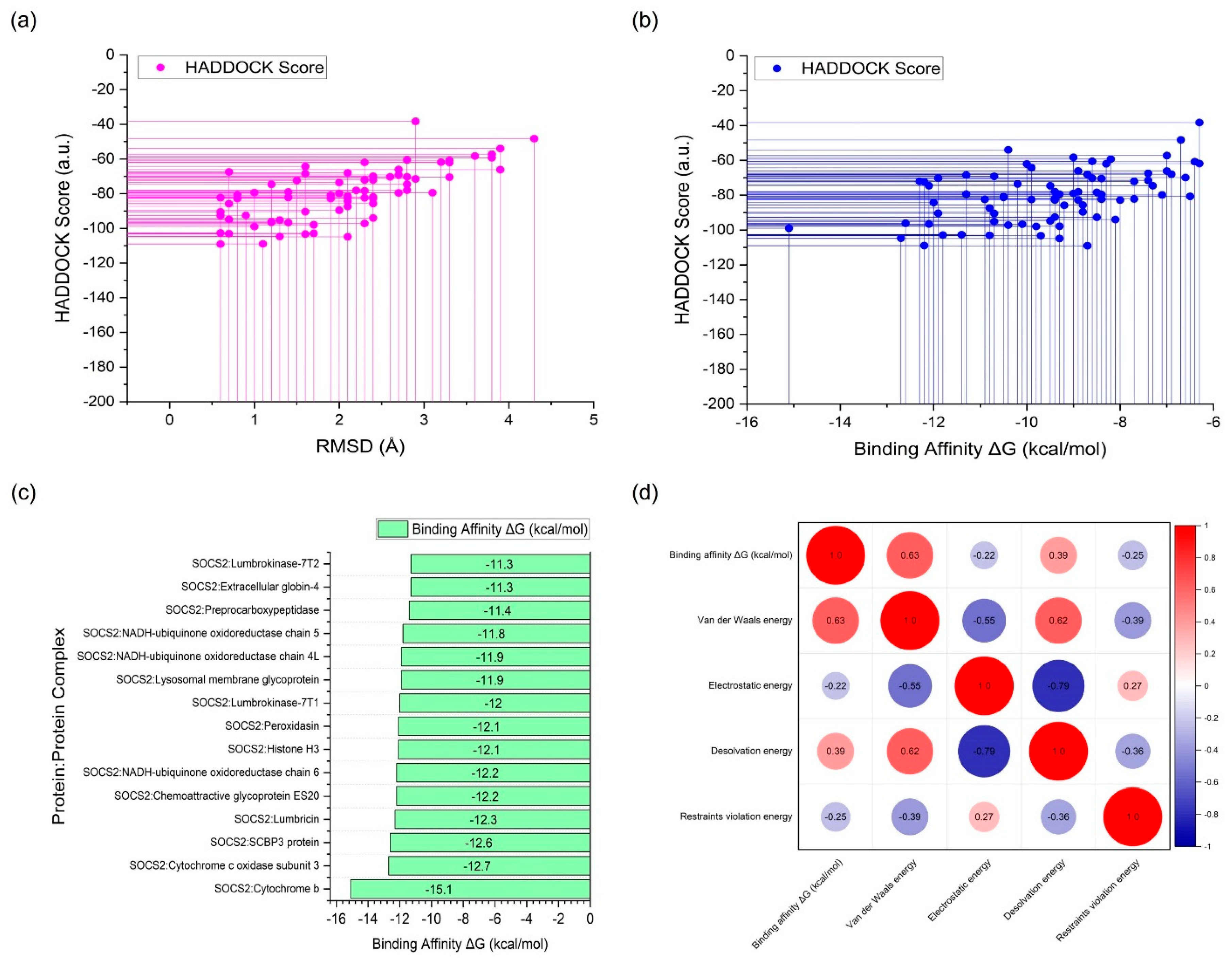
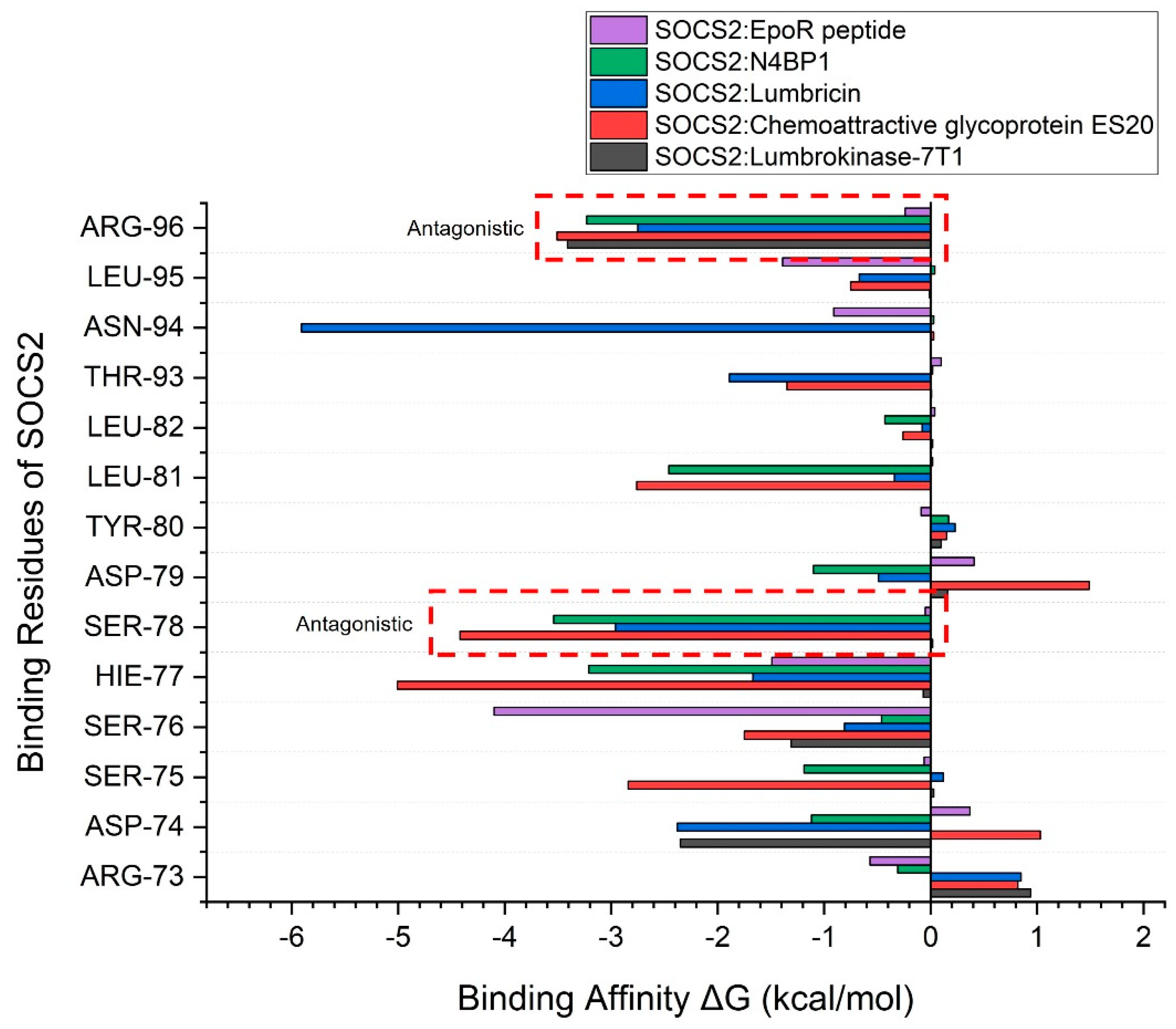
| Complex | HADDOCK Score (a.u.) |
Binding Affinity ΔG (kcal/mol) | Kd (M) |
Cluster size | RMSD (Å) |
|---|---|---|---|---|---|
| Standard | |||||
| SOCS2: EpoR peptide (Standard Agonist) | -81.0 +/- 3.5 | -8.8 | 6.50e-7 | 22 | 1.7 +/- 0.1 |
| SOCS2:N4BP1 (Standard Antagonist) |
-94.3 +/- 11.4 | -8.3 | 1.50e-6 | 17 | 1.2 +/- 0.3 |
| Protein and peptide derived from earthworm (Lumbricus genus) | |||||
| SOCS2:Cytochrome b | -99.0 +/- 8.2 | -15.1 | 2.30e-11 | 27 | 1.0 +/- 0.6 |
| SOCS2:Cytochrome c oxidase subunit 3 | -104.7 +/- 6.3 | -12.7 | 1.10e-09 | 15 | 1.3 +/- 0.5 |
| SOCS2:SCBP3 protein | -96.2 +/- 6.2 | -12.6 | 1.30e-09 | 41 | 1.2 +/- 0.9 |
| SOCS2:Lumbricin | -72.1 +/- 3.7 | -12.3 | 2.10e-09 | 43 | 2.3 +/- 0.3 |
| SOCS2:Chemoattractive glycoprotein ES20 | -72.4 +/- 5.3 | -12.2 | 2.30e-09 | 20 | 1.5 +/- 1.0 |
| SOCS2:NADH-ubiquinone oxidoreductase chain 6 | -109.0 +/- 7.1 | -12.2 | 2.30e-09 | 14 | 1.1 +/- 0.7 |
| SOCS2:Histone H3 | -96.6 +/- 4.0 | -12.1 | 3.10e-09 | 8 | 1.2 +/- 0.2 |
| SOCS2:Peroxidasin | -74.6 +/- 7.6 | -12.1 | 2.70e-09 | 8 | 2.8 +/- 0.0 |
| SOCS2:Lumbrokinase-7T1 | -84.3 +/- 2.8 | -12.0 | 3.70e-09 | 43 | 2.1 +/- 0.2 |
| SOCS2:Lysosomal membrane glycoprotein | -70.3 +/- 2.3 | -11.9 | 4.30e-09 | 5 | 2.6 +/- 0.9 |
| SOCS2:NADH-ubiquinone oxidoreductase chain 4L | -90.4 +/- 5.4 | -11.9 | 4.10e-09 | 12 | 1.6 +/- 1.0 |
| SOCS2:NADH-ubiquinone oxidoreductase chain 5 | -102.9 +/- 11.6 | -11.8 | 4.70e-09 | 7 | 1.7 +/- 0.3 |
| SOCS2:Preprocarboxypeptidase | -102.7 +/- 9.1 | -11.4 | 9.70e-09 | 19 | 0.6 +/- 0.4 |
| SOCS2:Extracellular globin-4 | -79.4 +/- 10.8 | -11.3 | 1.20e-08 | 6 | 1.0 +/- 0.6 |
| SOCS2:Lumbrokinase-7T2 | -68.5 +/- 2.5 | -11.3 | 1.10e-08 | 35 | 1.6 +/- 0.5 |
| Complex |
ICs charged-charged | ICs charged-polar | ICs charged-apolar | ICs polar-polar | ICs polar-apolar | ICs apolar-apolar | NIS charged | NIS apolar |
|---|---|---|---|---|---|---|---|---|
| Standard | ||||||||
| SOCS2: EpoR peptide (Standard Agonist) | 3 | 8 | 12 | 2 | 11 | 12 | 25.55 | 38.69 |
| SOCS2:N4BP1 (Standard Antagonist) |
5 | 11 | 20 | 6 | 11 | 8 | 29.71 | 39.49 |
| Protein and peptide derived from earthworm (Lumbricus genus) | ||||||||
| SOCS2:Cytochrome b | 0 | 0 | 25 | 0 | 38 | 33 | 15.13 | 52.85 |
| SOCS2:Cytochrome c oxidase subunit 3 | 2 | 7 | 22 | 6 | 30 | 26 | 14.71 | 49.41 |
| SOCS2:SCBP3 protein | 2 | 3 | 24 | 2 | 27 | 16 | 27.83 | 41.74 |
| SOCS2:Lumbricin | 7 | 9 | 20 | 0 | 22 | 12 | 26.34 | 40.98 |
| SOCS2:Chemoattractive glycoprotein ES20 | 8 | 15 | 24 | 5 | 23 | 16 | 22.98 | 42.39 |
| SOCS2:NADH-ubiquinone oxidoreductase chain 6 | 0 | 3 | 21 | 7 | 32 | 29 | 14.29 | 52.01 |
| SOCS2:Histone H3 | 7 | 6 | 27 | 0 | 20 | 14 | 27.56 | 42.67 |
| SOCS2:Peroxidasin | 13 | 20 | 24 | 5 | 22 | 6 | 26.23 | 41.86 |
| SOCS2:Lumbrokinase-7T1 | 3 | 15 | 12 | 4 | 24 | 10 | 17.72 | 41.14 |
| SOCS2:Lysosomal membrane glycoprotein | 12 | 17 | 18 | 5 | 18 | 11 | 20.49 | 39.02 |
| SOCS2:NADH-ubiquinone oxidoreductase chain 4L | 0 | 8 | 21 | 2 | 24 | 14 | 17.73 | 46.82 |
| SOCS2:NADH-ubiquinone oxidoreductase chain 5 | 0 | 3 | 17 | 1 | 25 | 29 | 14.12 | 50.10 |
| SOCS2:Preprocarboxypeptidase | 7 | 17 | 23 | 5 | 20 | 28 | 26.18 | 40.05 |
| SOCS2:Extracellular globin-4 | 15 | 18 | 28 | 2 | 12 | 6 | 31.28 | 37.04 |
| SOCS2:Lumbrokinase-7T2 | 5 | 10 | 16 | 9 | 24 | 11 | 19.03 | 41.69 |
- ICs: Number of intermolecular contacts
- NIS: Non-interacting surface
| Complex | Residue (Receptor) | Protein Atom (Receptor) |
Residue (Interacting Protein/Peptide) |
Protein Atom (Interacting Protein/Peptide) |
Interaction Distance (Å) |
|---|---|---|---|---|---|
| SOCS2: EpoR peptide (Standard Agonist) | Val55 | N | Asp8 | OD1 | 2.76 |
| Ser76 | OG | Asp8 | OD1 | 3.09 | |
| Arg96 | NH1 | Glu3 | O | 2.69 | |
| Arg96 | NH2 | Glu3 | O | 3.33 | |
| Lys113 | NZ | Ser1 | OG | 2.73 | |
| SOCS2:N4BP1 (Standard Antagonist) | Gln32 | NE2 | Glu178 | OE1 | 2.77 |
| Arg41 | NH1 | Glu118 | OE1 | 2.65 | |
| Arg41 | NH2 | Glu118 | OE2 | 2.57 | |
| Tyr49 | OH | Lys132 | NZ | 2.78 | |
| Asp74 | OD2 | Lys132 | NZ | 2.55 | |
| Ser75 | O | Asn136 | ND2 | 3.04 | |
| Ser78 | O | Ser174 | OG | 2.63 | |
| Asp79 | OD2 | Lys145 | NZ | 2.62 | |
| Arg96 | NH2 | Glu138 | OE1 | 2.67 | |
| SOCS2:Cytochrome b | His77 | NE2 | Leu122 | O | 2.74 |
| Arg96 | NH2 | Ile119 | O | 2.72 | |
| SOCS2:SCBP3 protein | Lys59 | NZ | Asp50 | O | 2.58 |
| Arg96 | NH1 | Leu68 | O | 2.66 | |
| SOCS2:Lumbricin | Arg41 | NH1 | Glu56 | O | 3.08 |
| Arg41 | NH1 | Glu56 | OE1 | 2.60 | |
| Arg41 | NH2 | Glu56 | O | 2.98 | |
| Tyr49 | OH | Lys53 | NZ | 2.83 | |
| Ser52 | OG | Glu63 | OE1 | 2.59 | |
| Asp74 | OD2 | Lys53 | NZ | 2.65 | |
| Ser78 | OG | Ile46 | O | 2.67 | |
| Asp79 | OD1 | Arg45 | NH1 | 2.65 | |
| Thr93 | OG1 | Glu72 | OE2 | 2.70 | |
| Asn94 | N | Glu72 | OE2 | 2.73 | |
| Asn94 | ND2 | Glu72 | OE1 | 2.64 | |
| Arg96 | NE | Glu72 | O | 2.74 | |
| Arg96 | NH2 | Glu72 | O | 2.87 | |
| SOCS2:Lumbrokinase-7T1 | Glu57 | OE2 | Lys20 | NZ | 2.55 |
| Asp74 | OD2 | Arg17 | NH1 | 2.69 | |
| Asp74 | OD2 | Arg17 | NH2 | 2.72 | |
| Ser76 | O | Gln19 | NE2 | 3.20 | |
| Asp101 | OD1 | Asn249 | ND2 | 2.66 | |
| Asp101 | OD2 | Ser170 | OG | 2.66 | |
| Cys111 | O | Lys168 | NZ | 2.91 | |
| Lys113 | NZ | Gly32 | O | 2.87 | |
| Lys113 | NZ | Gln35 | OE1 | 2.66 | |
| Leu116 | O | Asn214 | ND2 | 2.98 |
| Complex | Average RMSD (Å) | Average RMSF (Å) | Average RoG (Å) |
Number of Hydrogen Bonds Between the Two Proteins | Potential Energy (kcal/mol) |
|---|---|---|---|---|---|
| Standard | |||||
| SOCS2 (Apo-protein) | 2.417 | 1.089 | 1.674 | N/A | -158,603.87 |
| SOCS2:EpoR peptide (Standard Agonist) | 2.423 | 1.397 | 2.101 | 11 | -159,708.72 |
| SOCS2:N4BP1 (Standard Antagonist) | 2.496 | 1.179 | 2.162 | 24 | -459,214.66 |
| Protein and peptide derived from earthworm (Lumbricus genus) | |||||
| SOCS2:Cytochrome b | 2.467 | 0.876 | 2.182 | 39 | -471,304.36 |
| SOCS2:Cytochrome c oxidase subunit 3 | 2.504 | 1.122 | 2.176 | 30 | -450,869.63 |
| SOCS2:SCBP3 protein | 2.587 | 1.301 | 2.198 | 22 | -316,369.57 |
| SOCS2:Lumbricin | 2.495 | 1.031 | 2.148 | 18 | -694,628.89 |
| SOCS2:Chemoattractive glycoprotein ES20 | 2.512 | 0.941 | 2.176 | 22 | -621,068.14 |
| SOCS2:NADH-ubiquinone oxidoreductase chain 6 | 2.487 | 1.001 | 2.178 | 20 | -466,577.25 |
| SOCS2:Histone H3 | 2.413 | 0.917 | 2.287 | 16 | -629,402.13 |
| SOCS2:Peroxidasin | 2.599 | 1.106 | 2.678 | 35 | -2,208,424.10 |
| SOCS2:Lumbrokinase-7T1 | 2.523 | 1.123 | 2.213 | 27 | -1,065,029.39 |
| SOCS2:Lysosomal membrane glycoprotein | 2.511 | 1.115 | 2.298 | 31 | -1,645,741.31 |
| SOCS2:NADH-ubiquinone oxidoreductase chain 4L | 2.498 | 1.178 | 2.199 | 26 | -406,049.87 |
| SOCS2:NADH-ubiquinone oxidoreductase chain 5 | 2.487 | 1.101 | 2.190 | 21 | -1,187,770.16 |
| SOCS2:Preprocarboxypeptidase | 2.596 | 1.259 | 2.188 | 30 | -560,689.83 |
| SOCS2:Extracellular globin-4 | 2.543 | 1.028 | 2.287 | 23 | -473,808.36 |
| SOCS2:Lumbrokinase-7T2 | 2.524 | 1.060 | 2.214 | 24 | -764,436.84 |
| Complex | MM/PBSA Calculation Results ΔGbinding (kcal/mol) | Average (kcal/mol) | ||
|---|---|---|---|---|
| I | II | III | ||
| Standard | ||||
| SOCS2:EpoR peptide (Standard Agonist) | -42.84 | -43.48 | -41.48 | -42.60 |
| SOCS2:N4BP1 (Standard Antagonist) | -42.34 | -42.34 | -42.85 | -42.51 |
| Protein and peptide derived from earthworm (Lumbricus genus) | ||||
| SOCS2:Cytochrome b | -50.57 | -49.93 | -49.82 | -50.11 |
| SOCS2:Cytochrome c oxidase subunit 3 | -44.93 | -44.93 | -44.87 | -44.91 |
| SOCS2:SCBP3 protein | -29.33 | -29.45 | -29.45 | -29.41 |
| SOCS2:Lumbricin | -59.22 | -59.26 | -59.26 | -59.25 |
| SOCS2:Chemoattractive glycoprotein ES20 | -53.69 | -57.76 | -53.60 | -55.02 |
| SOCS2:NADH-ubiquinone oxidoreductase chain 6 | -52.81 | -51.48 | -53.15 | -52.48 |
| SOCS2:Histone H3 | -44.05 | -44.84 | -45.82 | -44.90 |
| SOCS2:Peroxidasin | -42.91 | -42.91 | -42.91 | -42.91 |
| SOCS2:Lumbrokinase-7T1 | -69.22 | -69.35 | -69.27 | -69.28 |
| SOCS2:Lysosomal membrane glycoprotein | -34.42 | -34.89 | -34.75 | -34.69 |
| SOCS2:NADH-ubiquinone oxidoreductase chain 4L | -48.14 | -48.35 | -48.35 | -48.28 |
| SOCS2:NADH-ubiquinone oxidoreductase chain 5 | -37.74 | -36.11 | -36.11 | -36.65 |
| SOCS2:Preprocarboxypeptidase | -39.55 | -38.03 | -39.58 | -39.05 |
| SOCS2:Extracellular globin-4 | -50.09 | -49.68 | -50.30 | -50.02 |
| SOCS2:Lumbrokinase-7T2 | -46.18 | -45.80 | -47.11 | -46.36 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





