Submitted:
20 September 2024
Posted:
23 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Global Impact of Foodborne Pathogens
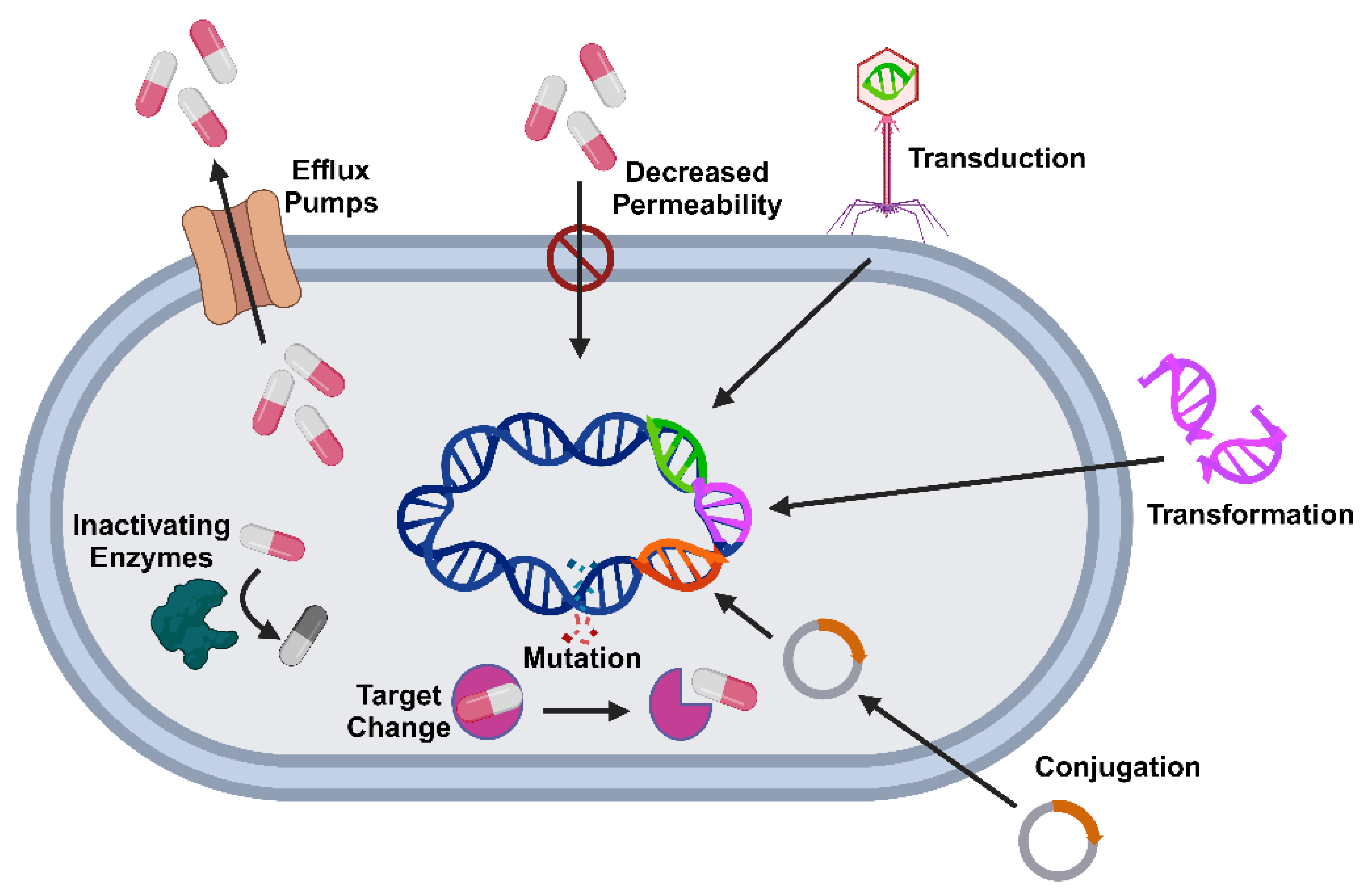
3. Mechanisms of Antibiotic Resistance in Foodborne Pathogens
4. Resistome Mapping: Concepts and Techniques
4.1. Definition and Importance of Resistome Mapping
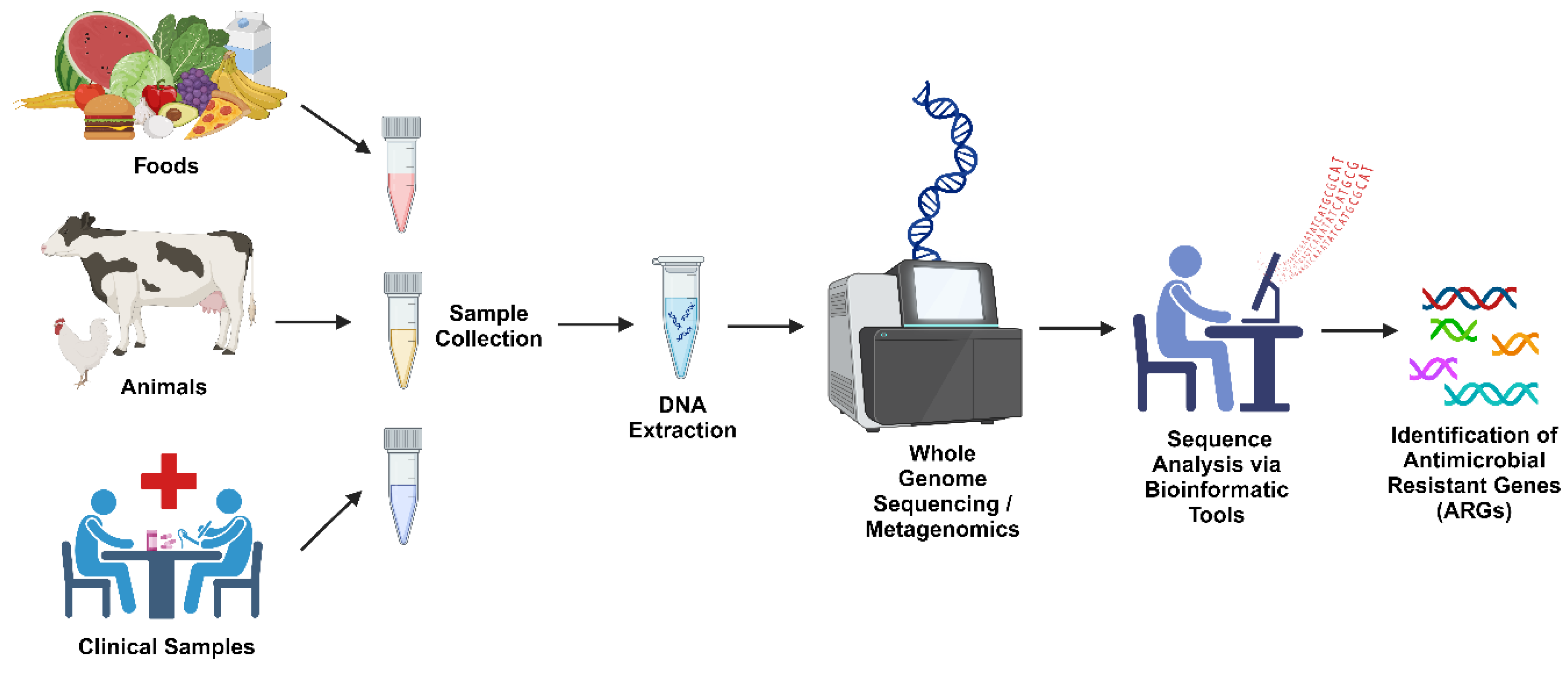
4.2. Techniques for Resistome Mapping
5. Resistome Mapping Studies in Food Borne Pathogens
5.1. Salmonella
5.2. Escherichia coli
5.3. Listeria monocytogenes
5.4. Campylobacter spp.
5.5. Vibrio cholerae
5.6. Clostridium perfringens
5.7. Shigella
5.8. Clostridium botulinum
5.9. Yersinia enterocolitica
6. Impact of Agricultural Practices on the Resistome
8. Challenges in Resistome Mapping
9. Future Directions to Overcome the Challenges
9.1. Integrative Approaches
9.2. Policy and Global Collaboration
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Almansour, A.M.; Alhadlaq, M.A.; Alzahrani, K.O.; Mukhtar, L.E.; Alharbi, A.L.; Alajel, S.M. The Silent Threat: Antimicrobial-Resistant Pathogens in Food-Producing Animals and Their Impact on Public Health. Microorganisms 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis#:~:text=If%20no%20action%20is%20taken, food%20systems%20are%20increasingly%20precarious New Report Calls for Urgent Action to Avert Antimicrobial Resistance Crisis.
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare (Switzerland) 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Balbin, M.M.; Hull, D.; Guest, C.; Nichols, L.; Dunn, R.; Thakur, S. Antimicrobial Resistance and Virulence Factors Profile of Salmonella Spp. and Escherichia Coli Isolated from Different Environments Exposed to Anthropogenic Activity. J Glob Antimicrob Resist 2020, 22, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Rafiq, K.; Islam, M.R.; Siddiky, N.A.; Samad, M.A.; Chowdhury, S.; Hossain, K.M.M.; Rume, F.I.; Hossain, M.K.; Mahbub-E-Elahi, A.T.M.; Ali, M.Z.; et al. Antimicrobial Resistance Profile of Common Foodborne Pathogens Recovered from Livestock and Poultry in Bangladesh. Antibiotics 2022, 11. [Google Scholar] [CrossRef]
- Kim, D.W.; Cha, C.J. Antibiotic Resistome from the One-Health Perspective: Understanding and Controlling Antimicrobial Resistance Transmission. Exp Mol Med 2021, 53, 301–309. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Andremont, A.; Bengtsson-Palme, J.; Brandt, K.K.; de Roda Husman, A.M.; Fagerstedt, P.; Fick, J.; Flach, C.F.; Gaze, W.H.; Kuroda, M.; et al. Critical Knowledge Gaps and Research Needs Related to the Environmental Dimensions of Antibiotic Resistance. Environ Int 2018, 117, 132–138. [Google Scholar] [CrossRef]
- Vezeau, N.; Kahn, L. Current Understanding and Knowledge Gaps Regarding Wildlife as Reservoirs of Antimicrobial Resistance. Am J Vet Res 2024, 85. [Google Scholar] [CrossRef]
- Sauerborn, E.; Corredor, N.C.; Reska, T.; Perlas, A.; Vargas da Fonseca Atum, S.; Goldman, N.; Wantia, N.; Prazeres da Costa, C.; Foster-Nyarko, E.; Urban, L. Detection of Hidden Antibiotic Resistance through Real-Time Genomics. Nat Commun 2024, 15. [Google Scholar] [CrossRef]
- Dufailu, O.A.; Yaqub, M.O.; Owusu-Kwarteng, J.; Addy, F. Prevalence and Characteristics of Listeria Species from Selected African Countries. Trop Dis Travel Med Vaccines 2021, 7. [Google Scholar] [CrossRef]
- https://www.who.int/news-room/fact-sheets/detail/food-safety Food Safety.
- Batz, M.B.; Henke, E.; Kowalcyk, B. Long-Term Consequences of Foodborne Infections. Infect Dis Clin North Am 2013, 27, 599–616. [Google Scholar] [CrossRef]
- Elbehiry, A.; Abalkhail, A.; Marzouk, E.; Elmanssury, A.E.; Almuzaini, A.M.; Alfheeaid, H.; Alshahrani, M.T.; Huraysh, N.; Ibrahem, M.; Alzaben, F.; et al. An Overview of the Public Health Challenges in Diagnosing and Controlling Human Foodborne Pathogens. Vaccines (Basel) 2023, 11. [Google Scholar] [CrossRef]
- Hussain, M.A.; Dawson, C.O. Economic Impact of Food Safety Outbreaks on Food Businesses. Foods 2013, 2, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zhang, A.; van Klinken, R.D.; Schrobback, P.; Muller, J.M. Consumer Trust in Food and the Food System: A Critical Review. Foods 2021, 10. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Yu, S.; Li, D.; Gillings, M.R.; Ren, H.; Mao, D.; Guo, J.; Luo, Y. Inter-Plasmid Transfer of Antibiotic Resistance Genes Accelerates Antibiotic Resistance in Bacterial Pathogens. ISME Journal 2024, 18. [Google Scholar] [CrossRef] [PubMed]
- Emamalipour, M.; Seidi, K.; Zununi Vahed, S.; Jahanban-Esfahlan, A.; Jaymand, M.; Majdi, H.; Amoozgar, Z.; Chitkushev, L.T.; Javaheri, T.; Jahanban-Esfahlan, R.; et al. Horizontal Gene Transfer: From Evolutionary Flexibility to Disease Progression. Front Cell Dev Biol 2020, 8. [Google Scholar] [CrossRef]
- Virolle, C.; Goldlust, K.; Djermoun, S.; Bigot, S.; Lesterlin, C. Plasmid Transfer by Conjugation in Gram-Negative Bacteria: From the Cellular to the Community Level. Genes (Basel) 2020, 11, 1–33. [Google Scholar] [CrossRef]
- Hasegawa, H.; Suzuki, E.; Maeda, S. Horizontal Plasmid Transfer by Transformation in Escherichia Coli: Environmental Factors and Possible Mechanisms. Front Microbiol 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.N.; Penadés, J.R.; Chen, J. Genetic Transduction by Phages and Chromosomal Islands: The New and Noncanonical. PLoS Pathog 2019, 15. [Google Scholar] [CrossRef]
- Muteeb, G.; Rehman, M.T.; Shahwan, M.; Aatif, M. Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals 2023, 16. [Google Scholar] [CrossRef]
- Gaurav, A.; Bakht, P.; Saini, M.; Pandey, S.; Pathania, R. Role of Bacterial Efflux Pumps in Antibiotic Resistance, Virulence, and Strategies to Discover Novel Efflux Pump Inhibitors. Microbiology (United Kingdom) 2023, 169. [Google Scholar] [CrossRef] [PubMed]
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Yang, L.; Huang, Y.; Jia, J.; Dou, T. Antibiotic Resistance Genes in Bacteria: Occurrence, Spread, and Control. J Basic Microbiol 2021, 61, 1049–1070. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Cheng, W. The Mechanism of Bacterial Resistance and Potential Bacteriostatic Strategies. Antibiotics 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Mulchandani, R.; Zhao, C.; Tiseo, K.; Pires, J.; Van Boeckel, T.P. Predictive Mapping of Antimicrobial Resistance for Escherichia Coli, Salmonella, and Campylobacter in Food-Producing Animals, Europe, 2000-2021. Emerg Infect Dis 2024, 30, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Guitor, A.K.; Raphenya, A.R.; Klunk, J.; Kuch, M.; Alcock, B.; Surette, M.G.; McArthur, A.G.; Poinar, H.N.; Wright, G.D. Capturing the Resistome: A Targeted Capture Method to Reveal Antibiotic Resistance Determinants in Metagenomes. Antimicrob Agents Chemother 2020, 64. [Google Scholar] [CrossRef] [PubMed]
- A PUBLIC HEALTH PERSPECTIVE ON ANTIMICROBIAL RESISTANCE DIAGNOSTICS; 2016;
- Rahman, M.M.; Alam Tumpa, M.A.; Zehravi, M.; Sarker, M.T.; Yamin, M.; Islam, M.R.; Harun-Or-rashid, M.; Ahmed, M.; Ramproshad, S.; Mondal, B.; et al. An Overview of Antimicrobial Stewardship Optimization: The Use of Antibiotics in Humans and Animals to Prevent Resistance. Antibiotics 2022, 11. [Google Scholar] [CrossRef]
- Baquero, F.; Martínez, J.L.; Lanza, V.F.; Rodríguez-Beltrán, J.; Galán, J.C.; San Millán, A.; Cantón, R.; Coque, T.M. Evolutionary Pathways and Trajectories in Antibiotic Resistance. Clin Microbiol Rev 2021, 34. [Google Scholar] [CrossRef]
- Xia, Y.; Li, X.; Wu, Z.; Nie, C.; Cheng, Z.; Sun, Y.; Liu, L.; Zhang, T. Strategies and Tools in Illumina and Nanopore-Integrated Metagenomic Analysis of Microbiome Data. iMeta 2023, 2. [Google Scholar] [CrossRef]
- Setubal, J.C. Metagenome-Assembled Genomes: Concepts, Analogies, and Challenges. Biophys Rev 2021, 13, 905–909. [Google Scholar] [CrossRef]
- Usyk, M.; Peters, B.A.; Karthikeyan, S.; McDonald, D.; Sollecito, C.C.; Vazquez-Baeza, Y.; Shaffer, J.P.; Gellman, M.D.; Talavera, G.A.; Daviglus, M.L.; et al. Comprehensive Evaluation of Shotgun Metagenomics, Amplicon Sequencing, and Harmonization of These Platforms for Epidemiological Studies. Cell Reports Methods 2023, 3. [Google Scholar] [CrossRef]
- Anjum, M.F.; Zankari, E.; Hasman, H. Molecular Methods for Detection of Antimicrobial Resistance. Microbiol Spectr 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.; Fasolino, T.; Davis, N.J.; Ivankovic, D.; Brownlee, N. Multiplex Detection of Antimicrobial Resistance Genes for Rapid Antibiotic Guidance of Urinary Tract Infections. Microbiol Res (Pavia) 2023, 14, 591–602. [Google Scholar] [CrossRef]
- de Abreu, V.A.C.; Perdigão, J.; Almeida, S. Metagenomic Approaches to Analyze Antimicrobial Resistance: An Overview. Front Genet 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Apjok, G.; Számel, M.; Christodoulou, C.; Seregi, V.; Vásárhelyi, B.M.; Stirling, T.; Eszenyi, B.; Sári, T.; Vidovics, F.; Nagrand, E.; et al. Characterization of Antibiotic Resistomes by Reprogrammed Bacteriophage-Enabled Functional Metagenomics in Clinical Strains. Nat Microbiol 2023, 8, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Köser, C.U.; Ellington, M.J.; Peacock, S.J. Whole-Genome Sequencing to Control Antimicrobial Resistance. Trends in Genetics 2014, 30, 401–407. [Google Scholar] [CrossRef]
- Burnard, D.; Gore, L.; Henderson, A.; Ranasinghe, A.; Bergh, H.; Cottrell, K.; Sarovich, D.S.; Price, E.P.; Paterson, D.L.; Harris, P.N.A. Comparative Genomics and Antimicrobial Resistance Profiling of Elizabethkingia Isolates Reveal Nosocomial Transmission and in Vitro Susceptibility to Fluoroquinolones, Tetracyclines, and Trimethoprim-Sulfamethoxazole. J Clin Microbiol 2020, 58. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob Agents Chemother 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Florensa, A.F.; Kaas, R.S.; Clausen, P.T.L.C.; Aytan-Aktug, D.; Aarestrup, F.M. ResFinder – an Open Online Resource for Identification of Antimicrobial Resistance Genes in next-Generation Sequencing Data and Prediction of Phenotypes from Genotypes. Microb Genom 2022, 8. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-Annot, a New Bioinformatic Tool to Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob Agents Chemother 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Bonin, N.; Doster, E.; Worley, H.; Pinnell, L.J.; Bravo, J.E.; Ferm, P.; Marini, S.; Prosperi, M.; Noyes, N.; Morley, P.S.; et al. MEGARes and AMR++, v3.0: An Updated Comprehensive Database of Antimicrobial Resistance Determinants and an Improved Software Pipeline for Classification Using High-Throughput Sequencing. Nucleic Acids Res 2023, 51, D744–D752. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Rensing, C.; Kristiansson, E.; Larsson, D.G.J. BacMet: Antibacterial Biocide and Metal Resistance Genes Database. Nucleic Acids Res 2014, 42. [Google Scholar] [CrossRef] [PubMed]
- Arango-Argoty, G.; Garner, E.; Pruden, A.; Heath, L.S.; Vikesland, P.; Zhang, L. DeepARG: A Deep Learning Approach for Predicting Antibiotic Resistance Genes from Metagenomic Data. Microbiome 2018, 6. [Google Scholar] [CrossRef]
- Liu, B.; Pop, M. ARDB - Antibiotic Resistance Genes Database. Nucleic Acids Res 2009, 37. [Google Scholar] [CrossRef]
- Gibson, M.K.; Forsberg, K.J.; Dantas, G. Improved Annotation of Antibiotic Resistance Determinants Reveals Microbial Resistomes Cluster by Ecology. ISME Journal 2015, 9, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Míguez, A.; Beghini, F.; Cumbo, F.; McIver, L.J.; Thompson, K.N.; Zolfo, M.; Manghi, P.; Dubois, L.; Huang, K.D.; Thomas, A.M.; et al. Extending and Improving Metagenomic Taxonomic Profiling with Uncharacterized Species Using MetaPhlAn 4. Nat Biotechnol 2023, 41, 1633–1644. [Google Scholar] [CrossRef] [PubMed]
- Franzosa, E.A.; McIver, L.J.; Rahnavard, G.; Thompson, L.R.; Schirmer, M.; Weingart, G.; Lipson, K.S.; Knight, R.; Caporaso, J.G.; Segata, N.; et al. Species-Level Functional Profiling of Metagenomes and Metatranscriptomes. Nat Methods 2018, 15, 962–968. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog Facilitate Examination of the Genomic Links among Antimicrobial Resistance, Stress Response, and Virulence. Sci Rep 2021, 11. [Google Scholar] [CrossRef]
- Gillespie, J.J.; Wattam, A.R.; Cammer, S.A.; Gabbard, J.L.; Shukla, M.P.; Dalay, O.; Driscoll, T.; Hix, D.; Mane, S.P.; Mao, C.; et al. Patric: The Comprehensive Bacterial Bioinformatics Resource with a Focus on Human Pathogenic Species. Infect Immun 2011, 79, 4286–4298. [Google Scholar] [CrossRef]
- Inouye, M.; Dashnow, H.; Raven, L.A.; Schultz, M.B.; Pope, B.J.; Tomita, T.; Zobel, J.; Holt, K.E. SRST2: Rapid Genomic Surveillance for Public Health and Hospital Microbiology Labs. Genome Med 2014, 6. [Google Scholar] [CrossRef]
- Bharat, A.; Petkau, A.; Avery, B.P.; Chen, J.; Folster, J.; Carson, C.A.; Kearney, A.; Nadon, C.; Mabon, P.; Thiessen, J.; et al. Correlation between Phenotypic and In Silico Detection of Antimicrobial Resistance in Salmonella Enterica in Canada Using Staramr. Microorganisms 2022, 10. [Google Scholar] [CrossRef]
- Seemann T Abricate, Https://Github.Com/Tseemann/Abricate.
- Carattoli, A.; Zankari, E.; Garciá-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico Detection and Typing of Plasmids Using Plasmidfinder and Plasmid Multilocus Sequence Typing. Antimicrob Agents Chemother 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Che, Y.; Dang, C.; Zhang, M.; Zhang, X.; Sun, Y.; Li, X.; Zhang, T.; Xia, Y. Nanopore-Based Long-Read Metagenomics Uncover the Resistome Intrusion by Antibiotic Resistant Bacteria from Treated Wastewater in Receiving Water Body. Water Res 2022, 226. [Google Scholar] [CrossRef] [PubMed]
- Kleinheinz, K.A.; Joensen, K.G.; Larsen, M.V. Applying the ResFinder and VirulenceFinder Web-Services for Easy Identification of Acquired Antibiotic Resistance and E. Coli Virulence Genes in Bacteriophage and Prophage Nucleotide Sequences. Bacteriophage 2014, 4, e27943. [Google Scholar] [CrossRef]
- Samtiya, M.; Matthews, K.R.; Dhewa, T.; Puniya, A.K. Antimicrobial Resistance in the Food Chain: Trends, Mechanisms, Pathways, and Possible Regulation Strategies. Foods 2022, 11. [Google Scholar] [CrossRef]
- Husna, A.; Rahman, M.M.; Badruzzaman, A.T.M.; Sikder, M.H.; Islam, M.R.; Rahman, M.T.; Alam, J.; Ashour, H.M. Extended-Spectrum β-Lactamases (ESBL): Challenges and Opportunities. Biomedicines 2023, 11. [Google Scholar] [CrossRef]
- Sah, A.K.; Feglo, P.K. Plasmid-Mediated Quinolone Resistance Determinants in Clinical Bacterial Pathogens Isolated from the Western Region of Ghana: A Cross-Sectional Study. Pan Afr Med J 2022, 43, 207. [Google Scholar] [CrossRef]
- Davies, A.R.; Chisnall, T.; Akter, S.; Afrad, M.M.H.; Sadekuzzaman, M.; Badhy, S.C.; Hasan, M.Z.; Rahman, M.T.; Smith, R.P.; Card, R.M.; et al. Genomic Characterisation of Escherichia Coli Isolated from Poultry at Retail through Sink Surveillance in Dhaka, Bangladesh Reveals High Levels of Multi-Drug Resistance. Front Microbiol 2024, 15. [Google Scholar] [CrossRef]
- Mondal, A.H.; Khare, K.; Saxena, P.; Debnath, P.; Mukhopadhyay, K.; Yadav, D. A Review on Colistin Resistance: An Antibiotic of Last Resort. Microorganisms 2024, 12. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Chen, H.; Li, N.; Wang, T.; Liang, W. The Spread of Antibiotic Resistance Genes In Vivo Model. Canadian Journal of Infectious Diseases and Medical Microbiology 2022, 2022. [Google Scholar] [CrossRef]
- Assar, S.; Hassanshahi, G.; Darehkordi, A.; Falahati-Pour, S.K.; Zarandi, E.R. Resistance Pattern of Escherichia Coli to Levofloxacin in Iran: A Narrative Review; 2020; Vol. 12;
- Mmatli, M.; Mbelle, N.M.; Osei Sekyere, J. Global Epidemiology, Genetic Environment, Risk Factors and Therapeutic Prospects of Mcr Genes: A Current and Emerging Update. Front Cell Infect Microbiol 2022, 12. [Google Scholar] [CrossRef]
- Osek, J.; Lachtara, B.; Wieczorek, K. Listeria Monocytogenes – How This Pathogen Survives in Food-Production Environments? Front Microbiol 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Tóth, A.G.; Csabai, I.; Krikó, E.; Tőzsér, D.; Maróti, G.; Patai, Á. V.; Makrai, L.; Szita, G.; Solymosi, N. Antimicrobial Resistance Genes in Raw Milk for Human Consumption. Sci Rep 2020, 10. [Google Scholar] [CrossRef]
- Kayode, A.J.; Okoh, A.I. Assessment of Multidrug-Resistant Listeria Monocytogenes in Milk and Milk Product and One Health Perspective. PLoS One 2022, 17. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, R.D.; Agunos, A.; Gow, S.P.; Varga, C. Assessing Antimicrobial Resistance in Campylobacter Jejuni and Campylobacter Coli and Its Association with Antimicrobial Use in Canadian Turkey Flocks. Epidemiol Infect 2023, 151. [Google Scholar] [CrossRef] [PubMed]
- NARMS Integrated Report: 2014 The National Antimicrobial Resistance Monitoring System: Enteric Bacteria;
- Liu, D.; Liu, W.; Lv, Z.; Xia, J.; Li, X.; Hao, Y.; Zhou, Y.; Yao, H.; Liu, Z.; Wang, Y.; et al. Emerging Erm(B)-Mediated Macrolide Resistance Associated with Novel Multidrug Resistance Genomic Islands in Campylobacter. 2019. [CrossRef]
- Premarathne, J.M.K.J.K.; Anuar, A.S.; Thung, T.Y.; Satharasinghe, D.A.; Jambari, N.N.; Abdul-Mutalib, N.A.; Yew Huat, J.T.; Basri, D.F.; Rukayadi, Y.; Nakaguchi, Y.; et al. Prevalence and Antibiotic Resistance against Tetracycline in Campylobacter Jejuni and C. Coli in Cattle and Beef Meat from Selangor, Malaysia. Front Microbiol 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- De, R. Mobile Genetic Elements of Vibrio Cholerae and the Evolution of Its Antimicrobial Resistance. Frontiers in Tropical Diseases 2021, 2. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiology and Molecular Biology Reviews 2001, 65, 232–260. [Google Scholar] [CrossRef]
- Schmidt, K.; Scholz, H.C.; Appelt, S.; Michel, J.; Jacob, D.; Dupke, S. Virulence and Resistance Patterns of Vibrio Cholerae Non-O1/Non-O139 Acquired in Germany and Other European Countries. Front Microbiol 2023, 14. [Google Scholar] [CrossRef]
- Koutsoumanis, K.; Allende, A.; Álvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; Hilbert, F.; et al. Role Played by the Environment in the Emergence and Spread of Antimicrobial Resistance (AMR) through the Food Chain. EFSA Journal 2021, 19. [Google Scholar] [CrossRef]
- Kiu, R.; Hall, L.J. An Update on the Human and Animal Enteric Pathogen Clostridium Perfringens. Emerg Microbes Infect 2018, 7. [Google Scholar] [CrossRef]
- Lina, G.; Quaglia, A.; Reverdy, M.-E.; Leclercq, R.; Vandenesch, F.O.; Etienne, J. Distribution of Genes Encoding Resistance to Macrolides, Lincosamides, and Streptogramins among Staphylococci; 1999; Vol. 43;
- Beres, C.; Colobatiu, L.; Tabaran, A.; Mihaiu, R.; Mihaiu, M. Prevalence and Characterisation of Clostridium Perfringens Isolates in Food-Producing Animals in Romania. Microorganisms 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J. xin; Zheng, H. ran; Wang, Y. yuan; Bai, L. lu; Du, X. li; Wu, Y.; Lu, J. xing Molecular Characteristics and Phylogenetic Analysis of Clostridium Perfringens from Different Regions in China, from 2013 to 2021. Front Microbiol 2023, 14. [Google Scholar] [CrossRef]
- Asad, A.; Jahan, I.; Munni, M.A.; Begum, R.; Mukta, M.A.; Saif, K.; Faruque, S.N.; Hayat, S.; Islam, Z. Multidrug-Resistant Conjugative Plasmid Carrying MphA Confers Increased Antimicrobial Resistance in Shigella. Sci Rep 2024, 14. [Google Scholar] [CrossRef]
- Hussain, H.I.; Aqib, A.I.; Seleem, M.N.; Shabbir, M.A.; Hao, H.; Iqbal, Z.; Kulyar, M.F. e. A.; Zaheer, T.; Li, K. Genetic Basis of Molecular Mechanisms in β-Lactam Resistant Gram-Negative Bacteria. Microb Pathog 2021, 158. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J. Transferable Mechanisms of Quinolone Resistance from 1998 Onward; 2019;
- Ma, Q.; Zhu, C.; Yao, M.; Yuan, G.; Sun, Y. Correlation between the Sulfamethoxazole-Trimethoprim Resistance of Shigella Flexneri and the Sul Genes. Medicine (United States) 2021, 100, E24970. [Google Scholar] [CrossRef] [PubMed]
- Sadeghabadi, A.F.; Ajami, A.; Fadaei, R.; Zandieh, M.; Heidari, E.; Sadeghi, M.; Ataei, B.; Hoseini, S.G. Widespread Antibiotic Resistance of Diarrheagenic Escherichia Coli and Shigella Species; 2014;
- Rawson, A.M.; Dempster, A.W.; Humphreys, C.M.; Minton, N.P. Pathogenicity and Virulence of Clostridium Botulinum. Virulence 2023, 14. [Google Scholar] [CrossRef]
- Roberts, M.C. Environmental Macrolide-Lincosamide-Streptogramin and Tetracycline Resistant Bacteria. Front Microbiol 2011, 2. [Google Scholar] [CrossRef]
- Swenson, J.M.; Thornsberry, C.; Mccroskey, L.M.; Hatheway, C.L.; Dowell, V.R. Susceptibility of Clostridium Botulinum to Thirteen Antimicrobial Agents; 1980; Vol. 18;
- Douillard, F.P.; Derman, Y.; Woudstra, C.; Selby, K.; Mäklin, T.; Dorner, M.B.; Saxén, H.; Dorner, B.G.; Korkeala, H.; Lindström, M. Genomic and Phenotypic Characterization of Clostridium Botulinum Isolates from an Infant Botulism Case Suggests Adaptation Signatures to the Gut. mBio 2022, 13. [Google Scholar] [CrossRef]
- Karlsson, P.A.; Tano, E.; Jernberg, C.; Hickman, R.A.; Guy, L.; Järhult, J.D.; Wang, H. Molecular Characterization of Multidrug-Resistant Yersinia Enterocolitica From Foodborne Outbreaks in Sweden. Front Microbiol 2021, 12. [Google Scholar] [CrossRef]
- Gkouletsos, T.; Patas, K.; Lambrinidis, G.; Neubauer, H.; Sprague, L.D.; Ioannidis, A.; Chatzipanagiotou, S. Antimicrobial Resistance of Yersinia Enterocolitica and Presence of Plasmid PYV Virulence Genes in Human and Animal Isolates. New Microbes New Infect 2019, 32. [Google Scholar] [CrossRef]
- Fredriksson-Ahomaa, M.; Grönthal, T.; Heljanko, V.; Johansson, V.; Rantala, M.; Heikinheimo, A.; Laukkanen-Ninios, R. Enteropathogenic Yersinia with Public Health Relevance Found in Dogs and Cats in Finland. Pathogens 2024, 13. [Google Scholar] [CrossRef]
- Ray, L.C.; Payne, D.C.; Rounds, J.; Trevejo, R.T.; Wilson, E.; Burzlaff, K.; Garman, K.N.; Lathrop, S.; Rissman, T.; Wymore, K.; et al. Syndromic Gastrointestinal Panel Diagnostic Tests Have Changed Our Understanding of the Epidemiology of Yersiniosis - Foodborne Diseases Active Surveillance Network, 2010-2021. Open Forum Infect Dis 2024, 11. [Google Scholar] [CrossRef]
- Perry, J.A.; Westman, E.L.; Wright, G.D. The Antibiotic Resistome: What’s New? Curr Opin Microbiol 2014, 21, 45–50. [Google Scholar] [CrossRef]
- Khmelevtsova, L.; Azhogina, T.; Karchava, S.; Klimova, M.; Polienko, E.; Litsevich, A.; Chernyshenko, E.; Khammami, M.; Sazykin, I.; Sazykina, M. Effect of Mineral Fertilizers and Pesticides Application on Bacterial Community and Antibiotic-Resistance Genes Distribution in Agricultural Soils. Agronomy 2024, 14. [Google Scholar] [CrossRef]
- Zhang, W.G.; Wen, T.; Liu, L.Z.; Li, J.Y.; Gao, Y.; Zhu, D.; He, J.Z.; Zhu, Y.G. Agricultural Land-Use Change and Rotation System Exert Considerable Influences on the Soil Antibiotic Resistome in Lake Tai Basin. Science of the Total Environment 2021, 771. [Google Scholar] [CrossRef]
- Durso, L.M.; Cook, K.L. Impacts of Antibiotic Use in Agriculture: What Are the Benefits and Risks? Curr Opin Microbiol 2014, 19, 37–44. [Google Scholar] [CrossRef]
- Barathan, M.; Ng, S.L.; Lokanathan, Y.; Ng, M.H.; Law, J.X. Unseen Weapons: Bacterial Extracellular Vesicles and the Spread of Antibiotic Resistance in Aquatic Environments. Int J Mol Sci 2024, 25. [Google Scholar] [CrossRef]
- Zhu, S.; Yang, B.; Yu, F.; Zhang, J.; Wang, Z.; Liu, Y. Investigation of the Impact of Widely Used Pesticides on Conjugative Transfer of Multidrug Resistance Plasmids. J Hazard Mater 2024, 478. [Google Scholar] [CrossRef]
- Chen, T.; Wang, Z.; Ruan, X. Antibiotic Resistome Dynamics in Agricultural River Systems: Elucidating Transmission Mechanisms and Associated Risk to Water Security. Science of the Total Environment 2024, 951. [Google Scholar] [CrossRef]
- Oulas, A.; Pavloudi, C.; Polymenakou, P.; Pavlopoulos, G.A.; Papanikolaou, N.; Kotoulas, G.; Arvanitidis, C.; Iliopoulos, I. Metagenomics: Tools and Insights for Analyzing next-Generation Sequencing Data Derived from Biodiversity Studies. Bioinform Biol Insights 2015, 9, 75–88. [Google Scholar] [CrossRef]
- Roumpeka, D.D.; Wallace, R.J.; Escalettes, F.; Fotheringham, I.; Watson, M. A Review of Bioinformatics Tools for Bio-Prospecting from Metagenomic Sequence Data. Front Genet 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Inda-Díaz, J.S.; Lund, D.; Parras-Moltó, M.; Johnning, A.; Bengtsson-Palme, J.; Kristiansson, E. Latent Antibiotic Resistance Genes Are Abundant, Diverse, and Mobile in Human, Animal, and Environmental Microbiomes. Microbiome 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Lanza, V.F.; Baquero, F.; Martínez, J.L.; Ramos-Ruíz, R.; González-Zorn, B.; Andremont, A.; Sánchez-Valenzuela, A.; Ehrlich, S.D.; Kennedy, S.; Ruppé, E.; et al. In-Depth Resistome Analysis by Targeted Metagenomics. Microbiome 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Van Der Helm, E.; Imamovic, L.; Ellabaan, M.M.H.; Van Schaik, W.; Koza, A.; Sommer, M.O.A. Rapid Resistome Mapping Using Nanopore Sequencing. Nucleic Acids Res 2017, 45. [Google Scholar] [CrossRef] [PubMed]
- Martiny, H.M.; Munk, P.; Brinch, C.; Aarestrup, F.M.; Petersen, T.N. A Curated Data Resource of 214K Metagenomes for Characterization of the Global Antimicrobial Resistome. PLoS Biol 2022, 20. [Google Scholar] [CrossRef]
- Lee, K.; Kim, D.W.; Cha, C.J. Overview of Bioinformatic Methods for Analysis of Antibiotic Resistome from Genome and Metagenome Data. Journal of Microbiology 2021, 59, 270–280. [Google Scholar] [CrossRef]
- Liu, F.; Luo, Y.; Xu, T.; Lin, H.; Qiu, Y.; Li, B. Current Examining Methods and Mathematical Models of Horizontal Transfer of Antibiotic Resistance Genes in the Environment. Front Microbiol 2024, 15. [Google Scholar] [CrossRef]
- Yarygin, K.S.; Kovarsky, B.A.; Bibikova, T.S.; Melnikov, D.S.; Tyakht, A. V.; Alexeev, D.G. ResistoMap-Online Visualization of Human Gut Microbiota Antibiotic Resistome. In Proceedings of the Bioinformatics; Oxford University Press, July 15 2017; Vol. 33; pp. 2205–2206. [Google Scholar]
- Munkholm, L.; Rubin, O. The Global Governance of Antimicrobial Resistance: A Cross-Country Study of Alignment between the Global Action Plan and National Action Plans. Global Health 2020, 16. [Google Scholar] [CrossRef]
- Edelstein, M.; Lee, L.M.; Herten-Crabb, A.; Heymann, D.L.; Harper, D.R. Strengthening Global Public Health Surveillance through Data and Benefit Sharing. Emerg Infect Dis 2018, 24, 1324–1330. [Google Scholar] [CrossRef]
| Database/Tool | Role in Resistome Analysis | Website | Reference |
|---|---|---|---|
| CARD (Comprehensive Antibiotic Resistance Database) | A curated database containing information on resistance genes and their mechanisms, using for the detection and annotation of AMR genes. | CARD | [40] |
| ResFinder | Identifies acquired ARGs from bacterial genome data. | ResFinder | [41] |
| ARG-ANNOT | Antibiotic Resistance Gene-ANNOTation tool providing ARG identification based on curated sequences. | ARG-ANNOT | [42] |
| MEGARes | Database of ARGs used in resistome analysis of metagenomic datasets. | MEGARes | [43] |
| BacMet | A database of antibacterial biocide- and metal-resistance genes. | BacMet | [44] |
| DeepARG | A tool that uses deep learning to predict ARGs from metagenomic data. | DeepARG | [45] |
| Antibiotic Resistance Genes Database (ARDB) | A comprehensive resource of ARGs and their associated phenotypes. | ARDB | [46] |
| Resfams | A collection of protein families associated with antibiotic resistance, for the functional annotation of ARGs. | Resfams | [47] |
| Antibiotic Resistance Ontology (ARO) | A standardized vocabulary of resistance genes, phenotypes, and mechanisms, using for AMR classification | ARO | [40] |
| MetaPhlAn | A tool for profiling microbial communities, including the identification of ARGs. | MetaPhlAn | [48] |
| Humann2 | Tool for functional profiling of microbial communities, with ARG identification. | Humann2 | [49] |
| AMRFinderPlus | Identifies AMR genes, virulence factors, and other genes from whole genome sequences. | AMRFinderPlus | [50] |
| RGI (Resistance Gene Identifier) | A tool integrated with CARD to detect and annotate ARGs from genomic and metagenomic sequences. | RGI | [40] |
| PATRIC | A bacterial bioinformatics platform comparative genomics and AMR gene detection. | PATRIC | [51] |
| SRST2 (Short Read Sequence Typing for Bacterial Pathogens) | A tool for strain typing and detecting ARGs from short-read sequence data. | SRST2 | [52] |
| StarAMR | A tool for identifying ARGs from assembled genome sequences. | StarAMR | [53] |
| ABRicate | A command-line tool that screens bacterial genomes for ARGs using multiple databases (e.g., CARD, ResFinder). | ABRicate | [54] |
| PlasmidFinder | Identifies plasmid replicons in whole-genome sequencing data. Tools helps to identify the detection of plasmids related to ARGs. | PlasmidFinder | [55] |
| ARGpore | Detects ARGs from long-read (nanopore) sequencing data. | ARGpore | [56] |
| VirulenceFinder | Identifies virulence genes in bacterial genomes that may be associated with antimicrobial resistance. | VirulenceFinder | [57] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





