Submitted:
20 September 2024
Posted:
20 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Characteristics of Biofilm Carriers
2.2. Preparation of Bio-Carriers
2.3. Crystal Violet Staining
2.4. LIVE/DEAD Staining and Confocal Laser-Scanning Microscopy (CLSM)
2.5. Analytical Methods
2.6. Analysis of Microbial Communities by 16S rRNA Gene Next-Generation Sequencing
3. Results and Discussion
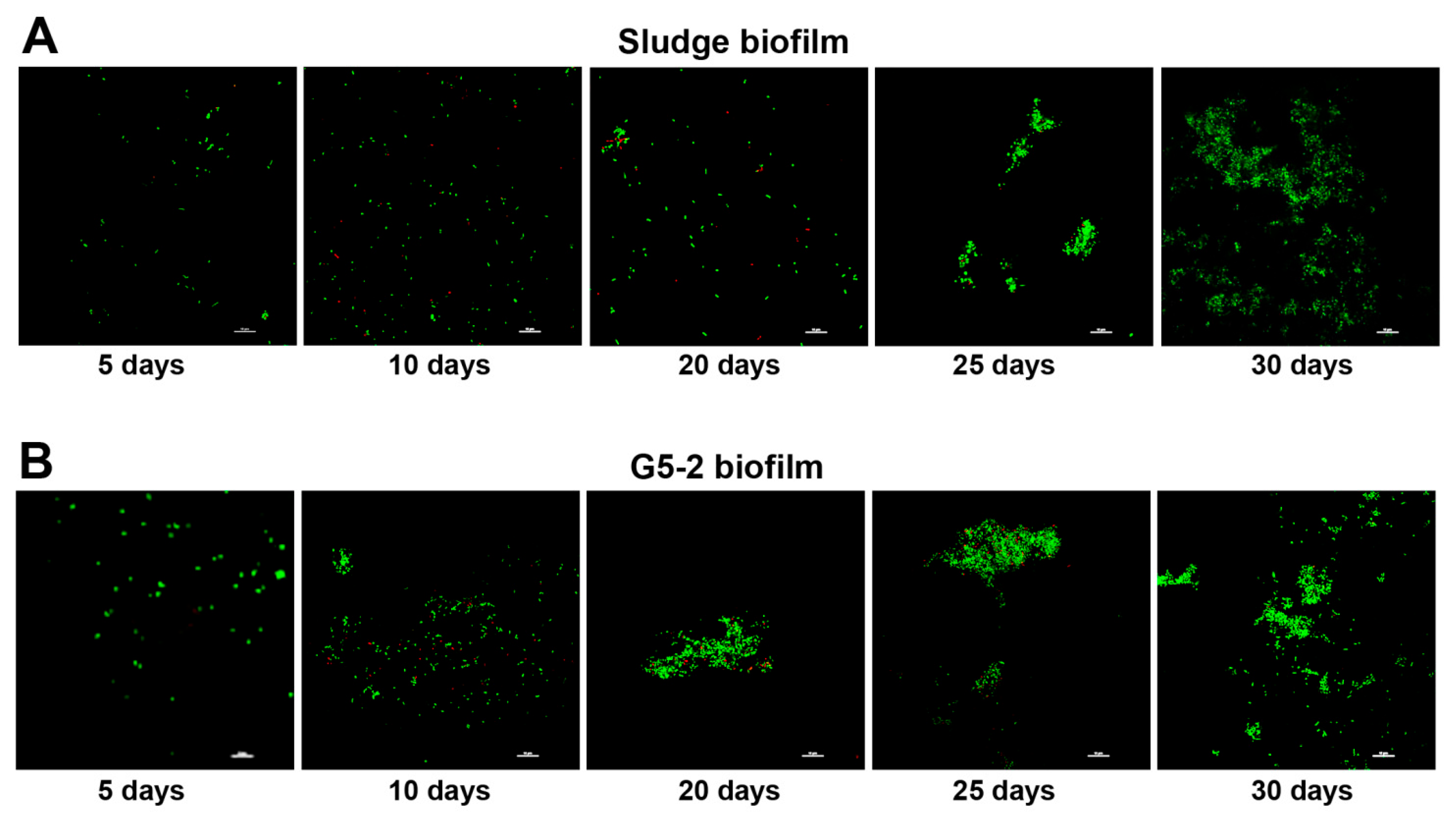
3.1. Monitoring and Quantification of Biofilm Formation
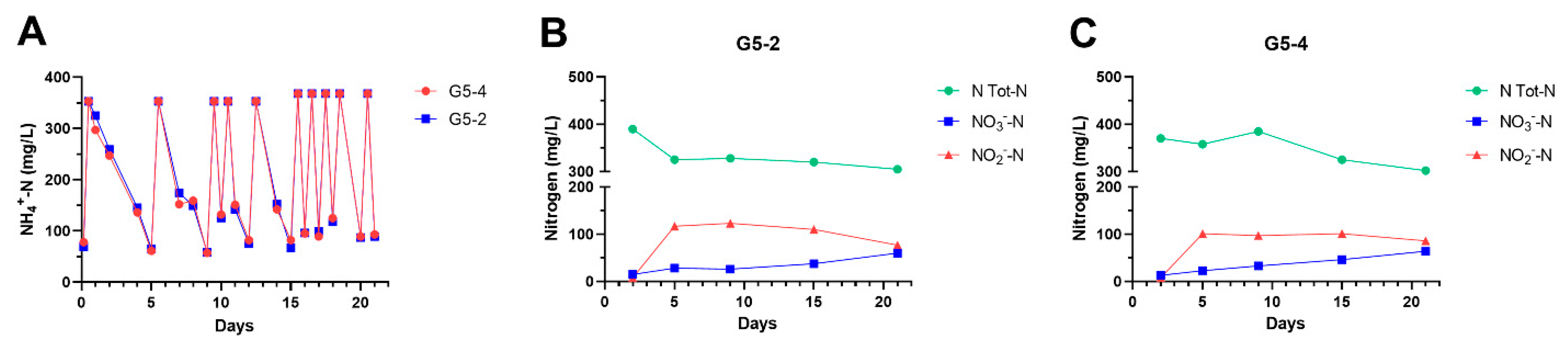
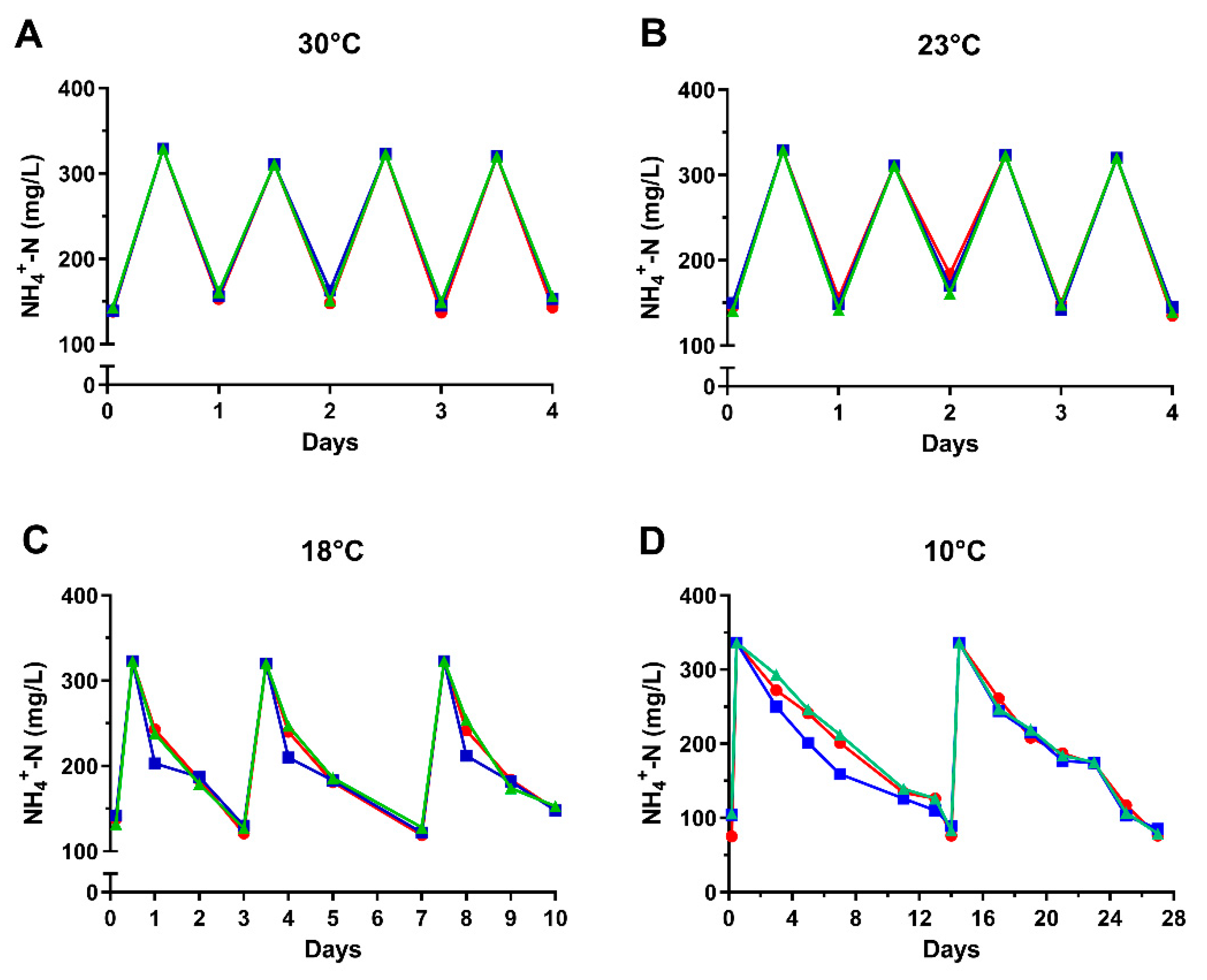
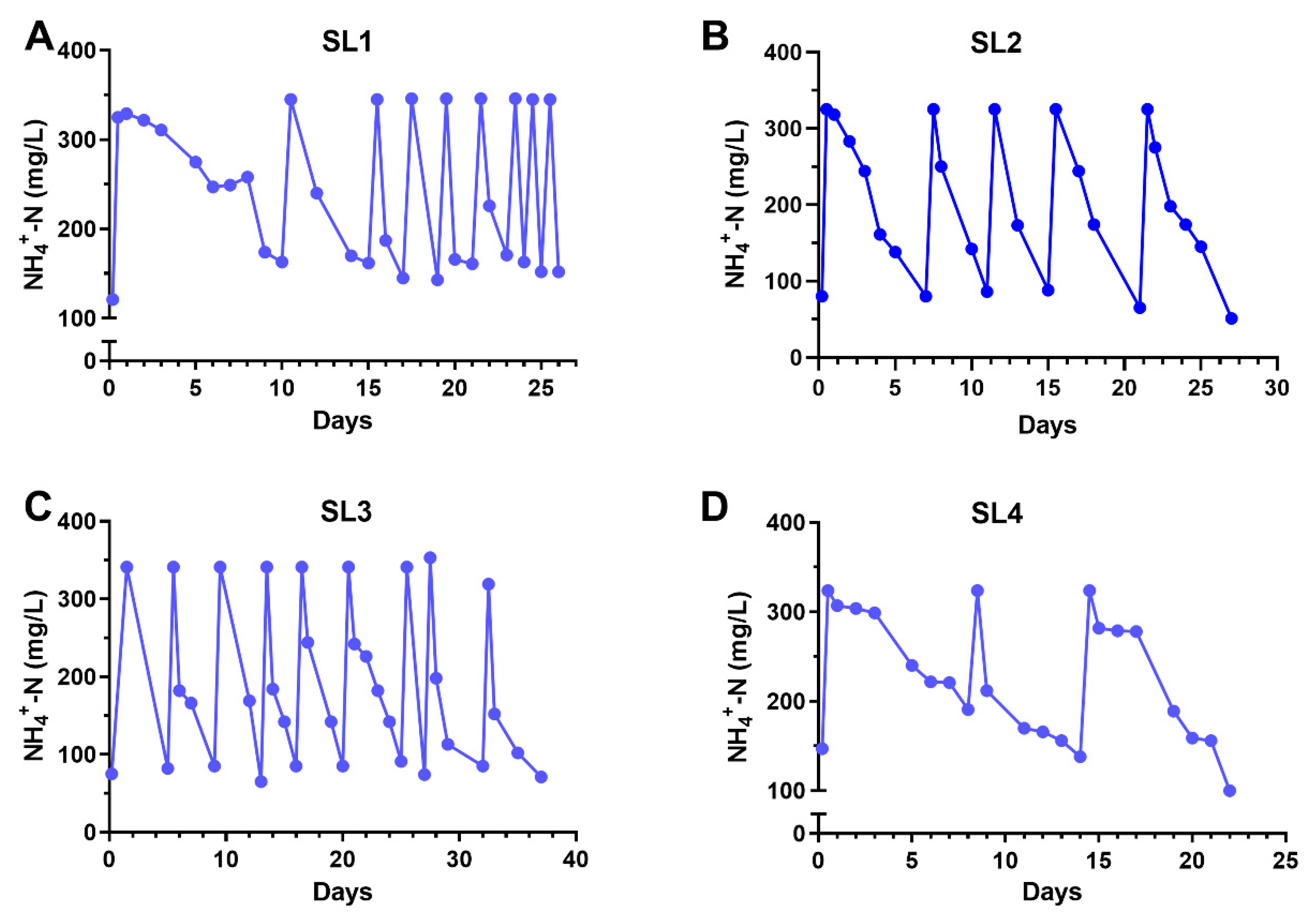
3.2. Ammonia Degradation by Biofilms Developed from Ammonia-Selected Bacterial Populations and Activated Sludges
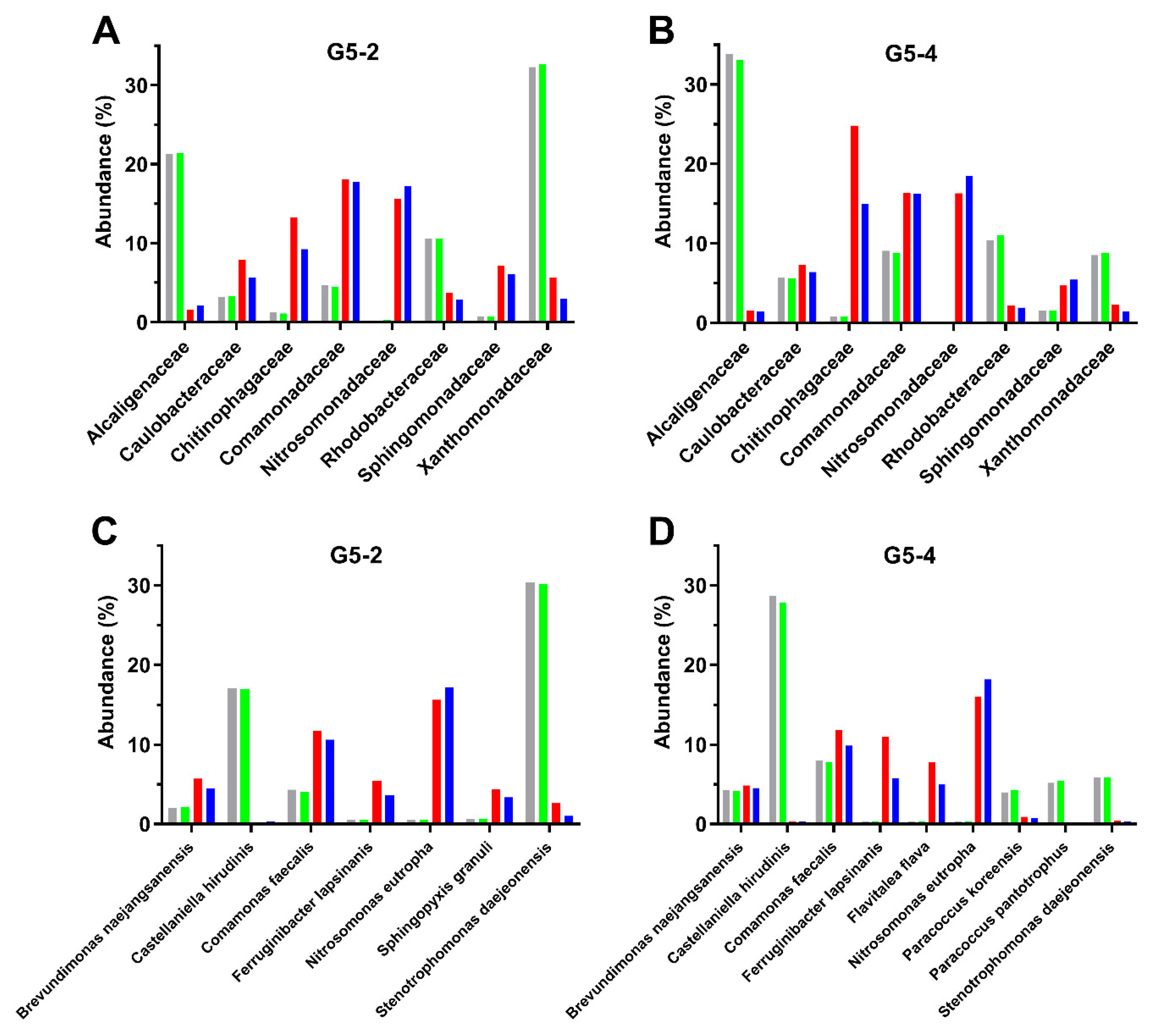
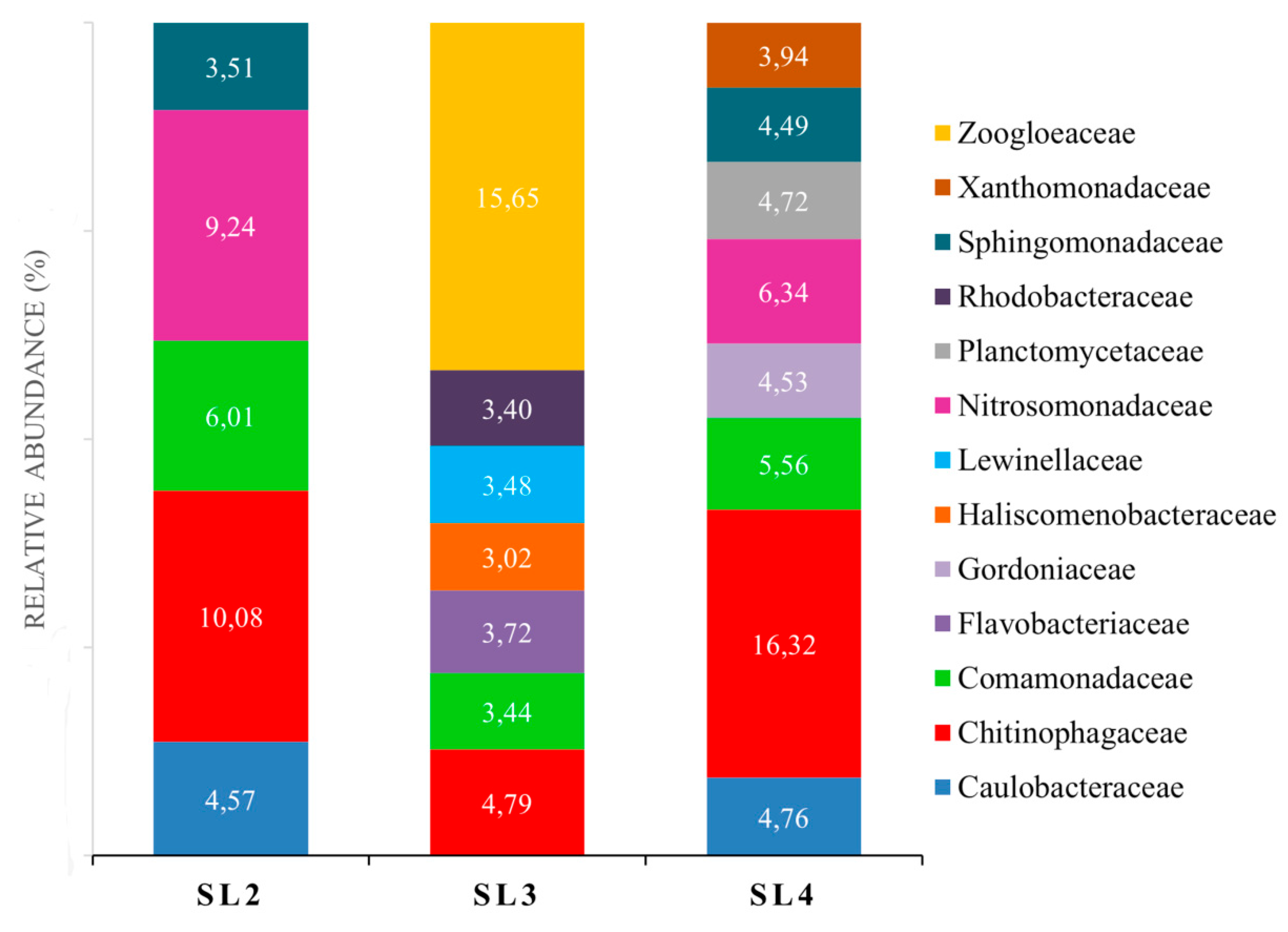
3.3. Composition of Biofilms Formed from Ammonia-Selected Microbial Communities and Activated Sludges
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Costa, A.M.; de, R.G.; Alfaia, S.M.; Campos, J.C. Landfill leachate treatment in Brazil–an overview. J. Environ. Manag. 2019, 232, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Yang, G.; Tao, T.; Peng, Y. Recent advances in nitrogen removal from landfill leachate using biological treatments—A review. J. Environ. Manag. 2019, 235, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Wijekoon, P.; Koliyabandara, P.A.; Cooray, A.T.; Lam, S.S.; Athapattu, B.C.L.; Vithanage, M. Progress and prospects in mitigation of landfill leachate pollution: Risk, pollution potential, treatment and challenges. J. Hazard Mater. 2022, 421, 126627. [Google Scholar] [CrossRef] [PubMed]
- Lebron, Y.A.R.; Moreira, V.R.; Brasil, Y.L.; Silva, A.F.R.; Santos, L.V.S.; Lange, L.C.; Amaral, M.C.S. A survey on experiences in leachate treatment: Common practices, differences worldwide and future perspectives. J. Environ. Manag. 2021, 288, 112475. [Google Scholar] [CrossRef]
- Ma, S.; Zhou, C.; Pan, J.; Yang, G.; Sun, C.; Liu, Y.; Chen, X.; Zhao, Z. Leachate from municipal solid waste landfills in a global perspective: Characteristics, influential factors and environmental risks. J. Clean. Prod. 2022, 333, 130234. [Google Scholar] [CrossRef]
- Mai, W.; Chen, J.; Liu, H.; Liang, J.; Tang, J.; Wei, Y. Advances in Studies on Microbiota Involved in Nitrogen Removal Processes and Their Applications in Wastewater Treatment. Front. Microbiol. 2021, 12, 746293. [Google Scholar] [CrossRef]
- Ren, Y.; Hao Ngo, H.; Guo, W.; Wang, D.; Peng, L.; Ni, B.J.; Wei, W.; Liu, Y. New perspectives on microbial communities and biological nitrogen removal processes in wastewater treatment systems. Bioresour. Technol. 2020, 297, 122491. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563. [Google Scholar] [CrossRef] [PubMed]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The biofilm life cycle: expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef]
- Shineh, G.; Mobaraki, M.; Perves Bappy, M.J.; Mills, D.K. Biofilm Formation, and RelatedImpacts on Healthcare, Food Processing and Packaging, Industrial Manufacturing, Marine Industries, and Sanitation–A Review. Appl. Microbiol. 2023, 3, 629–665. [Google Scholar] [CrossRef]
- Sharma, S.; Mohler, J.; Mahajan, S.D.; Schwartz, S.A.; Bruggemann, L.; Aalinkeel, R. Microbial Biofilm: A Review on Formation, Infection, Antibiotic Resistance, Control Measures, and Innovative Treatment. Microorganisms. 2023, 11, 1614. [Google Scholar] [CrossRef] [PubMed]
- Grinberg, M.; Orevi, T.; Kashtan, N. Bacterial surface colonization, preferential attachment and fitness under periodic stress. PLoS Comput. Biol. 2019, 15, e1006815. [Google Scholar] [CrossRef] [PubMed]
- McQuarrie, J.P.; Boltz, J.P. Moving bed biofilm reactor technology: process applications, design, and performance. Water Environ. Res. 2011, 83, 560–575. [Google Scholar] [CrossRef]
- Leyva-Díaz, J.C.; Martín-Pascual, J.; Poyatos, J.M. Moving bed biofilm reactor to treat wastewater. Int. J. Environ. Sci. Technol. 2017, 14, 881–910. [Google Scholar] [CrossRef]
- Di Capua, F.; Iannacone, F.; Sabba, F.; Esposito, G. Simultaneous nitrification-denitrification in biofilm systems for wastewater treatment: Key factors, potential routes, and engineered applications. Bioresour. Technol. 2022, 361, 127702. [Google Scholar] [CrossRef]
- Pan, D.; Shao, S.; Zhong, J.; Wang, M.; Wu, X. Performance and mechanism of simultaneous nitrification–denitrification and denitrifying phosphorus removal in long-term moving bed biofilm reactor (MBBR). Bioresour. Technol. 2022, 348, 126726. [Google Scholar] [CrossRef] [PubMed]
- Kulikowska, D.; Kaczówka, E.; Pokój, T.; Gusiatin, Z. Application of moving bed biofilm reactor (MBBR) for high-ammonium landfill leachate nitrification. New Biotechnol. 2009, 25, S351–S352. [Google Scholar] [CrossRef]
- Zulkifli, M.; Abu Hasan, H.; Sheikh Abdullah, SR.; Muhamad, M.H. A review of ammonia removal using a biofilm-based reactor and its challenges. J. Environ. Manage. 2022, 315, 115162. [Google Scholar] [CrossRef]
- Di Biase, A.; Kowalski, M.S.; Devlin, T.R.; Oleszkiewicz, J.A. Moving bed biofilm reactor technology in municipal wastewater treatment: A review. J. Environ. Manage. 2019, 247, 849–866. [Google Scholar] [CrossRef]
- Xiong, J.; Zheng, Z.; Yang, X.; He, J.; Luo, X.; Gao, B. Mature landfill leachate treatment by the MBBR inoculated with biocarriers from a municipal wastewater treatment plant. Process. Saf. Environ. Prot. 2018, 119, 304–310. [Google Scholar] [CrossRef]
- Madan, S.; Madan, R.; Hussain, A. Advancement in biological wastewater treatment using hybrid moving bed biofilm reactor (MBBR): a review. Appl. Water Sci. 2022, 12, 141. [Google Scholar] [CrossRef]
- Saini, S.; Tewari, S.; Dwivedi, J.; Sharma, V. Biofilm-mediated wastewater treatment: a comprehensive review. Mater. Adv. 2023, 4, 1415–1443. [Google Scholar] [CrossRef]
- Pratap, V.; Kumar, R.; Kumar, S.; Yadav, B.R. Optimization of moving bed biofilm reactors for the treatment of municipal wastewater. Environ. Res. 2024, 241, 117560. [Google Scholar] [CrossRef] [PubMed]
- Petrilli, R.; Fabbretti, A.; Cerretani, A.; Pucci, K.; Pagliaretta, G.; Picciolini, M.; Napolioni, V.; Falconi, M. ; Selection, Identification and Functional Performance of Ammonia-Degrading Microbial Communities from an Activated Sludge for Landfill Leachate Treatment. Microorganisms 2023, 11, 311. [Google Scholar] [CrossRef]
- Djordjevic, D.; Wiedmann, M.; McLandsborough, L. A. Microtiter plate assay for assessment of listeria monocytogenes biofilm formation. Appl.Environ.Microbiol. 2002, 68, 2950–2958. [Google Scholar] [CrossRef] [PubMed]
- Sabaeifard, P.; Abdi-Ali, A.; Soudi, M. R.; Dinarvand, R. Optimization of tetrazolium salt assay for pseudomonas aeruginosa biofilm using microtiter plate method. J. Microbiol. Methods 2014, 105, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Pantanella, F.; Valenti, P.; Natalizi, T.; Passeri, D.; Berlutti, F. Analytical techniques to study microbial biofilm on abiotic surfaces: Pros and cons of the main techniques currently in use. Ann. Ig. 2013, 25, 31–42. [Google Scholar] [CrossRef]
- Franklin, R.B.; Garland, J.L.; Bolster, C.H.; Mills, A.L. Impact of dilution on microbial community structure and functional potential: comparison of numerical simulations and batch culture experiments. Appl. Environ. Microbiol. 2001, 67, 702–712. [Google Scholar] [CrossRef]
- Doll, K.; Jongsthaphongpun, K.L.; Stumpp, N.S.; Winkel, A.; Stiesch, M. Quantifying implant-associated biofilms: Comparison of microscopic, microbiologic and biochemical methods. J. Microbiol. Methods. 2016, 130, 61–68. [Google Scholar] [CrossRef]
- Irankhah, S.; Abdi Ali, A.; Reza Soudi, M.; Gharavi, S.; Ayati, B. Highly efficient phenol degradation in a batch moving bed biofilm reactor: benefiting from biofilm-enhancing bacteria. World J. Microbiol. Biotechnol. 2018, 34, 164. [Google Scholar] [CrossRef]
- Berney, M.; Hammes, F.; Bosshard, F.; Weilenmann, H.-U. U.; Egli, T. Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight kit in combination with flow cytometry. Appl. Environ. Microbiol. 2007, 73, 3283–3290. [Google Scholar] [CrossRef] [PubMed]
- Stiefel, P.; Schmidt-Emrich, S.; Maniura-Weber, K.; Ren, Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 2015, 15, 36. [Google Scholar] [CrossRef] [PubMed]
- Chaali, M.; Naghdi, M.; Brar, S.K.; Avalos-Ramirez, A. A review on the advances in nitrifying biofilm reactors and their removal rates in wastewater treatment. J. Chem. Technol. Biotechnol. 2018, 93, 3113–3124. [Google Scholar] [CrossRef]
- Ren, J.; Cheng, W.; Jiao, M.; Wan, T.; Wang, M.; Li, D. Characteristics of oxygen mass transfer and its impact on pollutant removal performance and microbial community structure in an aerobic fluidized bed biofilm reactor for treatment of municipal wastewater. Bioresour. Technol. 2021, 323, 124552. [Google Scholar] [CrossRef]
- Wang, Y.; Du, Z.; Liu, Y.; Wang, H.; Xu, F.; Liu, B.; Zheng, Z. The nitrogen removal and sludge reduction performance of a multi-stage anoxic/oxic (A/O) biofilm reactor. Water Environ. Res. 2020, 92, 94–105. [Google Scholar] [CrossRef]
- Bassin, J.P.; Dias, I.N.; Cao, S.M.S.; Senra, E.; Laranjeira, Y.; Dezotti, M. Effect of increasing organic loading rates on the performance of moving-bed biofilm reactors filled with different support media: assessing the activity of suspended and attached biomass fractions. Process Saf. Environ. Protect. 2016, 100, 131–141. [Google Scholar] [CrossRef]
- Saidulu, D.; Majumder, A.; Gupta, A.K. A systematic review of moving bed biofilm reactor, membrane bioreactor, and moving bed membrane bioreactor for wastewater treatment: Comparison of research trends, removal mechanisms, and performance. J. Environ. Chem. Eng. 2021, 9, 106112. [Google Scholar] [CrossRef]
- Zhao, J.; Ni, G.; Piculell, M.; Li, J.; Hu, Z.; Wang, Z.; Guo, J.; Yuan, Z.; Zheng, M.; Hu, S. Characterizing and comparing microbial community and biofilm structure in three nitrifying moving bed biofilm reactors. J. Environ. Manage. 2022, 320, 115883. [Google Scholar] [CrossRef]
- Sun, H.W.; Peng, Y.Z.; Shi, X.N. Advanced treatment of landfill leachate using anaerobic-aerobic process: Organic removal by simultaneous denitritation and methanogenesis and nitrogen removal via nitrite. Bioresour. Technol. 2015, 177, 337–345. [Google Scholar] [CrossRef]
- Garcia, K.A.; McLee, P.; Schuler, A.J. Effects of media length on biofilms and nitrification in moving bed biofilm reactors. Biofilm. 2022, 4, 100091. [Google Scholar] [CrossRef]
- Janka, E.; Pathak, S.; Rasti, A.; Gyawali, S.; Wang, S. Simultaneous Heterotrophic Nitrification and Aerobic Denitrification of Water after Sludge Dewatering in Two Sequential Moving Bed Biofilm Reactors (MBBR). Int. J. Environ. Res. Public Health. 2022, 19, 1841. [Google Scholar] [CrossRef] [PubMed]
- Fujitani, H.; Kumagai, A.; Ushiki, N.; Momiuchi, K.; Tsuneda, S. Selective isolation of ammonia-oxidizing bacteria from autotrophic nitrifying granules by applying cellsorting and sub-culturing of microcolonies. Front. Microbiol. 2015, 6, 1159. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wang, Y.; Li, R.; Zhang, R.; Zhang, X.; Hua, S.; Peng, D. The differential proliferation of AOB and NOB during natural nitrifier cultivation and acclimation with raw sewage as seed sludge. RSC Adv. 2020, 10, 28277–28286. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Duan, H.; Zhang, Y.; Huang, X.; Yuan, Z.; Liu, Y.; Zheng, M. Adaptation of nitrifying community in activated sludge to free ammonia inhibition and inactivation. Sci. Total Environ. 2020, 728, 138713. [Google Scholar] [CrossRef]
- Phanwilai, S.; Kangwannarakul, N.; Noophan, P.; Kasahara, T.; Terada, A.; Munakata- Marr, J.; Figueroa, L.A. Nitrogen removal efficiencies and microbial communities in full-scale IFAS and MBBR municipal wastewater treatment plants at high COD:N ratio. Front. Environb. Sci. Eng. 2020, 14, 115. [Google Scholar] [CrossRef]
- Kruglova, A.; Kesulahti, J.; Minh Le, K.; Gonzalez-Martinez, A.; Mikola, A.; Vahala, R. Low-temperature adapted nitrifying microbial communities of Finnish wastewater treatment systems. Water 2020, 12, 2450. [Google Scholar] [CrossRef]
- Zhou, L.-J.; Han, P.; Yu, Y.; Wang, B.; Men, Y.; Wagner, M.; Wu, Q.L. Cometabolic biotransformation and microbial-mediated abiotic transformation of sulfonamides by three ammonia oxidizers. Water Res. 2019, 159, 444–453. [Google Scholar] [CrossRef]
- Wang, Z.; Yao, Y.; Steiner, N.; Cheng, H.; Wu, Y.; Woo, S.; Criddle, C.S. Impacts of nitrogen-containing coagulants on the nitritation/denitrification of anaerobic digester centrate. Environ. Sci.: Water Res. Technol. 2020, 6, 3451–3459. [Google Scholar] [CrossRef]
- Patureau, D.; Davison, J.; Bernet, N.; Moletta, R. Denitrification under various aeration conditions in Comamonas sp., strain SGLY2. FEMS Microbiol. Ecol. 1994, 14, 71–78. [Google Scholar] [CrossRef]
- Wang, Z.; Woo, S.; Yao, Y.; Cheng, H.; Wu, Y.; Criddle, C.S. Nitrogen removal as nitrous oxide for energy recovery: Increased process stability and high nitrous yields at short hydraulic residence times. Water Res. 2020, 173, 115575. [Google Scholar] [CrossRef]
- Hassan, H.; Jin, B.; Donner, E.; Vasileiadis, S.; Saint, C.; Dai, S. Microbial community and bioelectrochemical activities in MFC for degrading phenol and producing electricity: microbial consortia could make differences. Chem. Eng. J. 2018, 332, 647–657. [Google Scholar] [CrossRef]
- Shi, J.; Xu, C.; Han, Y.; Han, H. Enhanced anaerobic biodegradation efficiency and mechanism of quinoline, pyridine, and indole in coal gasification wastewater. Chem. Eng. J. 2019, 361, 1019–1029. [Google Scholar] [CrossRef]
- Zheng, M.; Zhu, H.; Han, Y.; Xu, C.; Zhang, Z.; Han, H. Comparative investigation on carbon-based moving bed biofilm reactor (MBBR) for synchronous removal of phenols and ammonia in treating coal pyrolysis wastewater at pilot-scale. Bioresour. Technol. 2019, 288, 121590. [Google Scholar] [CrossRef]
- Spieck, E.; Hartwig, C.; McCormack, I.; Maixner, F.; Wagner, M.; Lipski, A.; Daims, H. Selective enrichment and molecular characterization of a previously uncultured Nitrospira-like bacterium from activated sludge. Environ. Microbiol. 2006, 8, 405–415. [Google Scholar] [CrossRef]
- Mehrani, M.J.; Sobotka, D.; Kowal, P.; Ciesielski, S.; Makinia, J. The occurrence and role of Nitrospira in nitrogen removal systems. Bioresour. Technol. 2020, 303, 122936. [Google Scholar] [CrossRef]
- Cao, Y.; van Loosdrecht, M.C.; Daigger, G.T. Mainstream partial nitritation-anammox in municipal wastewater treatment: status, bottlenecks, and further studies. Appl. Microbiol. Biotechnol. 2017, 101, 1365–1383. [Google Scholar] [CrossRef]
- Zhang, T.; Shao, M.F.; Ye, L. 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 2012, 6, 1137–1147. [Google Scholar] [CrossRef]
- Huang, T.L.; Zhou, S.L.; Zhang, H.H.; Bai, S.Y.; He, X.X.; Yang, X. Nitrogen Removal Characteristics of a Newly Isolated Indigenous Aerobic Denitrifier from Oligotrophic Drinking Water Reservoir, Zoogloea sp. N299. Int. J. Mol. Sci. 2015, 16, 10038–10060. [Google Scholar] [CrossRef]
- Sowani, H.; Kulkarni, M.; Zinjarde, S. Harnessing the catabolic versatility of Gordonia species for detoxifying pollutants. Biotechnol. Adv. 2019, 37, 382–402. [Google Scholar] [CrossRef]
- Tamura, T.; Saito, S.; Hamada, M.; Kang, Y.; Hoshino, Y.; Gonoi, T.; Mikami, Y.; Yaguchi, T. Gordonia crocea sp. nov. and Gordonia spumicola sp. nov. isolated from sludge of a wastewater treatment plant. Int. J. Syst. Evol. Microbiol. 2020, 70, 3718–3723. [Google Scholar] [CrossRef]
- Riesco, R.; Rose, J.J.A.; Batinovic, S.; Petrovski, S.; Sánchez-Juanes, F.; Seviour, R.J.; Goodfellow, M.; Trujillo, M.E. Gordonia pseudamarae sp. nov., a home for novel actinobacteria isolated from stable foams on activated sludge wastewater treatment plants. Int. J. Syst. Evol. Microbiol 2022, 72. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




