1. Introduction
Among all causes of death, cancer ranks sixth in the world,it is an essential pathological condition.Brain tumours are thought to be among the deadliest malignancies, with the meager survival rates.[
1] tumour shape, texture, location and other factors can significantly affect the shape tumours that occur in the brain include glioma, pituitary, and meningioma [
2].The rates of occurrence for tumours of brain during clinical monitoring are approximately 45 percent, 15 percent, and 15 percent, respectively, for brain tumours. Having a good doctor may make this diagnosis and predict survival of patients based on the type of tumour [
3]. Additionally, make a decision.Following appropriate care before surgery. Radiotherapy and chemotherapy adopt a "wait and see" strategy. Keep away of invasive procedures. Even so, a crucial part of treatment planning is tumour grading [
4].
Brain tumour can be detected in 2D and 3D using magnetic resonance imaging (MRI), a nonsurgical, quick and easy medical imaging technique. It is one of the most widely accepted techniques for identifying and detecting cancer which features ultra-high definition pictures of brain tissue. [
5].
But identifying the type of cancer using tomography images may be a challenging, incorrect,time-consuming ,and highly specialized physician’s experience and is a laborious process[
6]. tumours can appear in many different of shapes, and the image may not contain enough discernible landmarks to aid in a medical assessment. In summary, manual diagnosis is often inaccurate. Furthermore, treating brain tumours is frequently a severe problem because it can impair a patient’s ability to respond effectively to significant surgery and lower their chances of survival[
7]. However, a proper diagnosis could well benefit patients to begin the right course of treatment right away. Consequently, there is a critical need to develop innovative machine diagnosis systems in artificial intelligence (AI). These systems aim to alleviate the burden of diagnosing patients and tumour classification, providing a helpful radiologists’ and physicians’ tool [
8].By decreasing the impact associated with tumour diagnosis and classification, these systems hope to benefit doctors and radiologists[
8].
Among the most popular used convolutional neural networks (CNN), has several layers which is based on linear equations between matrices known as convolutions.In addition, it has fully connected, nonlinear, pooling, and conventional layers. While fully connected, traditional layers have parameters, nonlinear and pooling layers. [
9].
The EfficientNetV2 may be new type of CNN designed to improve speed and effectiveness than past structures. It conveyed foremost excellent comes around and the foremost lifted effectiveness in classifying many kinds of the ImageNet .In brief,the EfficientNetV2B3 may be a sensible model for medical image classification design that concedes an input of 32X32 image and 14.5M parameters with an accuracy of 95.8%.[
10].
EfficientNetV2 surpasses EfficientNet to improve speed and effectiveness parameter productivity and created by employing a set of scaling (profundity, determination,and width).
EfficientNetV2 design which is much quicker than past and more current states of art model and is much littler (up to 6.8x times)[
11].
The input image size and the regularization parameter are user defined,where EfficicentNetV2 employments three diverse sorts of regularization Dropout, RandAugment,and Mistake[
12].
The CNN model, which comprises five blocks of layers with two dropout layers and max pooling after each block, is combined with EfficientNetV2B3 in the proposed framework.with a primary block of 32 filters and individual blocks of 64, 128 and 256 filters.[
13].
EfficientNetV2B3 combines dropout, pooling, and batch normalization ,24 layer architecture consist of 1 fully connected layer,1 soft max layer ,and 22 convolutional layers.Eight blocks make up the convolutional layers, and the inputs of images is changed in size to 32 by 32 pixels. [
14]. When the output of each model is integrated from 2 layers, each having 64 and 16 units, the proposed model performs better. [
15]
Many algorithms employed in brain tumour classification such as KNN and deep learning methods.In this research,A hybrid framework combining (CNN and EfficientNetV2B3) using for extracting feature and employing KNN for classifying images.
The organization of This essay is organized as follows:
Section 2 presents related research on the classification of brain tumours.
Section 3 illustrates the Problem definition.
Section 4 illustrates proposed approach, including the data set, image prepossessing, data augmentation, feature extraction, classification models,implementation and evaluation metrics.
Section 5 presents results of our experiments.section 6 presents ablation study ,and discussion is presented in section 7.Finally conclusions of this proposed hybrid method is presented in section 8.
2. Related Work
Image processing techniques play a significant role in medical application through assisting detect anomalies and diagnose diseases.This covers the view of medical images from imaging techniques such as CT scans, MRIs, and X-rays. MRI, in particular, has a significant advantage over other disease diagnosis because it doesn’t expose patients to radiation.
Across several domains,including computer vision, robotics, and most computer-aided diagnosis,deep learning has become a powerful implement.This paves the way for its integration with biomedical image processing, which could result in significant improvements in medical care.
Deep learning models acquire the capability to learn several representation and abstraction levels from a large scale because they are fed with raw data.There is more advantages in similar to existing machine learning techniques because of they restricted to processing only image data through real life,and more slow and requires a lot of effort to adjust the functionality.[
16]
CNN are among these types. They are most commonly utilized for image,video,and speech recognition.The biological functions of the visual system in animals have served as an inspiration for CNN. CNN’s ability to identify patterns in images has allowed it to be successfully used primarily in image processing.[
18]
Convolutional layers Is Imposed of images extracting features like colors and edges,and relies on utilizing learnable kernel.Cores’ spatial dimension is small but distributed over input area depth.layers spread every filter horizontally create a feature map using the spatial dimensions of the input.It is the pooling layer which is Imposed of decreasing number of variables and mathematical model’s complexity while lowering the dimension of the feature.Many algorithms have been employed in brain tumour classification like KNN.
Cheng et al. [
34] proposed model of GLCM,and BOW which applied to a datasets of three different kinds of brain tumours and obtained an precision of 91.28 percent.
Ismael et al. [
36] presented model of neural network algorithm and using statistical features obtained 91.9% precision. Specificity for pituitary tumours was 95.66%, 96%, and 96.29% for meningioma or glioma,respectively.
Afshar et al. [
37] proposed model for categorizing brain tumour of rough boundaries of the tumour as additional pipeline of input to improve Cap’s Net focus with an accuracy of 90.89%.In [
40], Deepak et al. proposed a comprehensive framework that pre-trained Google Net to feature extraction on MRI dataset. Gull et al. [
41] proposed a model uitized AlexNet and VGG-19 for brain tumour classification and achieved an accuraccy of 98.50% ,and 97.25% respectively for VGG-19 and AlexNet.Mondal et al.[
42] compared results of various CNN models like (DenseNet201, and ResNet50) achieved an accuracy of 98.33%
3. Problem Definition
Determining tumour grade and type is essential,vital at the beginning of the treatment plan.Accurate detection of abnormal tissue is essential,vital for diagnosis.That’s fully confirmed by the availability of practical methods that employ classification, segmentation, or combining both to quantitatively and subjectively characterize the brain. Processing MR images can be done manually, semi-automatically, or through fully programmed processes based on human interaction.Accurate segmentation and classification are essential for medical image processing, but they must usually be done manually by specialists, which takes time. Conversely, however,an accurate diagnosis can help patients start appropriate receive treatment sooner and have longer lifespans. Therefore, in the area of artificial intelligence (AI), there is a pressing need for the development and design of fresh and creative framework to decrease the amount of work that radiologists and physicians must do to diagnose and characterize tumours.
This paper uses an automation system that was developed for the purpose of classifying and segmenting brain tumours. This new technology could significantly improve the diagnostic skills of neurosurgeons and other medical professionals, especially when it comes to analyzing tumours ectopic brain regions, which is crucial for early and precise diagnosis.One of the main principles of this research is to promote accessible and efficient communication. To decrease the knowledge gap between medical professionals, the system simplifies the presentation of Magnetic Resonance Imaging (MRI) results. This improved comprehension can enable well-informed decision-making across the board for clinical workflow.Furthermore, the proposed methodology champions a fully automated approach, minimizing the need for extensive pre-processing steps. This not only optimizes workflow efficiency but also mitigates the potential for human error during pre-processing, ultimately leading to more consistent and reliable results.
4. Proposed Approach
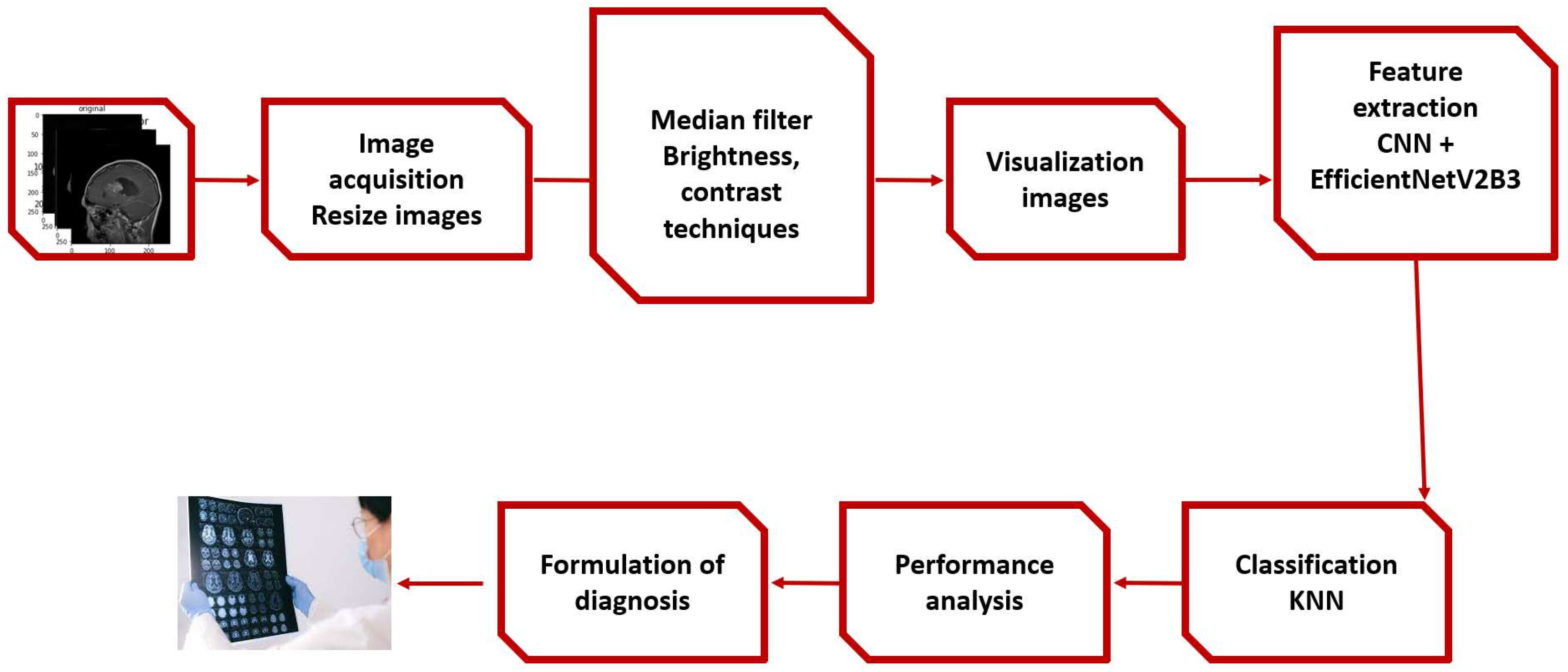
The proposed framework,and experimental stage described in
Figure 1 are discussed in this section.
4.1. The proposed Framework
Within the suggested framework, the input image shown in
Figure 2 and
Figure 3 from MRI data sets. [
26]
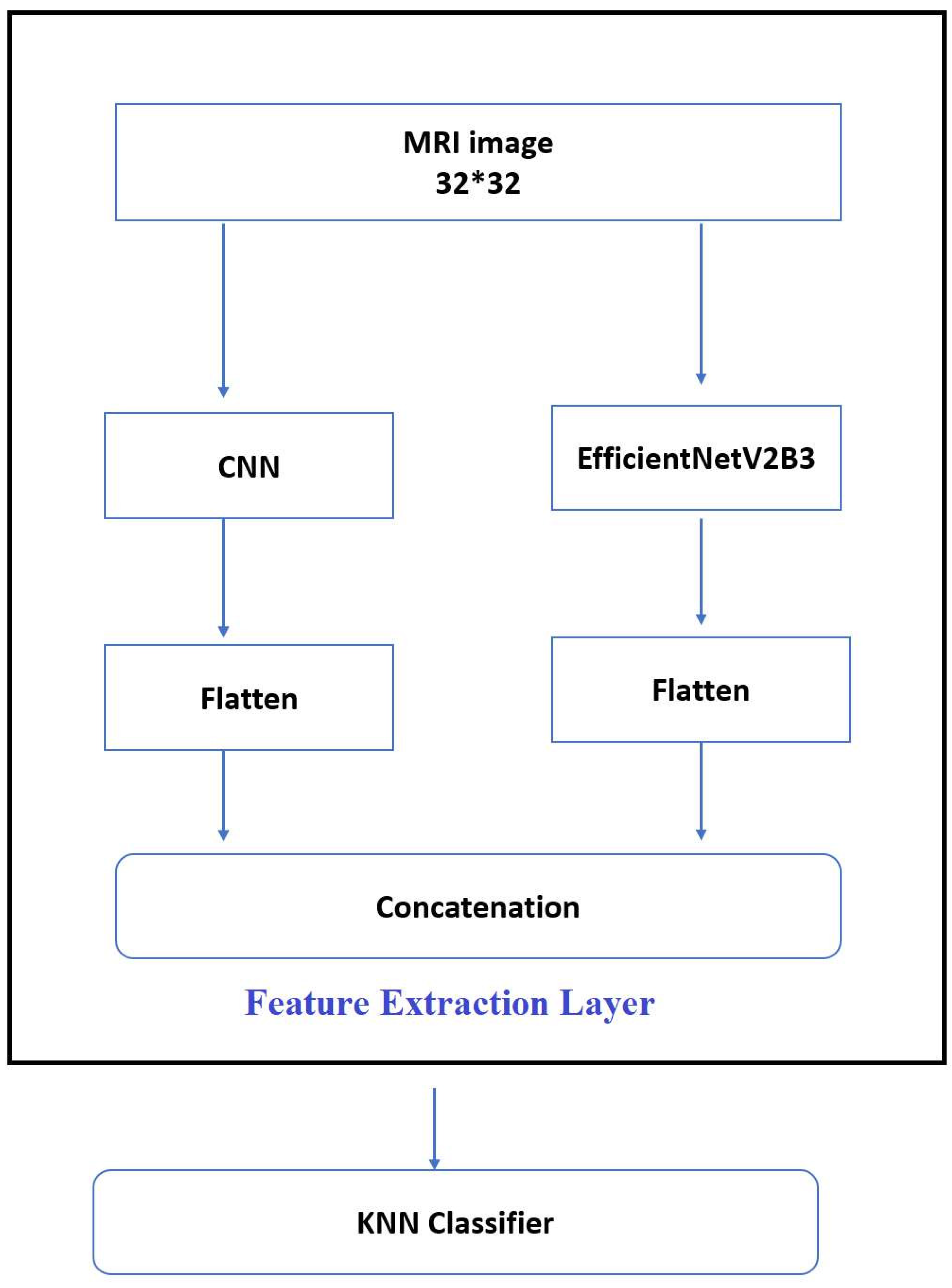
First,apply median filter to remove noise,then apply augmentation techniques such as brightness and contrast,and then transfer to extraction stage by combining EfficientNetV2B3 and the CNN and tested using KNN classifier models except for the deep CNN models’ standard soft-max classifier.Our studies used KNN because it was inspired by earlier research that showed KNN to perform well in CNN feature classification.
4.2. Dataset
We worked with two distinct publicly accessible brain MRI data sets to perform a experimental studies.
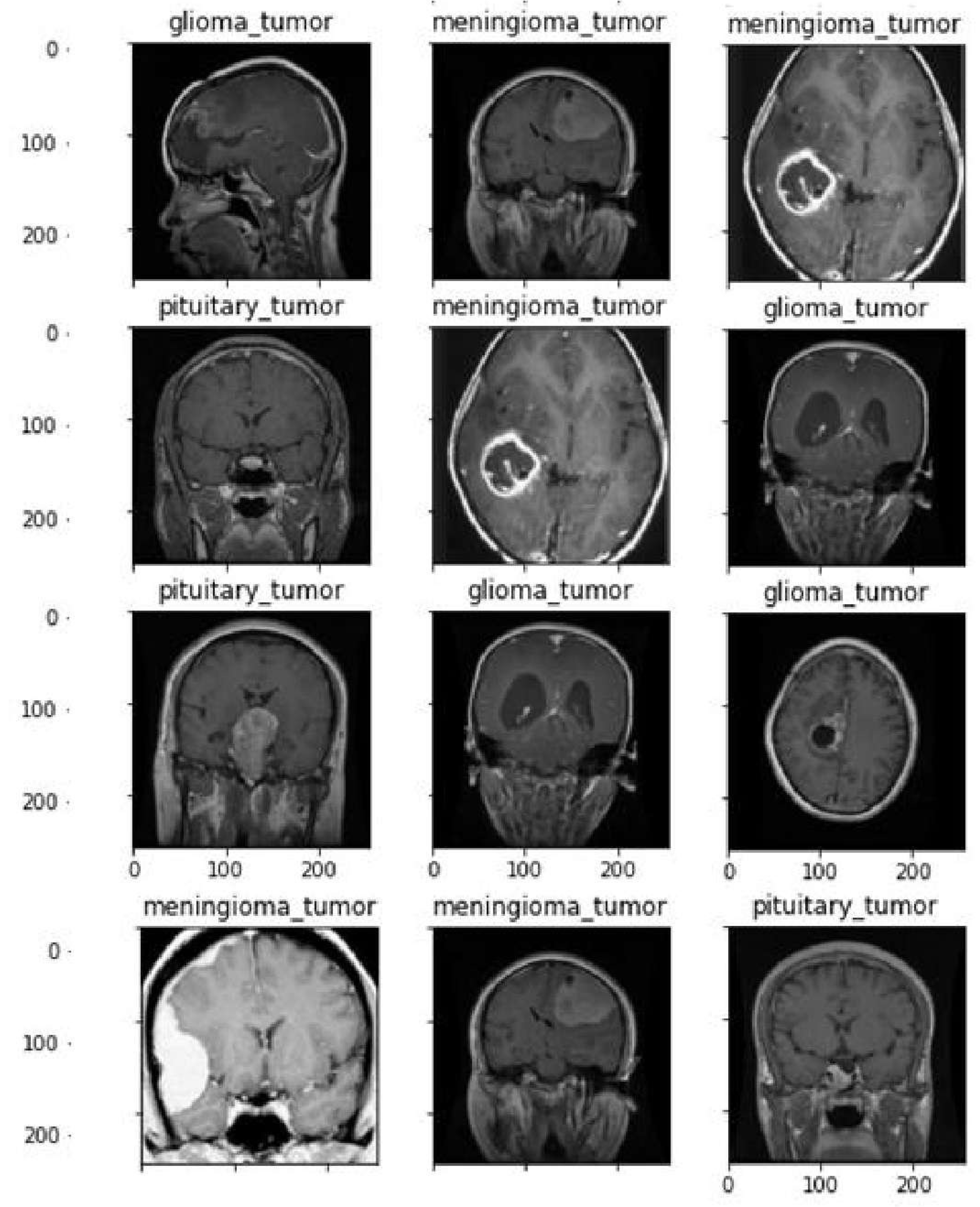
A first data set of three kinds of brain tumours MRI pictures of glioma,meningioma,and pituitary.Cheng et al.[
26] processed the data set initially which was obtained from Tianjin Medical University General Hospital and Nan Fang Hospital in Guangzhou, China.
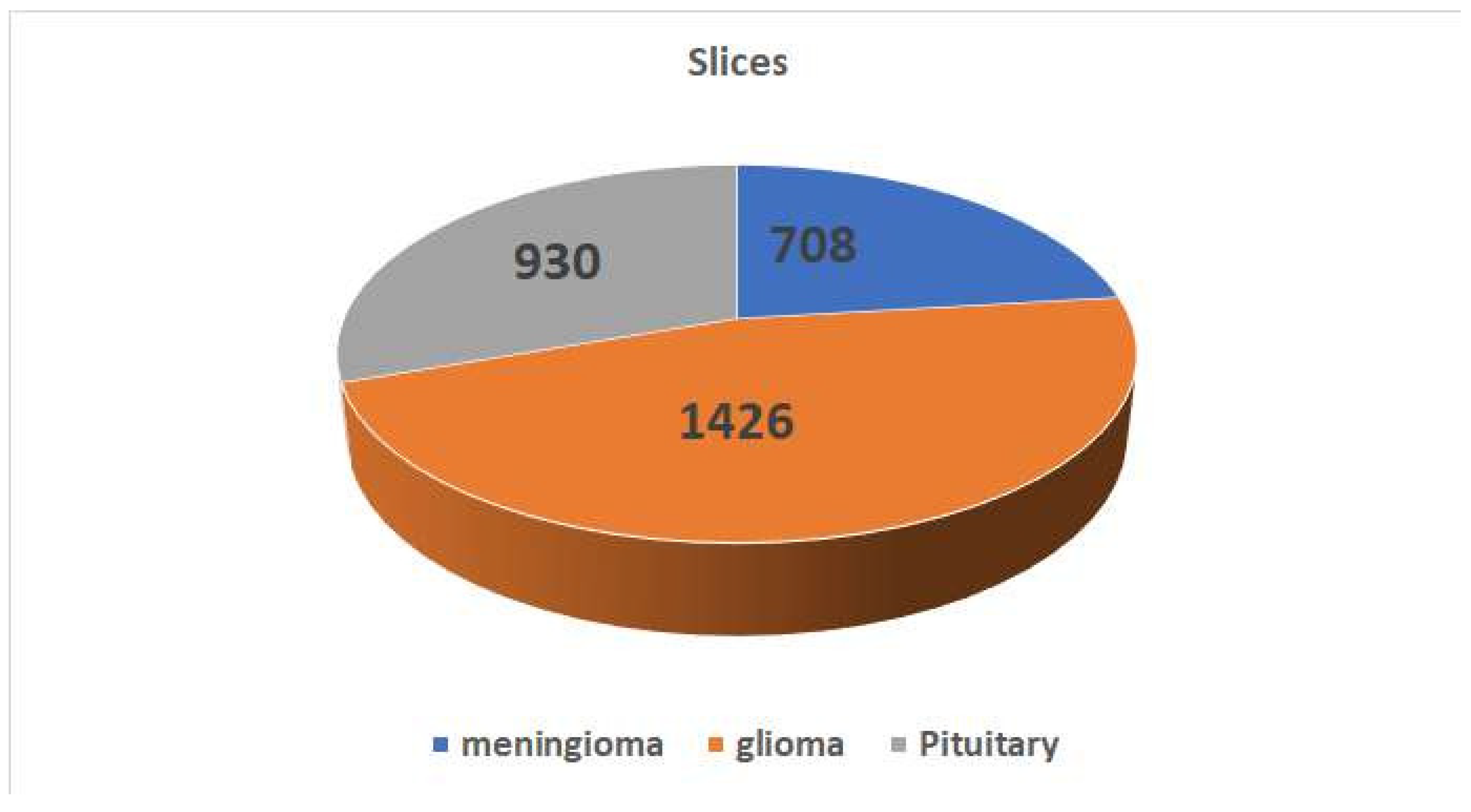
Figure 4 displays a selection of sample images from the data set along with their corresponding class labels. Ground truth for the tumour data set includes the following tumour types: Images of the pituitary, glioma, and meningioma total 930, 1426, and 708 respectively.
Figure 4 shows how the pictures are distributed within each category.The data set of brain CE-MRI images can be accessed at
1
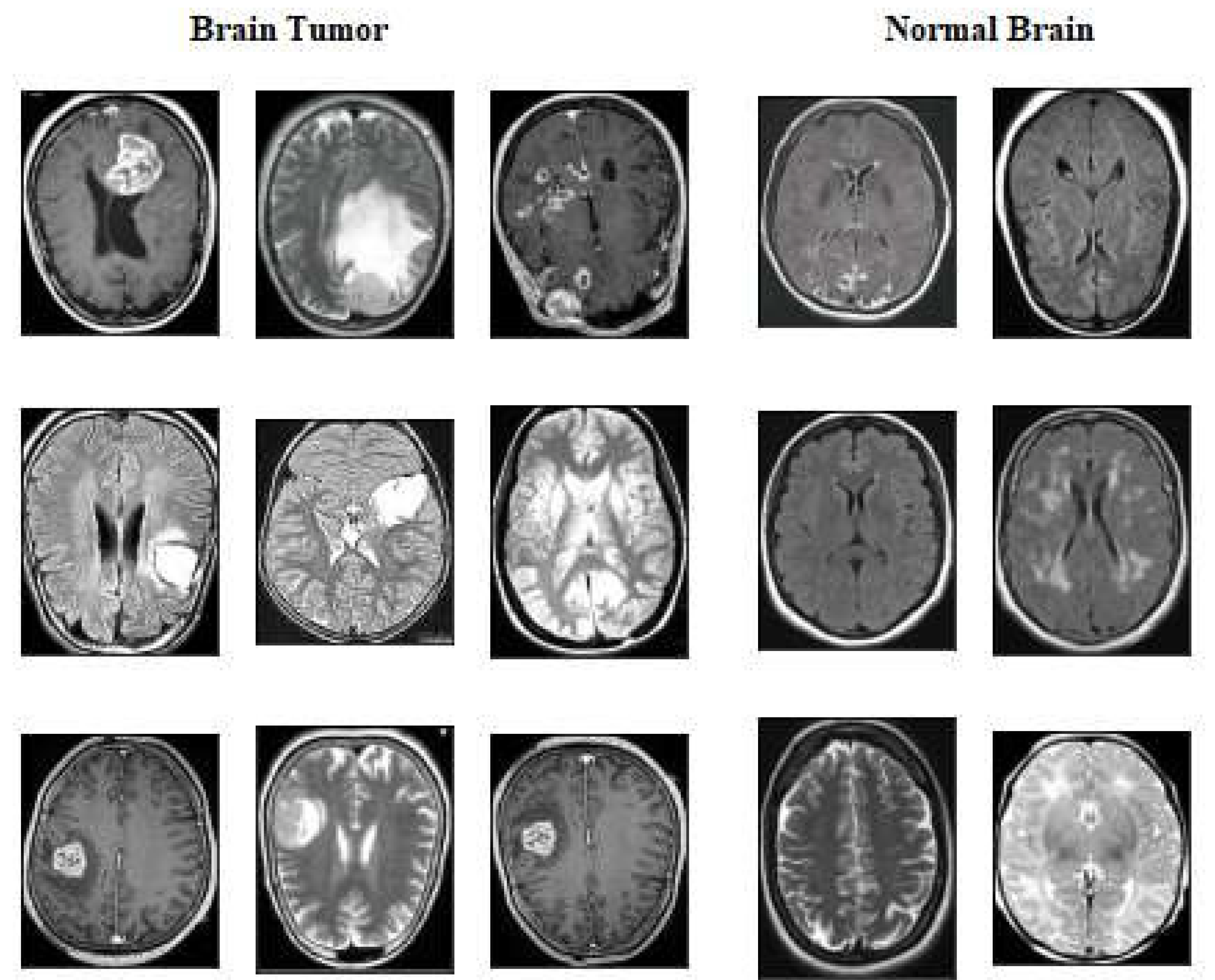
The second dataset Brain tumour Detection 2020 (BR35H) [
44] is MRI dataset obtained from Kaggle website at
2. We called this dataset as for our experimental work dataset2 of two classes which contains 1500 images of tumour class ,1500 images of normal or non tumour.
4.3. Images Pre-Processing
MRI images contain a significant amount of noise, which could be caused by the surroundings, instrumentation, or operator error.These could cause significant MRI scan inaccuracies. Thus, first stage is to clean up the MRI image from noise.two different kinds of noise reduction methods:linear and non-linear. Weight average of the neighborhood was used to update the pixel value in linear filters for noise reduction. The image quality is decreased by this process.Conversely, in non-linear methodology,the delicate structures are degraded but the sides remain intact[
27]. In this case, the noise from images was removed using a median filter. As illustrated in
Figure 5, described median filter technique.
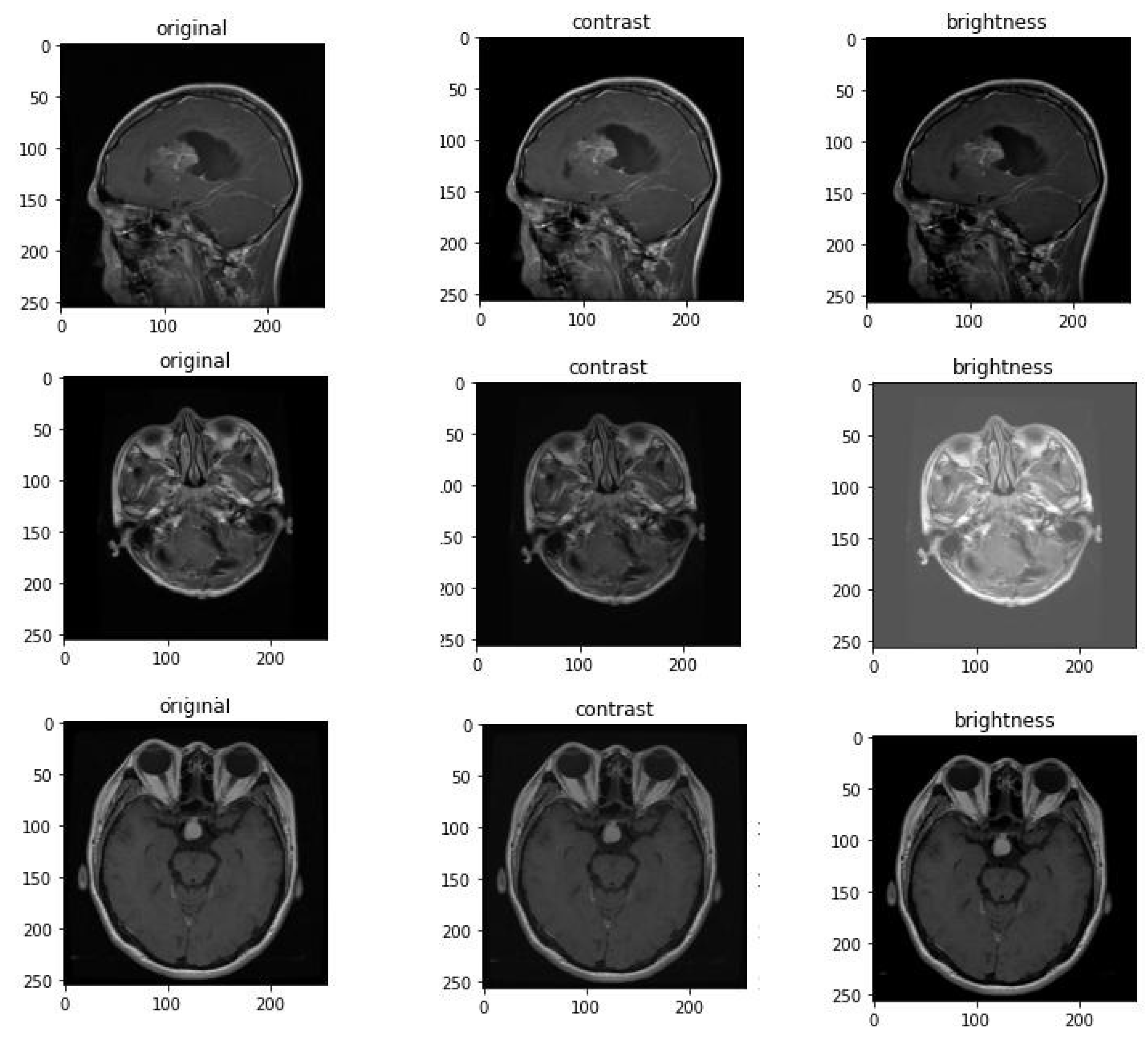
4.4. Data Augmentation
Data augmentation defined as a technique of artificially by creating updated versions of a dataset that increase the training set and use the available data.It includes making minor modifcation to the dataset to generate new data points. [
28]
Data augmentation has demonstrated strong performance in various clinical image issues and brain scan analysis [
29]. In this study, the Contrast and Brightness augmentation methods illustrated in
Figure 6 were applied to images during training.
4.5. Feature Extraction
In these experiments, automated feature extraction was obtained by integrating CNN and EfficientNetV2B3, as seen in
Figure 7:
4.6. Classification Methods
The algorithm known as KNN,is supervised learning algorithm ,a non-parametric,that relies on proximity to do predictions or classifications placement of a specific data , and can be used for classifying and regression tasks.The principle it operates on is that similar points are located close to each other.KNN algorithm was utilized in classification methods of brain tumours datasets .[
25].
K-nearest Neighbours — Pseudo-code is shown in algorithm one.
|
Algorithm 1: KNN Algorithm |
1.run dist(Z, ) where x =1,2,...., m; where dist is distance between points. 2. Order the Euclidean distances m . 3. Let k is +ve INT Get first k distance from list . 4. Get k-points correspond to kdistance . 5. Let be number of points belong to among k i.e., I ≥ 0 6.If > ∀ x ≠ y then put z in class x. |
4.7. Implementation
Image classification performance relies on dividing image features and classification as separate phase in framework.About the classification stage used in stages of the brain tumours classifying model.Framework code using python libraries and execute Python code on Google Co-laboratory (Colab).Tested on two different dataset using various augmentation techniques brightness, contrast and without augmenting the input image.
First, the median filter was used to minimize noise from input pictures, and the augmentation methods mentioned previously were applied. Then, the features were extracted using hybrid combining(CNN and EfficientNetV2B3) as feature extraction layer.The KNN algorithm was performed in classification stage. KNN parameters include k which is equal to 49,and defined as next neighborhood or distance metrics. A small k value can increase the system’s vulnerability to noise and over-fitting.A more excellent value of k increased importance calculations are done.Additionally, class-related data imbalances can occur. When k is given a high value, the results become dominant.The experiment was performed on five,and every experiment was performed as follows: five occasions validation process. The average result of 5 experiments is given as average values deviation format.
4.8. Evaluation Metrics
Precision measures how often the model correctly predicted the disease, and it is calculated using the following equation:
The following formula can be used to calculate recall using the following formula:
F1 score: Calculated using the following formula, it is a weighted average of true positive rates (recall rates).
5. Experimental Results
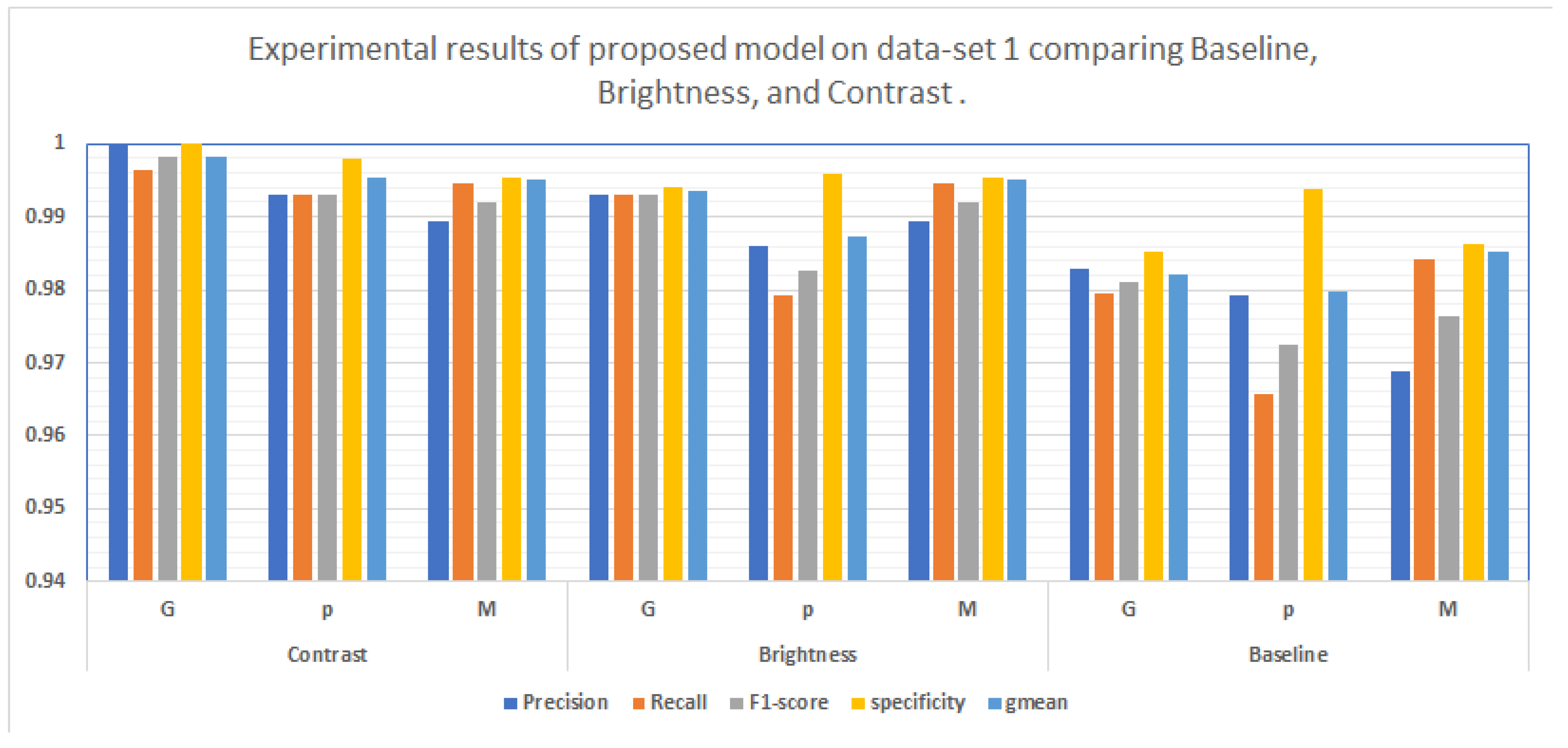
Multiple metrics have been established for the typical assessment of a classifier’s performance. The most widely used measure of quality is the accuracy of classification.The accuracy measures the percentage of corrected classified samples to the sum number of samples.
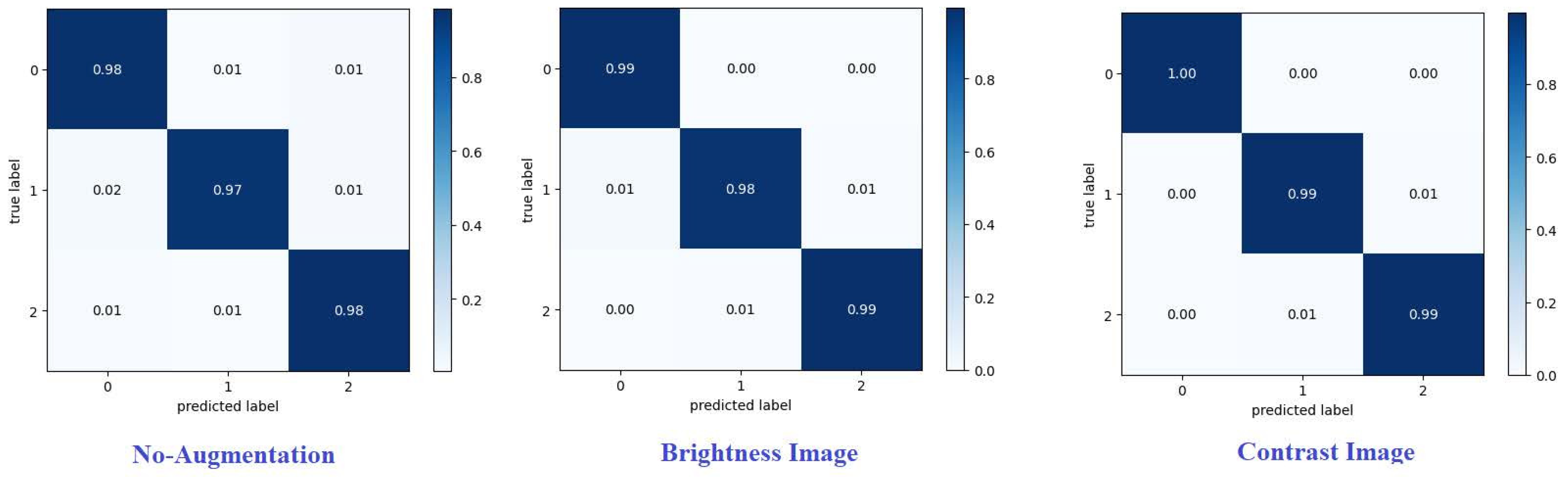
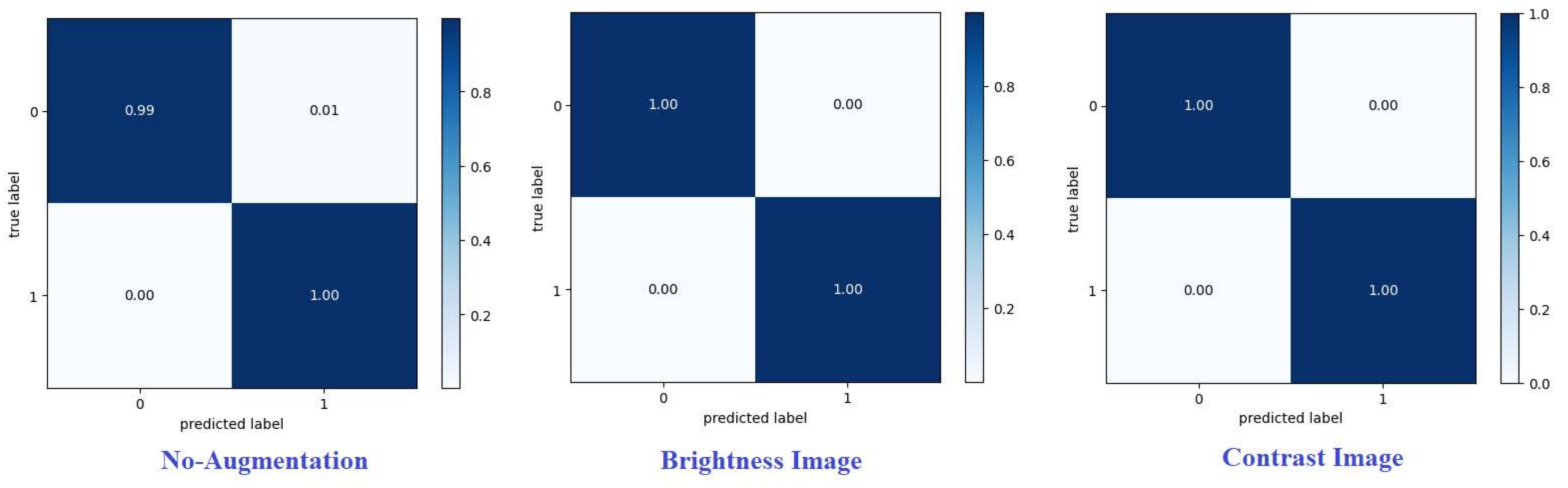
The accuracy of classifying achieved on each dataset is shown in
Table 1 ,and showed that the best method utilizing contrast images on dataset1 accuracy 99.51 % as shown in
Figure 8.
Confusion matrix summarizes predictions of the model.Every row corresponds to a real category and a single item. A confusion matrix is normalized by dividing each element value in every class,enhancing the visual representation of mis classification in each class[
30]
Normalized confusion matrix of the best technique are shown.Where G, P, and M or 0, 1, and 2, refer to glioma, pituitary, and meningioma tumours, respectively. From various metrics can be used from the confusion matrix to show the classifier is performing for each tumour category.Recall (or sensitivity) and precision are essential metrics. [
31,
32,
33].
Figure 9 and
Figure 10] present confusion matrices proposed framework using various augmentation techniques on each dataset.
6. Ablation Study
Contribution of significant of proposed approach, ablation experiments were performed with augmentation and baseline.In evaluating the hybrid classification approaches, the proposed hybrid models results presented in
Table 2,
Table 3 show the experimental performance of the proposed hybrid model.Performance of classification experiments conducted at the following locations if the hybrid approach implemented in conjunction with extensions compared to the basic model.
7. Discussion
This article introduced a precise and completely automated system, requiring minimal pre-processing, for categorizing brain tumours. The system being suggested utilized advanced transfer learning techniques to analyze features in brain MRI scans. KNN utilized the features.Classifiers using augmentation techniques for improving the performance.This framework achieved significantly higher precision when compared to all other relevant options research and it could further make use of being examined using a more significant number of images.
Typically, the primary metric in the majority of previous studies was accuracy criterion for evaluating system execution using sensitivity,precision and recall.comparing the obtained results with the past work conducted on the identical benchmark dataset1[
26] in
Table 4.Kaggle BR35H dataset in
Table 5
Authors in [
34,
35,
36,
37,
38,
39,
40,
41,
42] have achieved accuracy of 91.28%,84.19%, 91.90%, 93.68%, 90.89% , 97.1% , 97%, 97.25% and 97.91% respectively.The study has demonstrated that the suggested model has achieved high accuracy results.Tested on the same dataset1, achieving higher accuracy and exceeding the best-performing previous study by 1.5 %.
Authors in [
44,
45,
46,
47] have achieved accuracy of 97.5%,98.8%, 99.50%, 97.99% respectively.High accuracy has been attained in the proposed model tested on the same BR35H Kaggle dataset2, achieving a higher accuracy and exceeding the best-performing previous study by 0.3 %.
8. Conclusions
This paper introduced a system for categorizing brain tumours automatically, requiring only minimal processing.Training model of brain tumour MRI dataset consisting of 3064 images assessed by various performance metrics like accuracy, precision, and recall.Combining flatten layer of CNN with flatten layer of EfficientNETV2B3 as feature extraction layers and then using KNN classifiers using various augmentation techniques. That achieved 99.51% accuracy and improved cutting-edge technology increased it by 1.5%.We intend to expand proposed framework on more extensive data set and more brain tumour types. Moreover, the proposed framework becomes available in cloud to provides doctors fast and accurate a diagnosis from input MRI. Future research efforts will extend the proposed framework by incorporating additional types of brain tumours and larger datasets. With the help of the web applications that are recommended, medical professionals can diagnose MRI images with speed and precision.A range of medical imaging techniques, such as X-rays, ultrasound, endoscopic, thermos-copy, and histological images, can be employed with the proposed models.
Author Contributions
Eman M.G.Younis: Conceptualization, data collection, formal analysis, writing review and editing; Mahmoud N.Mahmoud and Ibrahim A.Ibrahim: conceptualization, methodology, writing-original draft, validation, supervision; N.A.: resources, investigation; Abdullah M. Albarrak: funding acquisition, validation, visualization; Mahmoud N.Mahmoud: writing draft, analysis; Eman M.G.Younis : investigation, data collection. All authors have read and agreed to the published version of the manuscript.
Funding
The Deanship of Scientific Research at Imam Mohammad Ibn Saud Islamic University (IMSIU) provided support and funding for this project (grant number IMSIU-RG23077).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data collected and analyzed during this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- El-Dahshan, El-Sayed A., et al. "Computer-aided diagnosis of human brain tumour through MRI: A survey and a new algorithm." Expert systems with Applications 41.11 (2014): 5526-5545.
- Gibbs P, Buckley DL, Blackband SJ, Horsman A (1996) Tumour volume determination from MR images by morphological segmentation. Physics in Medicine Biology 41(11):2437.
- Bhandarkar SM, Koh J, Suk M (1997) Multiscale image segmentation using a hierarchical selforganizing map. Neurocomputing 14(3):241–272.
- Benson C, Lajish V, Rajamani K (2015) Brain tumour extraction from MRI brain images using marker based watershed algorithm. In: IEEE international conference on advances in computing, communications and informatics (ICACCI), pp 318–323.
- Chen H, Qin Z, Ding Y, Lan T (2019) Brain tumour segmentation with generative adversarial nets. In: IEEE 2nd international conference on artificial intelligence and big data (ICAIBD), pp 301–305.
- Arunkumar, N. , Mohammed, M. A., Abd Ghani, M. K., Ibrahim, D. A., Abdulhay, E., Ramirez-Gonzalez, G., de Albuquerque, V. H. C. (2019). K-means clustering and neural network for object detecting and identifying abnormality of brain tumour. Soft Computing, 23(19), 9083-9096.
- Kumari, M., & Singh, V. (2018). Breast cancer prediction system. Procedia computer science, 132, 371-376.
- Ismael, S. A. A., Mohammed, A., & Hefny, H. (2020). An enhanced deep learning approach for brain cancer MRI images classification using residual networks. Artificial intelligence in medicine, 102, 101779.
- Albawi, S., Mohammed, T. A., & Al-Zawi, S. (2017, August). Understanding of a convolutional neural network. In 2017 international conference on engineering and technology (ICET) (pp. 1-6). Ieee.
- Tan, M., & Le, Q. (2021, July). Efficientnetv2: Smaller models and faster training. In International conference on machine learning (pp. 10096-10106). PMLR.
- Liu, D., Wang, W., Wu, X., & Yang, J. (2022, January). EfficientNetv2 model for breast cancer histopathological image classification. In 2022 3rd International Conference on Electronic Communication and Artificial Intelligence (IWECAI) (pp. 384-387). IEEE.
- Anagun, Y. (2023). Smart brain tumour diagnosis system utilizing deep convolutional neural networks. Multimedia Tools and Applications, 82(28), 44527-44553.
- D. Jiang, "Analysis on the Application of Artificial Intelligence in the Medical Field," 2020 8th International Conference on Orange Technology (ICOT), Daegu, Korea (South), 2020, pp. 1-4. [CrossRef]
- Wang, X., Ahmad, I., Javeed, D., Zaidi, S. A., Alotaibi, F. M., Ghoneim, M. E., ... & Eldin, E. T. (2022). Intelligent hybrid deep learning model for breast cancer detection. Electronics, 11(17), 2767.
- Dhillon, A., & Verma, G. K. (2020). Convolutional neural network: a review of models, methodologies and applications to object detection. Progress in Artificial Intelligence, 9(2), 85-112.
- LeCun, Y., Bengio, Y., & Hinton, G. (2015). Deep learning. nature, 521(7553), 436-444.
- Goodfellow, I., Bengio, Y., & Courville, A. (2016). Deep learning. MIT press.
- Wu, J. (2017). Introduction to convolutional neural networks. National Key Lab for Novel Software Technology. Nanjing University. China, 5(23), 495.
- Mascarenhas, S., & Agarwal, M. (2021, November). A comparison between VGG16, VGG19 and ResNet50 architecture frameworks for Image Classification. In 2021 International Conference on Disruptive Technologies for Multi-Disciplinary Research and Applications (CENTCON) (Vol. 1, pp. 96-99). IEEE.
- Theckedath, D., & Sedamkar, R. R. (2020). Detecting affect states using VGG16, ResNet50 and SE-ResNet50 networks. SN Computer Science, 1(2), 1-7.
- Szegedy, C., Ioffe, S., Vanhoucke, V., & Alemi, A. A. (2017, February). Inception-v4, inception-resnet and the impact of residual connections on learning. In Thirty-first AAAI conference on artificial intelligence.
- Vishwanathan, S. V. M., & Murty, M. N. (2002, May). SSVM: a simple SVM algorithm. In Proceedings of the 2002 International Joint Conference on Neural Networks. IJCNN’02 (Cat. No. 02CH37290) (Vol. 3, pp. 2393-2398). IEEE.
- Guo, G., Wang, H., Bell, D., Bi, Y., & Greer, K. (2003, November). KNN model-based approach in classification. In OTM Confederated International Conferences" On the Move to Meaningful Internet Systems" (pp. 986-996). Springer, Berlin, Heidelberg.
- Jakkula, V. (2006). Tutorial on support vector machine (svm). School of EECS, Washington State University, 37(2.5), 3.
- Guo, G., Wang, H., Bell, D., Bi, Y., & Greer, K. (2003, November). KNN model-based approach in classification. In OTM Confederated International Conferences" On the Move to Meaningful Internet Systems" (pp. 986-996). Springer, Berlin, Heidelberg.
- Cheng, J. (2017). Brain tumour dataset. figshare. dataset.
- Sonka, M., & Fitzpatrick, J. M. (2000). Handbook of medical imaging. Volume 2, Medical image processing and analysis. SPIE.
- Perumal, S., & Velmurugan, T. (2018). Preprocessing by contrast enhancement techniques for medical images. International Journal of Pure and Applied Mathematics, 118(18), 3681-3688.
- Van Dyk, D. A., & Meng, X. L. (2001). The art of data augmentation. Journal of Computational and Graphical Statistics, 10(1), 1-50.
- Visa, S., Ramsay, B., Ralescu, A. L., & Van Der Knaap, E. (2011). Confusion matrix-based feature selection. MAICS, 710(1), 120-127.
- Wise, M. N. (Ed.). (1997). The values of precision. Princeton University Press.
- Buckland, M., & Gey, F. (1994). The relationship between recall and precision. Journal of the American society for information science, 45(1), 12-19.
- Chicco, D., & Jurman, G. (2020). The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC genomics, 21(1), 1-13.
- Cheng, J., Huang, W., Cao, S., Yang, R., Yang, W., Yun, Z., ... & Feng, Q. (2015). Enhanced performance of brain tumour classification via tumour region augmentation and partition. PloS one, 10(10), e0140381.
- Paul, J. S., Plassard, A. J., Landman, B. A., & Fabbri, D. (2017, March). Deep learning for brain tumour classification. In Medical Imaging 2017: Biomedical Applications in Molecular, Structural, and Functional Imaging (Vol. 10137, pp. 253-268). SPIE.
- M. R. Ismael and I. Abdel-Qader, "Brain tumour Classification via Statistical Features and Back-Propagation Neural Network," 2018 IEEE International Conference on Electro/Information Technology (EIT), 2018, pp. 0252-0257. [CrossRef]
- A. Pashaei, H. Sajedi and N. Jazayeri, "Brain tumour Classification via Convolutional Neural Network and Extreme Learning Machines," 2018 8th International Conference on Computer and Knowledge Engineering (ICCKE), 2018, pp. 314-319. [CrossRef]
- P. Afshar, K. N. Plataniotis and A. Mohammadi, "Capsule Networks for Brain tumour Classification Based on MRI Images and Coarse tumour Boundaries," ICASSP 2019 - 2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), 2019, pp. 1368-1372. [CrossRef]
- Deepak S, Ameer PM. Brain tumour classification using deep CNN features via transfer learning. Comput Biol Med. 2019 Aug;111:103345. [CrossRef] [PubMed]
- Abdelaziz Ismael SA, Mohammed A, Hefny H. An enhanced deep learning approach for brain cancer MRI images classification using residual networks. Artif Intell Med. 2020 Jan;102:101779. [CrossRef] [PubMed]
- Gull, S., Akbar, S., & Shoukat, I. A. (2021, November). A Deep Transfer Learning Approach for Automated Detection of Brain tumour Through Magnetic Resonance Imaging. In 2021 International Conference on Innovative Computing (ICIC) (pp. 1-6). IEEE.
- Mondal, A., & Shrivastava, V. K. (2022). A novel Parametric Flatten-p Mish activation function based deep CNN model for brain tumour classification. Computers in Biology and Medicine, 150, 106183.
- Younis, E., Mahmoud, M. N., Ibrahim, I. A. (2023). Python Libraries Implementation for Brain tumour Detection Using MR Images Using Machine Learning Models. In S. Biju, A. Mishra, M. Kumar (Eds.), Advanced Interdisciplinary Applications of Machine Learning Python Libraries for Data Science (pp. 243-262). IGI Global. [CrossRef]
- hamada2020br35h, title=Br35h: Brain tumour Detection 2020, version 5, author=Hamada, A, year=2020.
- Naseer, A., Yasir, T., Azhar, A., Shakeel, T., & Zafar, K. (2021). Computer-aided brain tumour diagnosis: performance evaluation of deep learner CNN using augmented brain MRI. International Journal of Biomedical Imaging, 2021.
- Amran GA, Alsharam MS, Blajam AOA, Hasan AA, Alfaifi MY, Amran MH, Gumaei A, Eldin SM. Brain tumour Classification and Detection Using Hybrid Deep tumour Network. Electronics. 2022; 11(21):3457. [CrossRef]
- Brain tumour Detection Using Convolutional Neural Network, author=Chhatre, Falak and Deshpande, Sudhanva and Malik, Sidhant and Yan, Grace and Subramaniam, Suresh, journal=Journal of Student Research, volume=12, number=2, year=2023.
| 1 |
figshare.com/articles/brain_tumour_dataset/1512427/5) |
| 2 |
kaggle.com/datasets/ahmedhamada0/brain-tumour-detection |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).