1. Introduction
Functionalized magnetic nanoparticle (fMNP)-protein binding events, transduced via magnetic spectroscopy, enable biomolecule detection within a sample solution [
1,
2,
3]. Biomolecule binding increases fMNP hydrodynamic diameter (
) [
4,
5] and/or induces the formation of fMNP aggregates [
6]. The fMNP magnetic moment will have a delayed response to an external magnetic field due to
increases and aggregate formation. The magnetic moment response can be modeled via Brownian and Néel relaxation [
7,
8,
9]. fMNPs have been engineered to sense biomarkers, including SARS-CoV-2 [
10] and H1N1 nucleoprotein [
11],
in vitro. Salt induced changes in fMNP aggregation pose an obstacle to the clinical translation of fMNP-based aggregation assays for
in vivo applications. Additional factors impeding aggregation assay
in vivo applications include, protein corona formation [
12,
13] and non-specific bio-distribution. Changes in salt concentration can alter nanoparticle (NP) stability [
4,
14]. Which can cause changes in fMNP aggregation that are not proportional to the target biomolecule concentration. To this end, we have investigated the effects of salt concentration, in the biological range, on target induced fMNP aggregation. Our aggregation assay is a model system consisting of biotinylated MNPs (biotin-MNPs) that target streptavidin. We used Lodestone Biomedical’s Nanoparticle Characterization System (an AC magnetic spectrometer, NCS) [
15] to transduce biotin-MNP-streptavidin binding into a measurable signal. The NCS emits a 1 kHz AC magnetic field with 20 mT field strength and conveys the biotin-MNP harmonic response at 3 kHz [
15]. We used the phase of the 3rd harmonic (
) to assess the level of biotin-MNP aggregation [
16,
17] and the magnitude of the 3rd harmonic (
) to determine the Fe concentration present in our samples after biotinylation [
18]. We studied changes in streptavidin induced biotin-MNP aggregation in phosphate buffered saline (PBS) solutions. 1x PBS is a water-based solution with an approximate pH of 7.4 and salt contents that match those found in the human body. 1x PBS contains 137 mM NaCl, 10 mM phosphate, and 2.7 mM KCl. To alter the ionic strength of PBS we diluted it with deionized water (DI), resulting in the following PBS solutions: 1.00x, 0.70x, 0.50x, 0.30x, 0.05x, 0.045x, 0.04x, 0.035x, 0.025x, 0.02x, 0.015x, 0.01x, 0.005, and 0.00x (DI). We determined biotin-MNP zeta potential and
in each PBS solution. Additionally, we investigated differences in biosensor response characteristics in the 1.00x, 0.50x, 0.03x, 0.015x, 0.005x, and 0.00x PBS solutions. Here, we monitored changes in biotin-MNP harmonic response when in the presence of varying amounts of streptavidin. We refer to biotin-MNP
as a function of streptavidin concentration as a streptavidin-response curve. We compared the slope, maximum
, and linear range of each streptavidin-response curve prepared in the 1.00x, 0.0x, 0.03x, 0.015x, 0.005x, and 0.00x PBS solutions.
According to the extended Derjaguin-Landau-Verwey-Overbeek (xDLVO) theory, biotin-MNP stability will be dependent on the sum of the repulsive electrostatic double layer (EDL) force, which is dependent on biotin-MNP surface charge density and the electrolyte concentration of the diluent, and the attractive van der Waals and dipolar forces acting upon similarly charged biotin-MNPs [
8,
19]. A positive sum suggests stability, whereas a negative sum suggests instability and hence aggregation. If a nanoparticle (NP) has a high surface charge density and/or is in a low salt environment, particle interactions are repulsive and dominated by the EDL force [
20]. On the other hand, if a NP has a low surface charge density and/or is in a high salt environment, particle interactions are attractive and dominated by the van der Waals force [
20]. Jans et. al., 2009 demonstrated the effects of electrolyte concentration on gold NP (AuNP) stability [
4]. They found that the hydrodynamic diameter of citrate capped AuNPs increased with increasing NaCl concentration. Interestingly, larger (∼100nm) AuNPs were less stable in high salt environments compared to smaller (∼40 nm) AuNPs. MNPs are more apt to cluster, compared to NPs with non-magnetic cores, due to the attractive dipolar force acting amongst MNPs, in addition to the attractive van der Waals force [
21]. Nikam et. al., 2014 demonstrated the effects of electrolyte concentration and zeta potential on cobalt zinc ferrite (CZF) MNP stability [
14]. Their PEGylated CZF MNPs were stable in PBS for 2 weeks, although they underwent an approximate 75 nm hydrodynamic increase in double distilled water (DDW) for the same duration, indicating instability/aggregation in DDW. Moreover, they found a linear relationship between NaCl concentration and CZF MNP zeta potential. An increase in NaCl concentration led to a decrease in zeta potential absolute value. Zeta potential absolute value decreases with increasing electrolyte concentration given an increase in EDL thickness due to surface charge screening. Given previous work investigating salt induced changes in fMNP stability, we can expect a decrease in streptavidin induced biotin-MNP aggregation with decreasing salt concentration.
In this work we also show that receptor density can be used to tune the linear region of the streptavidin-response curve for our aggregation assay. The linear region of the streptavidin-response curve is commonly referred to as the dynamic range, which is the measurable concentration range of a biosensor [
22]. Takae et. al., 2005 demonstrated the tunability of AuNP-based aggregation assay dynamic range via ligand density [
23]. They investigated lactose functionalized AuNPs that target Ricinus communis agglutin (RCA120) lectin. Lactose-AuNP’s with 40% and 65% surface area lactose coverage had dynamic ranges spanning from 25 to 200
g/mL and 50 to 400
g/mL, respectively. A decrease in surface area lactose coverage led to an increase in assay sensitivity. Researchers have engineered biosensors targeting metal ions [
24], small molecules [
25], and proteins [
26] with tunable dynamic ranges. Li et. al., 2012 engineered a non-cross linking AuNP-based aggregation assay with a tunable dynamic range [
27]. Their target protein, ALP, altered the aggregation levels of AuNPs diluted in a Tris-HCL buffer solution containing adenosine triphosphate (ATP). The dynamic range of their AuNP-based aggregation assay was tuned via the addition of metal ions to the sensing solution.
Concerning fMNP ligand density modulation, Elias et. al., 2013 demonstrated a non-linear relationship between superparamagnetic MNP ligand density and cell targeting [
28]. They found that optimal cell targeting occurred when the MNPs had intermediate ligand densities. This relationship was held regardless of MNP core diameter or ligand type. Generally, MNPs with 1.7 ligands per 100
exhibited optimal cell targeting. This topic is expanded upon in Alkilany et. al., 2019 review [
29] of the relationship between a NP’s (including MNPs, AuNPs, and quantum dots) ligand density and its cellular uptake. The relationship between MNP ligand density and protein sensing dynamics, transduced via MPS, were reported by Wu et. al., 2021. They functionalized MNPs with polyclonal antibodies (pAbs), to lend the MNPs and affinity for SARs-CoV-2 spike or nucleocapsid proteins and monitored odd harmonic amplitudes of magnetization as the pAb-MNPs were exposed to varying concentrations of their target [
30]. MNPs were functionalized with 1, 2, 3, or 4 pAbs per MNP. For pAb-MNPs targeting spike protein, a pAb:MNP ratio of 3:1 yielded a linear
response from 0 to 12.5 nM spike protein. Additionally, for pAb-MNPs targeting nucleocapsid protein, a pAb:MNP ratio of 4:1 yielded a linear
response from 3.13 to 400 nM nucleocapsid protein. pAb-MNPs with 1:1, 2:1, or 3:1 pAb:MNP ratios did not yield linear
responses with respect to target concentration, regardless of target protein (splike or nucleocapsid). Our work differs from that of Wu et. al., 2021 in that we are studying changes in biotin-MNP biosensor response characteristics (the dynamic range, slope and
), via investigating changes in biotin-MNP
as a function of target concentration, instead of odd harmonic amplitudes. Given previous work investigating NP dynamic range tunability with ligand density, we expect an increase in biotin-MNP sensitivity with decreasing biotin density.
2. Materials and Methods
2.1. Materials
Amine functionalized 25 nm magnetite (SHA 25) particles were purchased from Ocean Nanotech, San Diego, CA, USA. N-hydroxysuccinide (NHS)--biotin was purchased from Thermo Fisher Scientific, USA. Streptavidin was purchased from Rockland Immunochemicals, USA.
2.2. Physical Characterization of SHA 25 MNPs
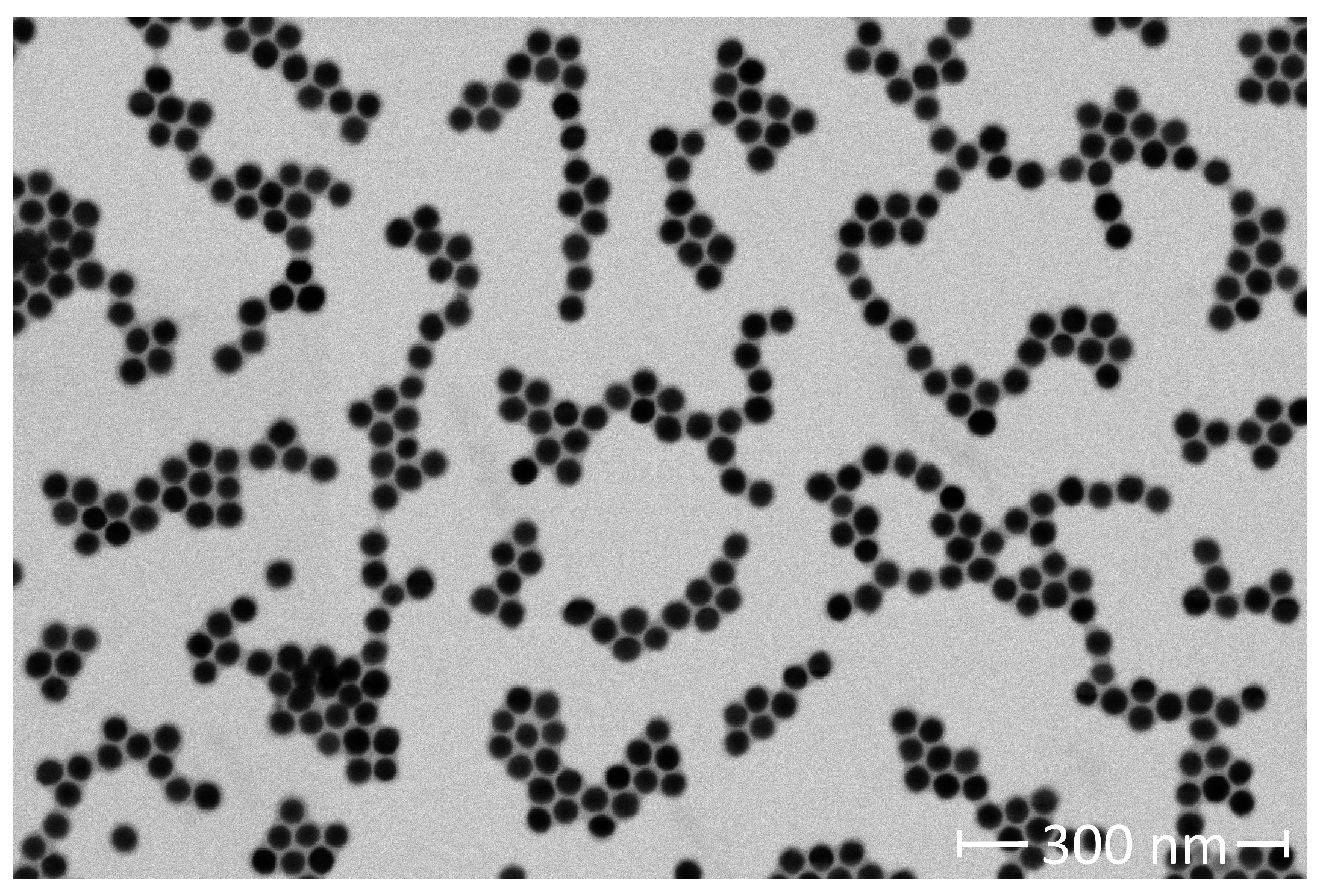
A carbon film mesh grid was dipped into a 0.5 mg Fe/mL SHA 25 solution. The SHA 25 MNPs adsorbed to the carbon film mesh grid were imaged with a scanning electron microscope (SEM, Helios 5 CX DualBeam for Materials Science, Thermo Fisher Scientific), to verify their size and shape distribution. The SEM had a current and voltage of 21 pA and 20 kV, respectively. The SEM images were analyzed with ImageJ (Fiji). ImageJ has an ‘Analyze Particles’ feature that measures the area of non-interacting particles. The ‘Analyze Particles’ feature is compatible with 8-bit binary images. We used minimum error thresholding to algorithmically threshold our image. The output of the thresholding operation was a binary image. The ‘Analyze Particles’ feature was applied to the 8-bit binary image to calculate the area of 371 SHA 25 MNPs. We algebraically manipulated the reported areas to calculate the SHA 25 MNP mean diameter and standard deviation. We conducted measurements of 10 mg Fe/L SHA 25 MNP samples. The SHA 25 MNP solution was diluted with PBS. The measurements were conducted via dynamic light scattering (DLS) with a Zetasizer Ultra Red (Malvern Panalytical). 1 mL of each SHA 25 solution was transferred to a disposable 10x10 plastic cell (DTS0012). The measurements were conducted in quintuplicate at 25°C with a light scattering collection angle of 174.7° (back scatter). To verify the zeta potential of the SHA 25 MNPs in PBS, 1 mL of a 10 mg Fe/L SHA 25 MNP solution was injected into a disposable folded capillary cell (DTS1070). The zeta potential measurements were conducted in quintuplicate, with measurements before and after each zeta potential measurement to monitor MNP stability.
2.3. Biotinylation of SHA 25 MNPs
Two Eppendorf tubes containing 300 L of 5 mg Fe/mL SHA 25 MNP solutions were centrifuged at 12,000 RPM for 30 minutes at 25°C. Once the centrifugation was complete, the diluent was aspirated and the SHA 25 MNPs were resuspended in 300 L of PBS. Quantities of 2 mg of NHS--biotin are stored in each vial of the Thermo ScientificTM No-WeighTM Format. To prevent moisture condensation, we allowed the NHS--biotin to equilibrate to room temperature before opening the vial. To produce biotin--SHA 25 MNPs with ∼10 available biotins for streptavidin binding per MNP (10 biotin-MNPs), the NHS--biotin was diluted to 0.50 mg/mL with DI, then 1.54 L of the 0.50 mg/mL NHS--biotin solution was added to one of the Eppendorf tubes containing the SHA 25 MNP solution. To produce biotin--SHA 25 MNPs with ∼90 available biotins for streptavidin binding per MNP (90 biotin-MNPs), the NHS--biotin was diluted to 2.86 mg/mL, then 158.80 L of the 2.86 mg/mL NHS--biotin solution was added to the SHA 25 MNP solution. Next, both mixtures were left to incubate at room temperature for 30 minutes. Once the SHA 25 MNPs were biotinylated, free biotin was removed from the solution via centrifugation. Each mixture was centrifuged at 12,000 RPM for 30 minutes at 25°C 8 times. After each centrifugation, the diluent was aspirated and the 10 biotin-MNP and 90 biotin-MNP solutions were resuspended in 300 L of DI. The of SHA 25 serial dilutions (0, 1, 2, 3, 4 and 5 g/L Fe concentrations) were measured with the NCS. The of the 10 biotin-MNP and 90 biotin-MNP solutions were compared with those of the SHA 25 MNP serial dilutions to determine the Fe concentration of the biotin-MNP solutions. Lastly, the 10 biotin-MNP and 90 biotin-MNP solutions were diluted to 1 mg Fe/mL based on their .
2.4. Physical Characterization of Biotin-MNPs
Biotin-MNP aliquots (10 mg Fe/L concentration) were submitted to DLS, to verify their
and zeta potential in the 1.00x to 0.00x PBS series. The DLS measurements were conducted in quintuplicate. The zeta potential measurements were conducted in quintuplicate as well, with
measurements before and after each zeta potential measurement to monitor biotin-MNP stability. The measurement parameters for the biotin-MNP samples were identical to those used for the physical characterization of the SHA 25 MNPs (
Section 2.2).
2.5. Magnetic Particle Spectroscopy Measurements
To obtain streptavidin-response curves for the 90 biotin-MNPs in 1.00x, 0.50x, 0.03x, 0.015x, 0.01x, 0.005x, and 0.00x PBS solutions, streptavidin serial dilutions spanning from 0.0 M to 2,694.8 M streptavidin were prepared in each PBS solution. Each MPS sample contained 2.5 L of a 1 mg Fe/mL 90 biotin-MNP solution mixed with 2.0 L of a streptavidin serial dilution sample. Additionally, streptavidin-response curves for the 10 biotin-MNPs were obtained in 1x PBS and DI. To obtain the streptavidin-response curve in 1.00x PBS for the 10 biotin-MNPs, a streptavidin serial dilution spanning from 0 M to 193 M streptavidin was prepared in 1.00x PBS. To obtain the streptavidin-response curve in DI for the 10 biotin-MNPs, a streptavidin serial dilution spanning from 0 M to 322 M streptavidin was prepared in DI. Each MPS sample was prepared in triplicate. The of each sample was measured in triplicate with the NCS after an approximate 40 minute incubation at room temperature. At the onset of each measurement, the NCS identifies the magnetic center of each sample. Next, the sample is measured at 7 points along the z-axis of the sample (from bottom to top). Where the center point is located at the magnetic center of the sample. The 7 point measurement is repeated 4 times during each NCS measurement.
We compared the 90 biotin-MNP streptavidin-response curves in 1.00x, 0.50x, 0.03x, 0.015x, 0.01x, 0.005x, and 0.00x PBS solutions to determine the effects of salt concentration of streptavidin induced biotin-MNP aggregation. We compared the 90 biotin-MNP and 10 biotin-MNP streptavidin response curves in 1.00x PBS to determine the effects of biotin density on streptavidin induced biotin-MNP aggregation. We compared the slope, maximum , and linear range of each streptavidin-response curve. We used analysis of covariance (ANCOVA) to determine whether the slopes of each of the curves was statistically significantly different. Specifically, Matlab’s “aoctool” command was used to perform the ANCOVA testing.
2.6. xDLVO Interparticle Energies
We modeled the repulsive EDL energy as well as the attractive van der Waals and dipolar energies acting amongst biotin-MNPs to gain further insights on particle stability in each PBS solution. The EDL energy between 2 NPs (sphere-sphere geometry) was calculated as [
31,
32]
where
is the permittivity of free space,
is the relative permittivity of the medium,
is the biotin-MNP magnetic core radius,
is the biotin-MNP surface potential, and
is the inverse Debeye length. EDL energy will decrease with increasing electrolyte concentration due to changes in biotin MNP Debeye length and surface potential. The zeta potential of the biotin MNPs in each PBS strength solution were used in place of the surface potential. Walker et. al., 2009 [
31] and Fiegel et. al., 2019 [
32] similarly substituted MNP zeta potential for surface potential in their EDL calculations. Our highest concentrated electrolyte solution, 1.000x PBS, has the following electrolyte contents: 157.00 mM
, 142.00 mM
, 4.45 mM
, and 7.30
, and 4.60 mM
. We assumed electrolyte concentration decreased linearly with decreasing PBS strength. Therefore, we assumed 0.500x PBS had 50% less electrolytes than 1.000x PBS, for example. However, we handled the EDL energy calculation for DI differently to agree with reported values of the Bjerrum length
in pH 7 DI at room temperature.
is the interparticle distance at which the EDL energy is comparable in magnitude to the thermal energy of the system [
33].
in pH 7 DI water at room temperature is 1
m, which corresponded to a Debye length (
) of 4.10
m for our biotin-MNPs in DI. van der Waals and dipolar energies remain constant with respect to electrolyte concentration. The van der Waals energy between 2 NPs was calculated as [
31]
where
is the Hamaker constant and
is the characteristic wavelength of the medium, which assumed to be 100 nm [
32]. van der Waals attractive energy is proportional to the particle Hamaker constant. Bergsrtom et. al., 2011 determined the Hamaker constant for magnetite NPs in water was 33 zJ [
34]. We utilized this Hamaker constant value for our van der Waals interparticle energy calculation. The dipolar energy between 2 NPs was calculated as [
19,
31]
where
is the permeability of free space and
is the magnetic saturation. Interparticle dipolar energy is proportional to the magnetic saturation of the biotin-MNPs. Wang et. al., 2021 reported that Ocean Nanotech SHA 25 MNPs had a magnetic moment per particle of 1.58 x
emu (1.99 x
) [
35]. We divided this value by the biotin-MNP core volume to calculate the magnetic saturation of our biotin-MNPs and ultimately the dipolar interaction energy. We modeled dipolar energy as an attarctive interaction. Dipolar energy can be either attractive or repulsive depending on the angle between the magnetic moment and the line connecting the particle centers [
36]. If the angle is
then the interaction is attractive, whereas if
then the interaction is repulsive. We used two-sample Kolmogorov-Smirnov (KS) tests to determine whether the total interation energy corresponding to each PBS solution had statistically significantly variations Specifically, Matlab’s “kstest2” command was used to perform the KS testing.
4. Discussion
SHA 25 MNPs diluted in PBS had a
of 60.19 ± 1.90 nm (
Figure 2C). Previous work from Wu et. al., 2021, reports SHA 25 MNPs had a
of 28.28 ± 10.38 nm in DI after 30 minutes of ultrasonication [
35]. Our SHA 25 MNP measurements were conducted without ultrasonication or other means of sample manipulation. It is important to note that the choice of diluent and Fe concentration can affect the accuracy of
measurements. DLS measurements conducted in DI typically yield
values 2-10 nm larger than the particle’s actual
[
40]. This inaccuracy is attributed to the absence of ions in solution to shield long distance interparticle interactions [
40]. Lim et al., 2013 reported an approximate 148 nm increase in MNP
due to an increase in the sample solution’s Fe concentration from 100 to 250 mg Fe/L for 18 nm superparamagnetic MNPs [
37]. Constant
data was acquired for the 18 nm superparamagnetic MNPs at Fe concentrations between 10 and 50 mg Fe/L. Therefore, we conducted our
measurements with 10 mg Fe/L MNP samples of our 25 nm core MNPs. Moreover, MNP core size must also be considered when determining an optimal Fe concentration for
measurements. At a constant Fe concentration, larger MNPs have a lower collision frequency, hence their
would more accurately reflect the physical properties of individual MNPs, compared to smaller MNPs. When comparing literature values of NP
one must consider the diluent, NP concentration, and core size to make an informed comparative analysis.
After biotinylating SHA 25 MNPs with ∼ 10 and 90 biotins/MNPs, we investigated the effects of ligand density on streptavidin induced biotin-MNP aggregation. We estimate that MNP biotin density is 2 times the streptavidin:biotin-MNP ratio at which the maximum
occurs. Traditional methods for NP ligand density quantification [
38] include nuclear magnetic resonance (NMR), fluorescently labelled ligands, and absorption spectroscopy. Previous works have used an absorption spectroscopy technique, wherein the ligand density of AuNPs was estimated by the shape of the absorbance versus added protein curve [
41,
42]. The protein concentration at which the absorbance curve plateaued indicated the amount of ligand bound to the AuNP surface. Our biotin-MNP ligand density quantification method is similar, however we multiply the peak value of our target response curve with 2. The multiplication factor is applied since our added protein is an aggregation inducing agent. We assume that each streptavidin is bound to 2 biotin-MNPs. Streptavidin-response curves were obtained for the 10 and 90 biotin-MNPs in 1.000x PBS. We compared the slope, maximum
, and range of the linear region of each streptavidin-response curve. Our results suggest an increase in streptavidin-response curve slope with decreasing biotin density. In other words, biotin-MNP streptavidin sensitivity increased with decreasing biotin density. Additionally, we found the streptavidin-response curve maximum
decreased with decreasing biotin-density. We suspect lower biotin density MNPs form smaller and more loosely packed clusters with streptavidin, compared to high biotin density MNPs. Our results can be leveraged for MNP-based aggregation assay development. fMNP target sensitivity and dynamic range can be tuned with ligand density. Another element of our data to consider for aggregation assay development pertains to the physical characterization of the SHA 25, 10 biotin-MNPs, and 90 biotin-MNPs. Our results suggest a non-linear relationship amongst ligand density and MNP
as well as zeta potential. NP
and zeta potential are commonly used to assess the level/success of NP functionalization. Our results suggest that
and zeta potential may not indicate the level/success of functionalization given that the SHA 25 and 90 biotin-MNPs showed no significant variation in
. An alternative method to assess the level of NP functionalization is to expose the bare and potentially functionalized NPs to varying amounts of target and compare transducer outputs. If the potentially functionalized NPs aggregate in the presence of their target, while the bare NPs remain fully dispersed, one can presume the NPs were successfully functionalized. This method could also be used to determine NP receptor density, wherein the target concentration corresponding to the maximum transducer output is an approximate indication of NP receptor density.
To determine the effects of electrolyte concentration on streptavidin induced biotin-MNP aggregation, streptavidin-response curves were obtained for the 90 biotin-MNPs in 1.00x, 0.50x, 0.03x, 0.015x, 0.005x, and 0.00x PBS solutions. We compared the slope, maximum , and range of the linear region of each streptavidin-response curve. There were no significant variations amongst linear region slope or range for the streptavidin-response curves prepared in 1.00x, 0.50x, 0.03x, 0.015x, and 0.005x PBS solutions. The biotin-MNPs formed aggregates with streptavidin in the 1.00x, 0.50x, 0.03x, 0.015x, and 0.005x PBS solutions, however the biotin-MNP were insusceptible to streptavidin induced aggregation in 0.00x PBS. The differences in biotin-MNP interparticle interactions were predicted with our xDLVO simulation. According to our simulation, interparticle interactions were dominated by the attractive van der Waals force in solutions with PBS strengths ranging from 0.005x to 1.00x PBS, while in 0.00x PBS interparticle interactions were dominated by the repulsive EDL force at particle separation distances between ∼23 nm and 50 nm. We have demonstrated that seemingly incremental changes in NP zeta potential and/or diluent electrolyte concentration can yield significant changes in NP-target interactions. For example, when suspended in 0.005x PBS (a relatively low electrolyte concentration diluent) the 90 biotin-MNPs had a zeta potential of 6.53 ± 0.72 mV and particle interactions were dominated by the attractive van der Waals force for separation distances (s) . However, after seemingly incremental changes in diluent to 0.00x PBS and 90 biotin-MNP zeta potential to 11.24 ± 3.88 mV, particle interactions were dominated by the repulsive EDL force for , resulting in no detectable levels of streptavidin induced biotin-MNP aggregation. Therefore, our study highlights the importance of xDLVO simulation during aggregation assay design to tune fMNP zeta potential to the diluent electrolyte concentration and pH for optimal target binding and colloidal stability. Concerning the effects of electrolyte concentration on the streptavidin-response curve maximum , no correlation was found.
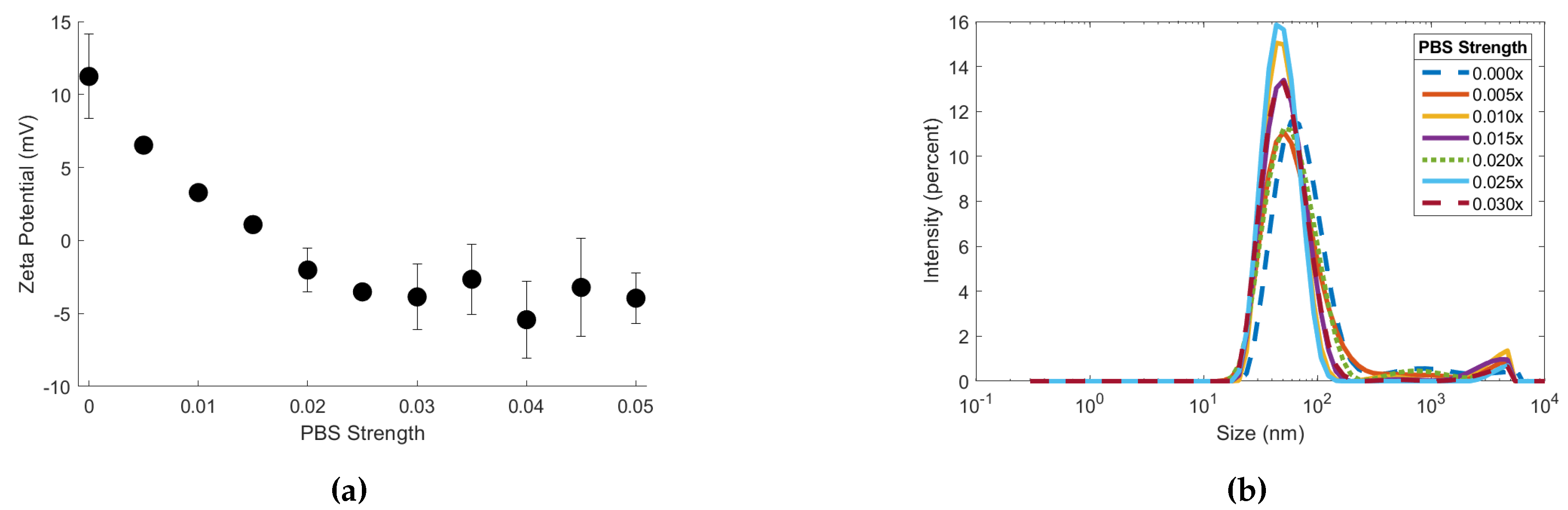
and zeta potential measurements were obtained for the 90 biotin-MNPs in the 1.00x to 0.00x PBS series to assess the effects of electrolyte concentration on colloidal stability. Ninety biotin-MNP zeta potential decreased linearly in solutions with PBS strengths ranging from 0.025x to 0.00x PBS (
Figure 3a). While 90 biotin-MNP zeta potential was constant in solutions with PBS strengths ranging from 1.00x to 0.03x PBS (
Figure 3a). Ninety biotin-MNP
showed no trend with electrolyte concentration, therefore 90 biotin-MNP
is independent of electrolyte concentration in solutions with PBS strengths ≤ 1.00x. Our 90 biotin-MNP
data as a function of diluent electrolyte concentration is limited because it does not account for aggregate growth over time. Our
data was acquired directly after the 90 biotin-MNPs were mixed with each PBS solution. Hence, our data reflect the initial 90 biotin-MNP size after exposure to diluents of varying PBS strength. Salt induced NP aggregates may grow in size over time depending on the salt concentration of the diluent [
39,
43,
44,
45]. To further verify whether 90 biotin-MNP
is independent of electrolyte concentration in solutions with PBS strengths ≤ 1.00x, one can assess whether MNP
changes with time in ≤ 1.00x PBS solutions.
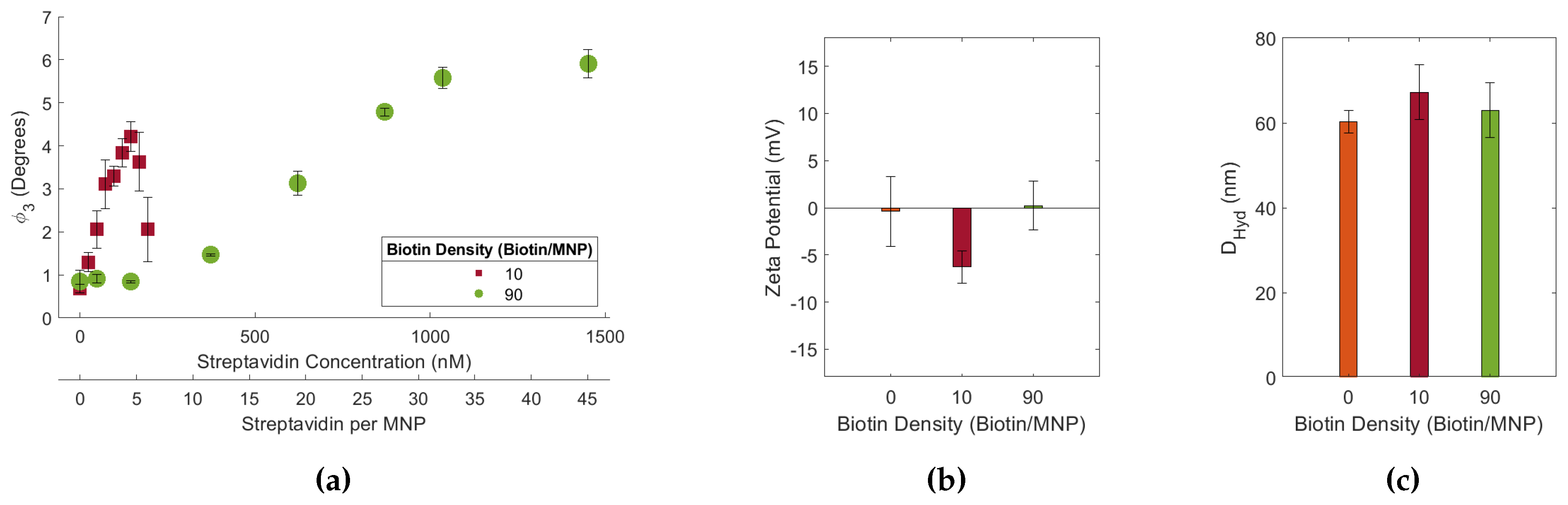
Figure 2.
(a) 10 and 90 biotin-MNP as a function of streptavidin concentration (streptavidin-response curve) in 1.00x PBS. Where 90 biotin-MNP in 0.00x PBS is regarded as zero. (b) Zeta potential and (c) of 10 and 90 biotin-MNPs in 1.00x PBS. The streptavidin response curve shows that the assay’s sensitivity increases as biotin density decreases.
Figure 2.
(a) 10 and 90 biotin-MNP as a function of streptavidin concentration (streptavidin-response curve) in 1.00x PBS. Where 90 biotin-MNP in 0.00x PBS is regarded as zero. (b) Zeta potential and (c) of 10 and 90 biotin-MNPs in 1.00x PBS. The streptavidin response curve shows that the assay’s sensitivity increases as biotin density decreases.
Figure 3.
(a) Zeta potential of 90 biotin-MNPs in 0.05x, 0.045x, 0.04x, 0.035x, 0.03x, 0.025x, 0.02x, 0.015x, 0.01x, 0.005, and 0.000x PBS solutions. (b) Hydrodynamic diameter of 90 biotin-MNPs in 0.03x, 0.025x, 0.02x, 0.015x, 0.01x, 0.005, and 0.00x PBS solutions.
Figure 3.
(a) Zeta potential of 90 biotin-MNPs in 0.05x, 0.045x, 0.04x, 0.035x, 0.03x, 0.025x, 0.02x, 0.015x, 0.01x, 0.005, and 0.000x PBS solutions. (b) Hydrodynamic diameter of 90 biotin-MNPs in 0.03x, 0.025x, 0.02x, 0.015x, 0.01x, 0.005, and 0.00x PBS solutions.
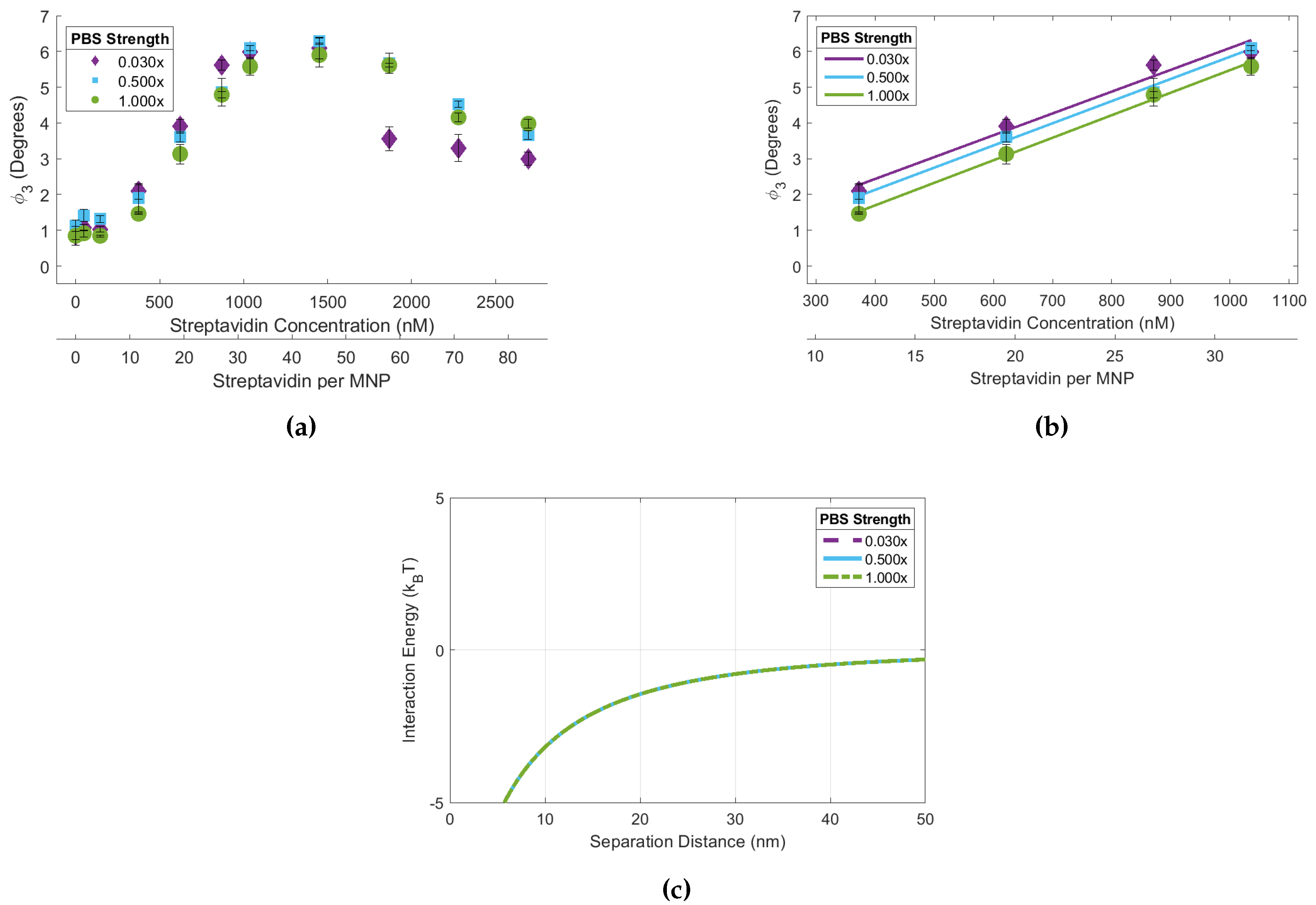
Figure 4.
(a) 90 biotin-MNP as a function of streptavidin concentration (streptavidin-response curve) in 1.00x, 0.50x, and 0.03x PBS solutions. Where 90 biotin-MNP in 0.000x PBS is regarded as zero. (b) The linear region of each curve fit with a linear regression, where there were no statistically significant variations amongst regression model slopes. (c) Total interparticle energy profiles for 90 biotin-MNPs in 1.00x, 0.50x, and 0.03x PBS solutions, where the energy profiles are indistinguishable and overlay each other. These data show that 90 biotin-MNP sensitivity to streptavidin induced aggregation is constant for 0.03x, 0.50x, and 1.00x PBS solutions. Our experimental results were supported by xDLVO simulations demonstrating that the total interparticle energy profile corresponding to each PBS solution came from the same continuous distribution.
Figure 4.
(a) 90 biotin-MNP as a function of streptavidin concentration (streptavidin-response curve) in 1.00x, 0.50x, and 0.03x PBS solutions. Where 90 biotin-MNP in 0.000x PBS is regarded as zero. (b) The linear region of each curve fit with a linear regression, where there were no statistically significant variations amongst regression model slopes. (c) Total interparticle energy profiles for 90 biotin-MNPs in 1.00x, 0.50x, and 0.03x PBS solutions, where the energy profiles are indistinguishable and overlay each other. These data show that 90 biotin-MNP sensitivity to streptavidin induced aggregation is constant for 0.03x, 0.50x, and 1.00x PBS solutions. Our experimental results were supported by xDLVO simulations demonstrating that the total interparticle energy profile corresponding to each PBS solution came from the same continuous distribution.
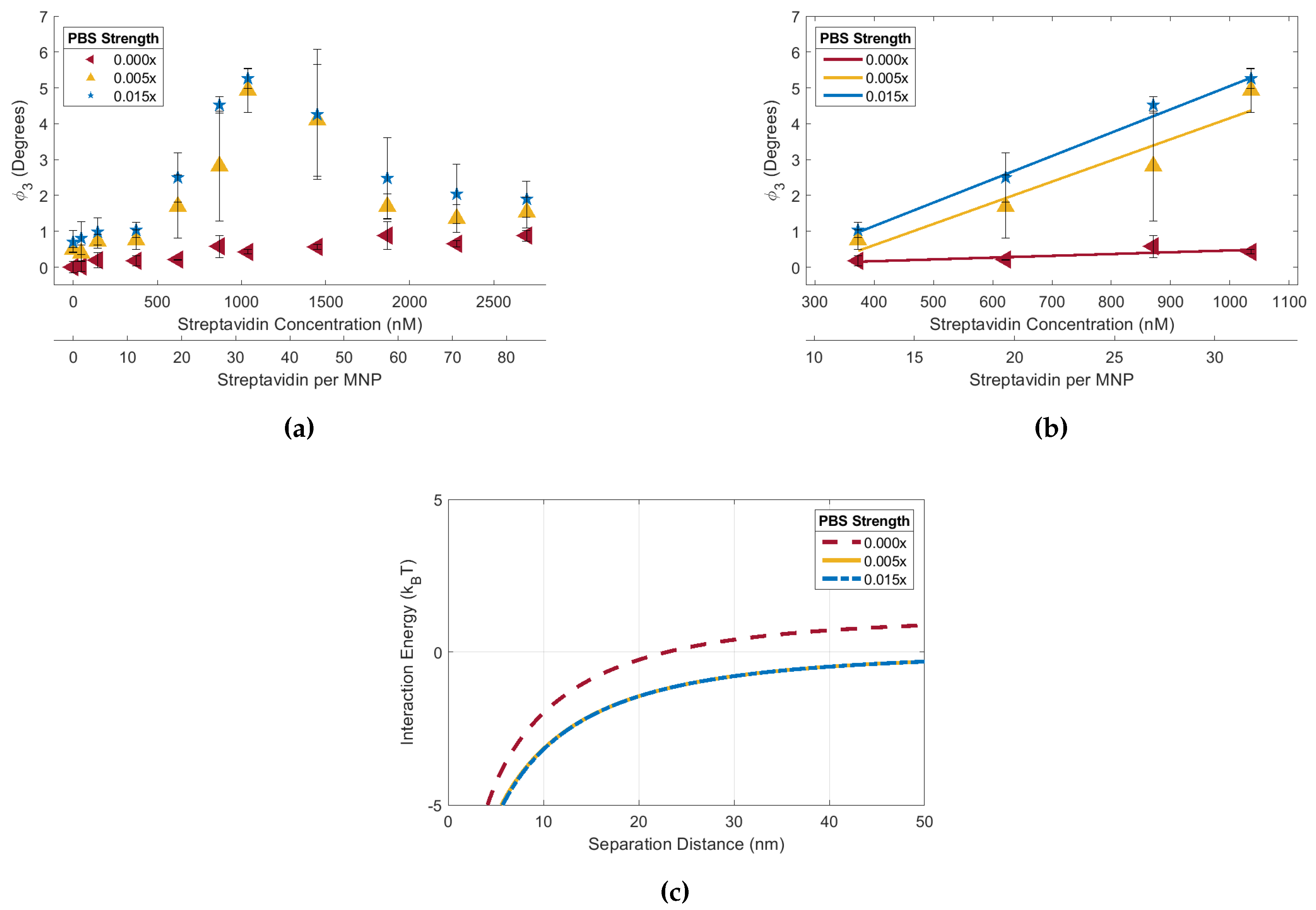
Figure 5.
(a) 90 biotin-MNP as a function of streptavidin concentration (streptavidin-response curve) in 0.015x, 0.005x, and 0.00x PBS solutions. Where 90 biotin-MNP in 0.000x PBS is regarded as zero. (b) The linear region of each curve fit with a regression, where the 0.000x regression model slope had statistically significant variations from the regression model slopes corresponding to the 0.015x and 0.005x solutions. (c) Total interparticle energy profiles for 90 biotin-MNPs in 0.015x, 0.005x, and 0.00x PBS solutions, where the 0.000x total interparticle energy profile had statistically significant variations from the energy profiles corresponding to the 0.015x and 0.005x solutions. These data show that 90 biotin-MNP senstivity to streptavidin induced aggregation is significantly decreased in 0.000x PBS. Our experimental result was supported by xDLVO simulation demonstrating that the total interparticle energy profile corresponding to the 0.00x is dominated by the attractive van der Waals force for separation distances between 0 and 22 nm, while interactions are dominated by the repulsive EDL force for separation distances between 23 and 50 nm. However, total interparticle energies corresponding to the 0.015x and 0.005x solutions are dominated by the attractive van der Waals force for separation distances between 0 and 50 nm.
Figure 5.
(a) 90 biotin-MNP as a function of streptavidin concentration (streptavidin-response curve) in 0.015x, 0.005x, and 0.00x PBS solutions. Where 90 biotin-MNP in 0.000x PBS is regarded as zero. (b) The linear region of each curve fit with a regression, where the 0.000x regression model slope had statistically significant variations from the regression model slopes corresponding to the 0.015x and 0.005x solutions. (c) Total interparticle energy profiles for 90 biotin-MNPs in 0.015x, 0.005x, and 0.00x PBS solutions, where the 0.000x total interparticle energy profile had statistically significant variations from the energy profiles corresponding to the 0.015x and 0.005x solutions. These data show that 90 biotin-MNP senstivity to streptavidin induced aggregation is significantly decreased in 0.000x PBS. Our experimental result was supported by xDLVO simulation demonstrating that the total interparticle energy profile corresponding to the 0.00x is dominated by the attractive van der Waals force for separation distances between 0 and 22 nm, while interactions are dominated by the repulsive EDL force for separation distances between 23 and 50 nm. However, total interparticle energies corresponding to the 0.015x and 0.005x solutions are dominated by the attractive van der Waals force for separation distances between 0 and 50 nm.