Submitted:
20 September 2024
Posted:
23 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Software and Network Databases
2.2. Prediction and Identification of Active Ingredients and Potential Targets of C. Japonicum
2.3. Construction of Networks and Pathway Analysis
2.4. Molecular Docking
2.5. Verification of Anti-inflammatory Activity of Active Ingredients of C. japonicum in RAW264.7 Cells
3. Results
3.1. The Identification of Active Ingredients in C. japonicum and the Prediction of Their Molecular Targets
3.2. Screening of Potential Anti-inflammatory Targets of C. japonicum
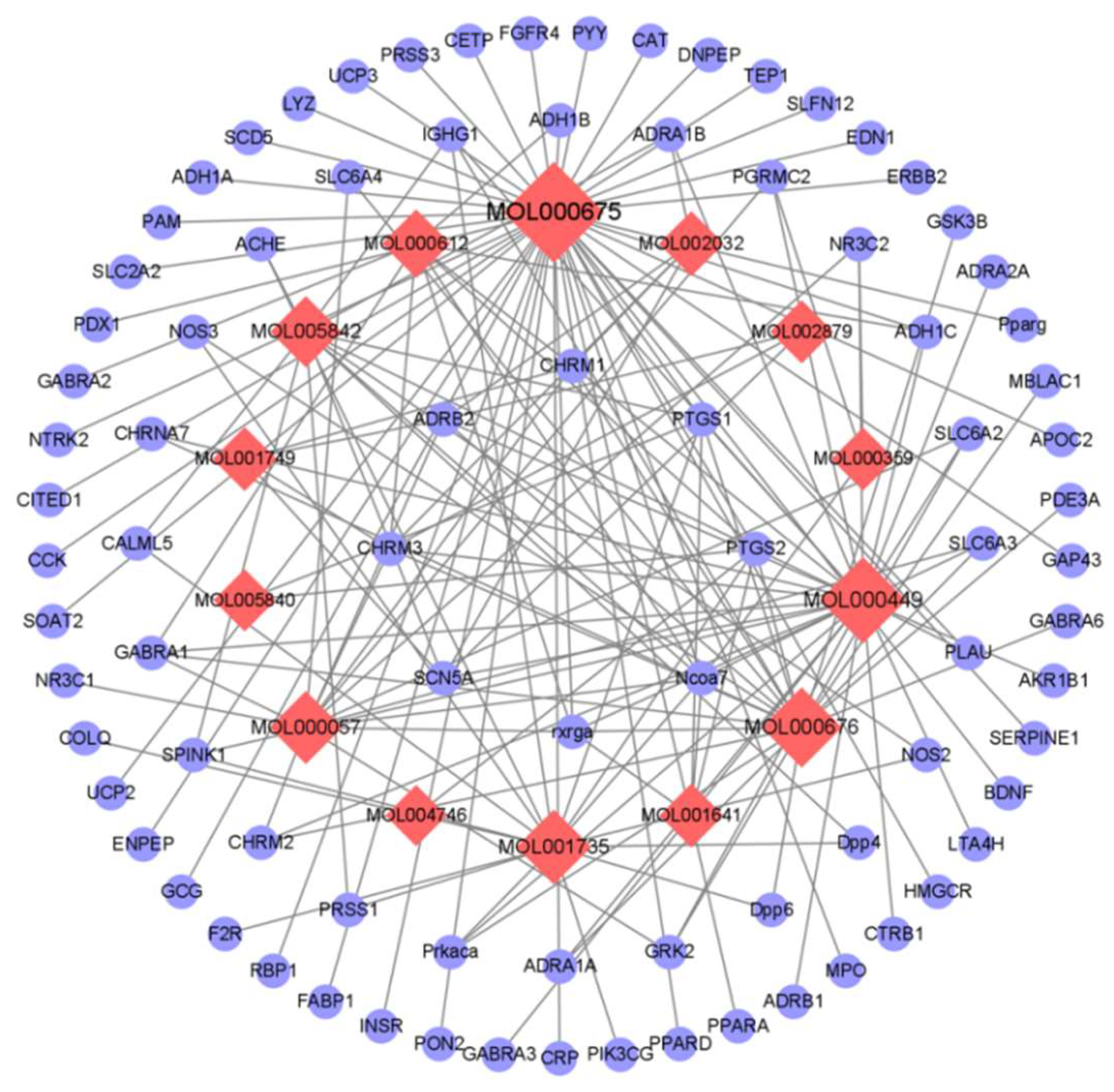
3.3. Construction of Protein-Protein Interaction (PPI) Network and Results of Core Gene Screening
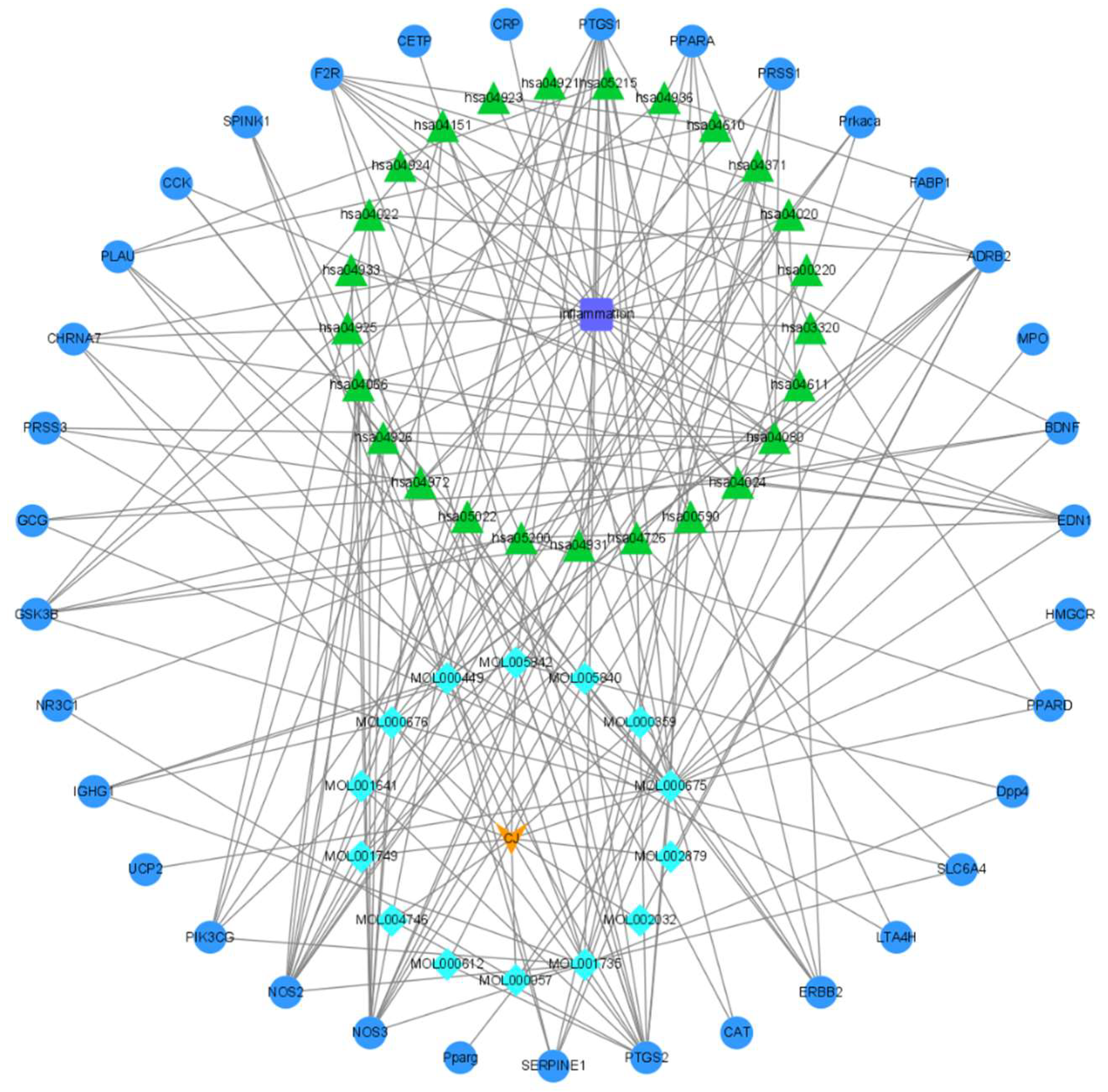
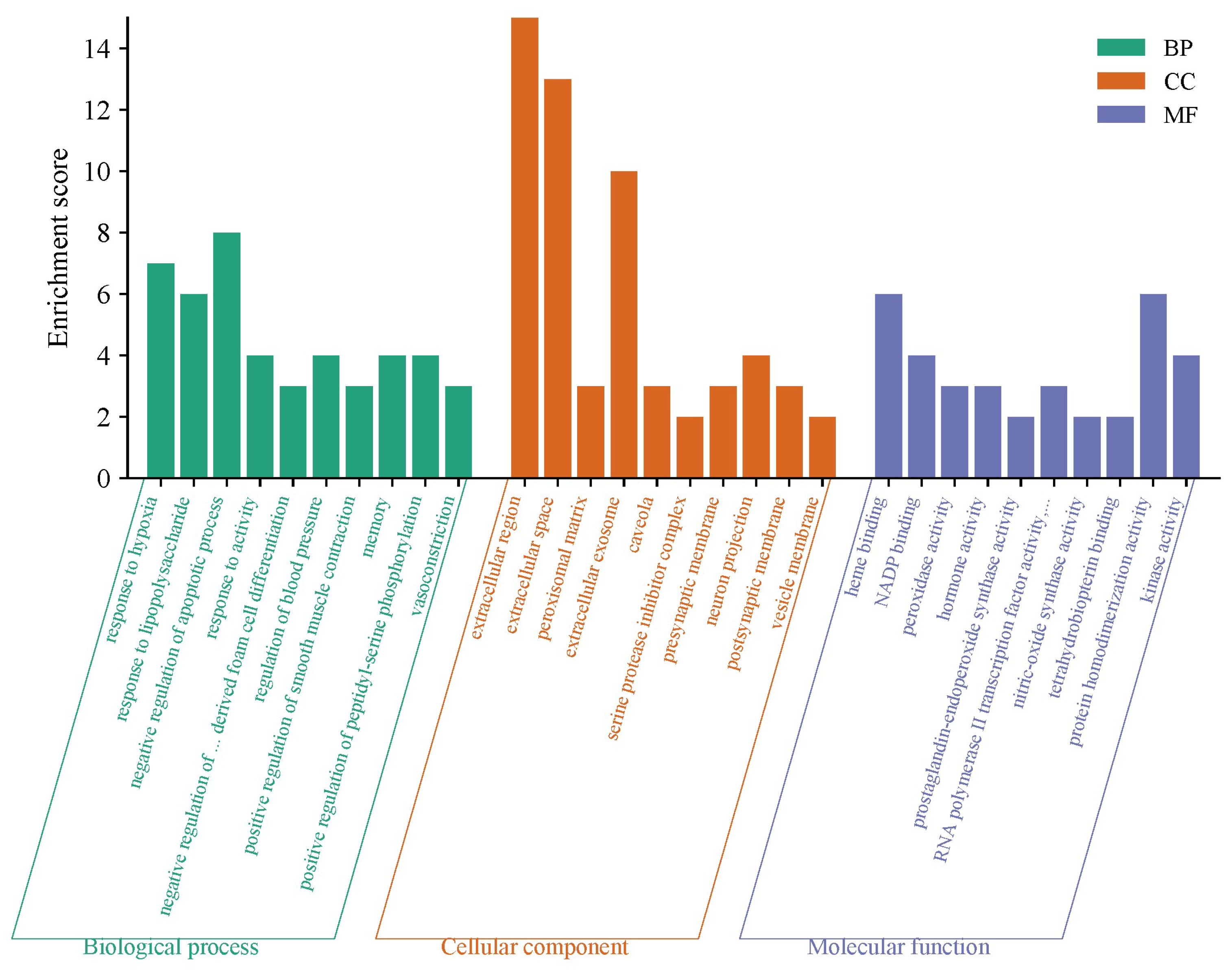
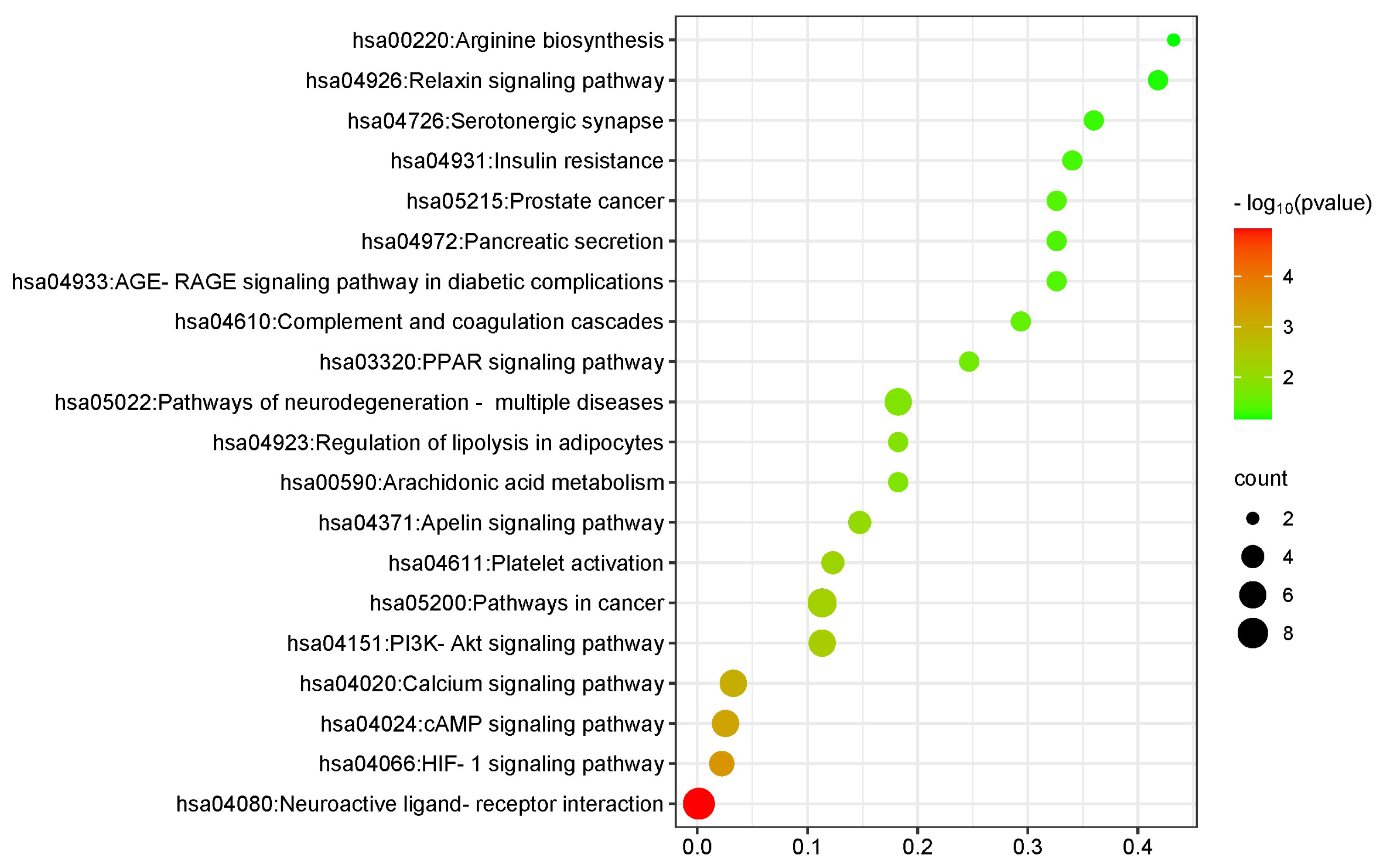
3.4. GO and KEGG Enrichment Function Analysis
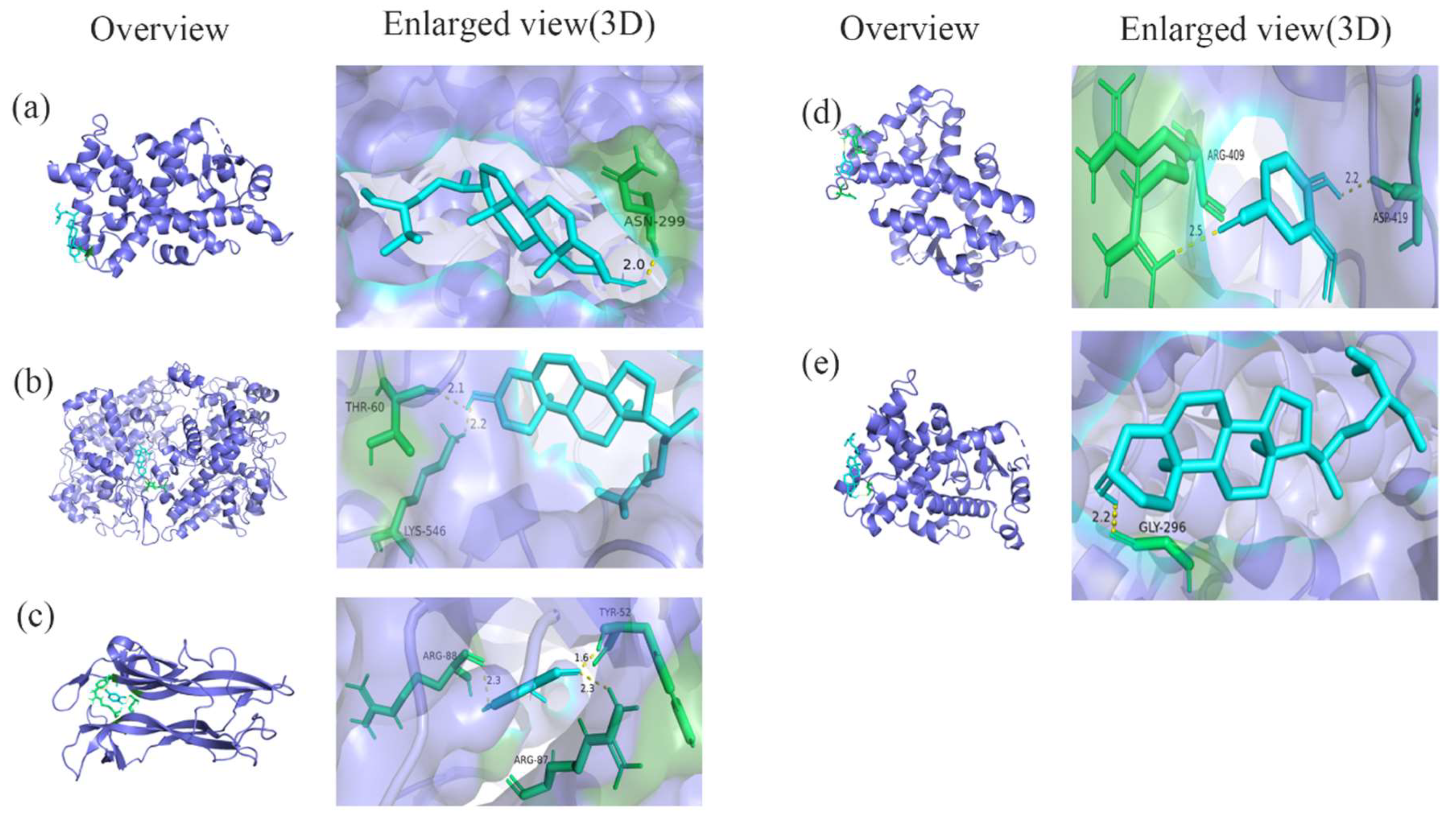
3.5. Molecular Docking Validation
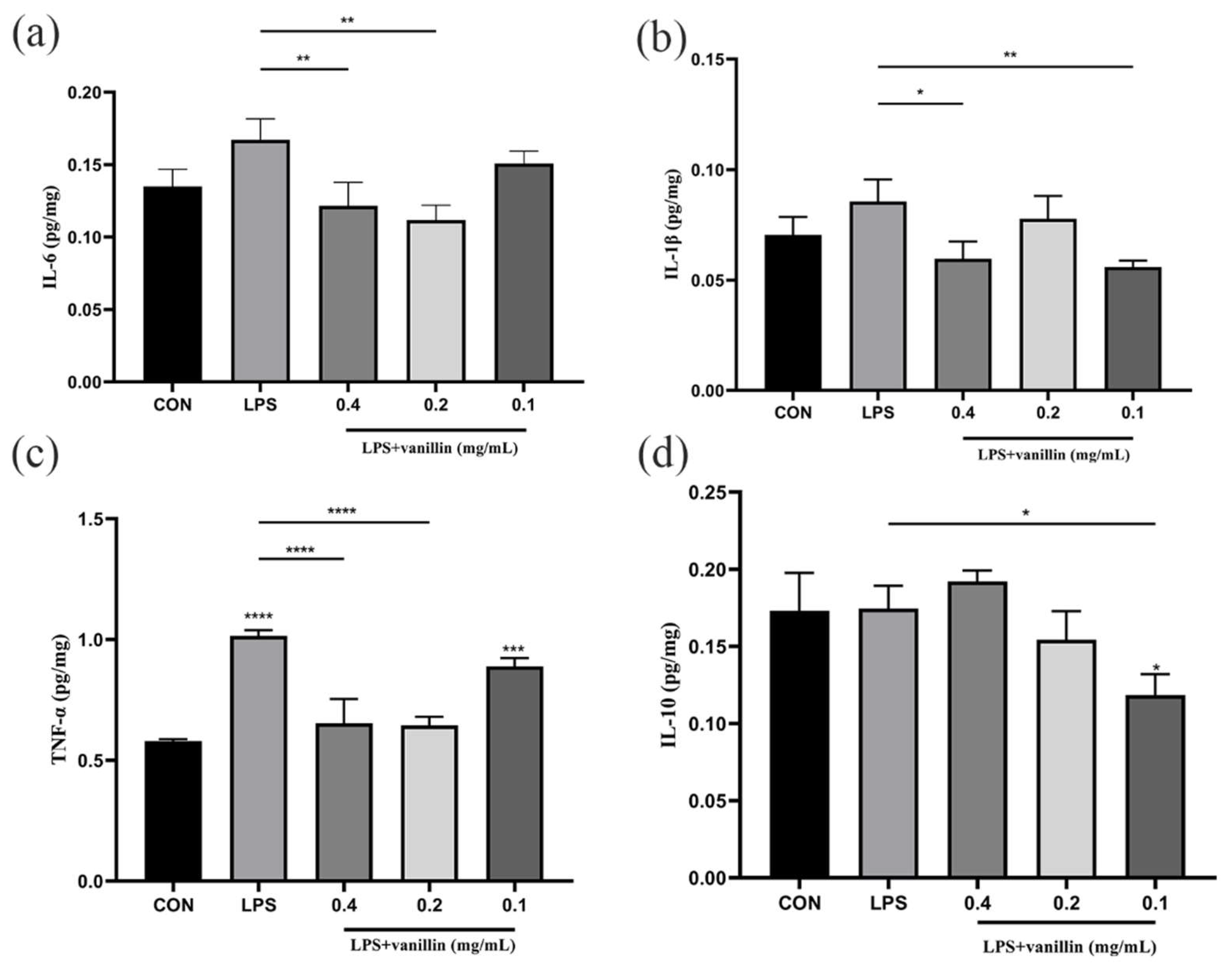
3.6. Effect of Vanillin on Inflammatory Factors in RAW264.7 Cells Challenged with LPS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, L.C.; Li, J.F.; Zhang, T.T.; Tao, F.F.; Liu, W.H. Discusses the Anti-inflammatory Effect of Stigmasterol Based on Network Pharmacology and Cell Experiments. Chin. Patent Med. 2022, 44, 7. [Google Scholar] [CrossRef]
- Park, J.Y.; Kim, H.Y.; Shibamoto, T.; Jang, T.S.; Lee, S.C.; Shim, J.S.; Hahm, D.H.; Lee, H.J.; Lee, S.; Kang, K.S. Beneficial Effects of a Medicinal Herb, Cirsium Japonicum var. Maackii, Extract and its Major Component, Cirsimaritin on Breast Cancer Metastasis in MDA-MB-231 Breast Cancer Cells. Bioorg. Med. Chem. Lett. 2017, 17, 3968–3973. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Shao, H.; Chen, Y.; Ding, N.; Yang, A.N.; Tian, J.; Jiang, Y.X.; Li, G.Z.; Jiang, Y.D. In Renal Hypertension, Cirsium Japonicum Strengthens Cardiac Function via the Intermedin/Nitric Oxide Pathway. Biomed. Pharmacother. 2018, 101, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Wagle, A.; Seong, S.H.; Shrestha, S.; J, H.A.; Choi, J.S. Korean Thistle (Cirsium Japonicum Var. Maackii (Maxim.) Matsum.): a Potential Dietary Supplement Against Diabetes and Alzheimer's Disease. Molecules 2019, 24, 649. [Google Scholar] [CrossRef]
- Yoon, S.; Kim, M.; Shin, S.; Woo, J.; Son, D.; Ryu, D.; Yoo, J.; Park, D.; Jung, E. Effect of Cirsium Japonicum Flower Extract on Skin Aging Induced by Glycation. Molecules 2022, 27, 2093. [Google Scholar] [CrossRef]
- Cho, C.; Kang, L.J.; Jang, D.; Jeon, J.; Lee, H.; Choi, S.; Han, S.J.; Oh, E.; Nam, J.; Kim, C.S.; Park, E.; Jeong, S.Y.; Park, C.H.; Shin, Y.S.; Eyun, S.I.; Yang, S. Cirsium Japonicum Var. Maackii and Apigenin Block Hif-2α-induced Osteoarthritic Cartilage Destruction. J. Cell. Mol. Med. 2019, 8, 5369–5379. [Google Scholar] [CrossRef]
- Meng, L.; Liu, R.; Sun, A.; Wu, S.J.; Liu, N.N. Separation and Purification of Rutin and Acaciin from the Chinese Medicinal Herb Herba Cirsii by Combination of Macroporous Absorption Resin and High-speed Counter-current Chromatography. J. Chromatogr. Sci. 2009, 47, 329–332. [Google Scholar] [CrossRef]
- Ye, L.; Yang, F.Q.; Liang, J.; Zhao, S.H.; Gao, Y.L. In Vitro Antibacterial Activity of Different Polar Parts of Cirsium Japonicum Extract from Ningxia Against Four Candida Species. Chin. J. Exper. Trad. Med. Form. 2011, 17, 222–223. [Google Scholar] [CrossRef]
- Кравченкo, О.В. Artichoke Extracts: Physiological Effects, Use in Obstetric Practice. Reprod. Endocrinol 2018, 40, 80–86. [Google Scholar] [CrossRef]
- Che, D.N.; Shin, J.Y.; Kang, H.J.; Cho, B.O.; Park, J.H.; Wang, F.; Hao, S.P.; Sim, J.S.; Sim, D.J.; Jang, S.I. Ameliorative Effects of Cirsium Japonicum Extract and Main Component Cirsimaritin in Mice Model of Hgh-fat Diet-induced Metabolic Dysfunction-associated Fatty Liver Disease. Food Sci. Nutr. 2021, 11, 6060–6068. [Google Scholar] [CrossRef]
- Li, T.; Zhang, W.; Hu, E.; Sun, Z.; Li, P.; Yu, Z.; Zhu, X.; Zheng, F.; Xing, Z.; Xia, Z.; He, F.; Luo, J.; Tang, T.; Wang, Y. Integrated Metabolomics and Network Pharmacology to Reveal the Mechanisms of Hydroxysafflor Yellow A Against Acute Traumatic Brain Injury Comput. Struct. Biotechnol. J 2021, 19, 1002–1013. [Google Scholar] [CrossRef]
- Shang, L.R.; Wang, Y.C.; Li, J.X.; Zhou, F.Y.; Xiao, K.M.; Liu, Y.H.; Zhang, M.Q.; Wang, S.H.; Yang, S.L. Mechanism of Sijunzi Decoction in the Treatment of Colorectal Cancer Based on Network Pharmacology and Experimental Validation. J. Ethnopharmacol. 2022, 302, 115876. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhang, D.F.; Zhou, W.A.; Wang, L.; Wang, B.Y.; Zhang, T.Y.; Li, S. Network Pharmacology: Towards the Artificial Intelligence-based Precision Traditional Chinese Medicine. Brief Bioinform. 2023, 25. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, H.; Li, N.; Chen, J.M.; Xu, H.; Wang, Y.J.; Liang, Q.Q. Network Pharmacology, A Promising Approach to Reveal the Pharmacology Mechanism of Chinese Medicine Formula. J. Ethnopharmacol. 2023, 116306. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Z.Q.; Liao, J.; Chen, Q.; Lu, X.Y.; Fan, X.H. Network Pharmacology Approaches for Research of Traditional Chinese Medicines. Chin. J Nat. Medicines. 2023, 21, 323–332. [Google Scholar] [CrossRef]
- Zhong, Y.; Luo, J.; Tang, T.; Li, P.; Liu, T.; Cui, H.; Wang, Y.; Huang, Z. Exploring Pharmacological Mechanisms of Xuefu Zhuyu Decoction in the Treatment of Traumatic Brain Injury via a Network Pharmacology Approach. Evid.-Based Compl. Alt. 2019, 2018, 7518374. [Google Scholar] [CrossRef]
- Ma, C.; Xu, T.; Sun, X.; Zhang, S.; Liu, S.; Fan, S.; Lei, C.; Tang, F.; Zhai, C.; Li, C.; Luo, J.; Wang, Q.; Wei, W.; Wang, X.; Cheng, F. Network Pharmacology and Bioinformatics Approach Reveals the Therapeutic Mechanism of Action of Baicalein in Hepatocellular Carcinoma. Evid.-Based Compl. Alt. 2019, 2019, 1–15. [Google Scholar] [CrossRef]
- Ma, Q.Y.; Liu, Y.C.; Zhang, Q.; Yi, W.D.; Sun, Y.; Gao, X.D.; Zhao, X.; Wang, H.W.; Lei, K.; Luo, W.J. Integrating Network Pharmacology, Molecular Docking and Experimental Verification to Reveal the Mechanism of Artesunate in Inhibiting Choroidal Melanoma. Front. Pharmacol. 2024, 151448381–1448381. [Google Scholar] [CrossRef]
- Y, J.H.; Li, L.; Hu, Z.X. Exploring the Molecular Mechanism of Action of Yinchen Wuling Powder for the Treatment of Hyperlipidemia, Using Network Pharmacology, Molecular Docking, and Molecular Dynamics Simulation. BioMed. Res. Int. 2021, 9965906. [Google Scholar] [CrossRef]
- Yuan, Z.Z.; Pan, Y.Y.; Leng, T.; Chu, Y.; Zhang, H.J.; Ma, J.R; Ma, X.J. Progress and Prospects of Research Ideas and Methods in the Network Pharmacology of Traditional Chinese Medicine. J. Pharm. Pharm. Sci. 2022, 25218–25226. [Google Scholar] [CrossRef]
- Aminu, K.S.; Uzairu, A.; Abechi, S.E.; Shallangwa, G.A.; Umar, A.B. Activity Prediction, Structure-based Drug Design, Molecular Docking, and Pharmacokinetic Studies of 1,4-dihydropyridines Derivatives as α-amylase inhibitors. J. Talbah Univ. Med. Sci. 2024, 19, 270–286. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.K.; Tao, B.; Xue, X.; Feng, C.X.; Ren, Y.T.; Ma, H.Y.; Zhang, J.L.; Si, Y.F.; Zhang, S.S.; Liu, S.; Li, H.; Zhou, J.H.; Li, G.; Wang, Z.F.; X, J.P.; Zhu, Z.L. Molecular Mechanism of Epicedium Treatment for Depression Based on Network Pharmacology and Molecular Docking Technology. BMC Complement Med. 2021, 21, 222. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.L.; Cui, Y.P. Study on the Mechanism of Action of RoucongrongTang in Treating Functional Constipation based on Network Pharmacology and Molecular Docking. J. Contemporary Med. 2024, 6, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Larisa, I.; Mati, K. The Impact of Software Used and the Type of Target Protein on Molecular Docking Accuracy. Molecules 2022, 27, 9041. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Narang, B.K.; Gupta, M.K.; Abbot, V.; Singh, V.; Rawal, R.K. Molecular Docking Studies on Isocytosine Analogues as Xanthine Oxidase Inhibitors. Drug. Res. 2018, 68, 395–402. [Google Scholar] [CrossRef]
- Zhu, W.T.; Fan, X.M.; Wei, H.; Wang, S.M. Mechanism Research of Apatinib-treated Breast Cancer Based on Network Pharmacology. Chin. Pharm. J. 2016, 51, 1860–1865. [Google Scholar] [CrossRef]
- Liu, Y.; Ju, Y.; Qin, X. Studies on the Compatibility Mechanism and Material Basis of Danggui Buxue Decoction Against Anemia Mice Using Metabonomics and Network Pharmacology. J. Pharm. Pharmacol. 2021, 73, 767–777. [Google Scholar] [CrossRef]
- Gong, P.Y.; Guo, Y.J.; Li, X.M.; Wang, N.; Gu, J. Study on the Potential Pharmacodynamic Substances of Jinhua Qinggan Granule for Prevention and Treatment of Novel Coronavirus Based on Network Pharmacology and Molecular Docking Technology. Chin. Herb Med. 2020, 51, 1685–1693. [Google Scholar] [CrossRef]
- Ding, H.X.; Zhao, Y.; Feng, J. Research progress of plant extracts in the field of feed additives based on network pharmacology. J. Anim. Nutr. 2021, 33, 3065–3071. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, Y.; Jiang, J.; Cui, X.; Cheng, S.; Liu, L.; Huang, Z.; Liao, R.; Zhao, P.; Yu, J.; Wang, J.; Jia, Y.; Jin, W.; Zhou, F. Stigmasterol Attenuates Hepatic Steatosis in Rats by Strengthening the Intestinal Barrier and Improving Bile Acid Metabolism. Npj. Sci. Food. 2022, 27, 38. [Google Scholar] [CrossRef]
- Liu, D.; Chen, Y.F.; Liu, T.J.; Tang, T.; Yu, S.J.; Liu, G.M.; Tang, Y.B. Z.Q. Network Pharmacology and Experimental Study of Jiawei Guizhi Fuling Pill in the Treatment of Benign Prostatic Hyperplasia. Res. Dev. Natur. Products. 2022, 34, 1234–1249. [Google Scholar] [CrossRef]
- Gao, M.M.; Chen, Y.L.; Hao, Y.K.; Guo, J. Metabonomics Study of Cirsium Japonicum Extract in Improving Hypercholesterolemia Model Mice. Chin. Ph. 2023, 34, 1590–1595. [Google Scholar] [CrossRef]
- Serafini, M.; Peluso, I.; Raguzzini, A. Flavonoids as Anti-inflammatory Agents. Proc. Nutr. Soc. 2010, 69, 27–28. [Google Scholar] [CrossRef] [PubMed]
- Mehrabani, M.; Sargazi, M.L.; Amirkhosravi, A.; Farhadi, S.; Vasei, S.; Raeiszadeh, M.; Mehrabani, M. The Influence of Harvest Time on Total Phenolic and Flavonoid Contents, Antioxidant, Antibacterial and Cytotoxicity of Rheum Khorasanicum Root Extract. Ann. Pharm. Fr. 2023, 81, 475–483. [Google Scholar] [CrossRef]
- Shi, M.Y.; Chen, Z.X.; Gong, H.; Peng, Z.L.; Sun, Q.; Luo, K.P.; Wu, B.Y.; Wen, C.B.; Lin, W. Luteolin, a Flavone Ingredient: Anticancer Mechanisms, Combined Medication Strategy, Pharmacokinetics, Clinical Trials, and Pharmaceutical Researches. Phytother. Res. 2024, 38, 880–911. [Google Scholar] [CrossRef]
- Naik, K.K.; Thangavel, S.; Alam, A.; Kumar, S. Flavone Analogues as Antimicrobial Agents. Recent Patents Infla. 2017, 11, 53–63. [Google Scholar] [CrossRef]
- Chen, Y.P.; Xie, T.; Zhang, H.; Zhou, Y.M. Physiological Function of β -sitosterol and Its Application in Animal Production. J. Anim. Physiol. An. N. 2022, 34, 11. [Google Scholar] [CrossRef]
- Liu, Y.X.; Wang, Z.W.; Yao, P.S.; Li, X.y.; Han, R.M.; Zhang, D.Q.; Zhao, Z.J.; Wang, Y.P.; Zhang, J.P. Antioxidation Activity Enhancement by Intramolecular Hydrogen Bond and Non-Browning Mechanism of Active Ingredients in Rosemary: Carnosic Acid and Carnosol. J. Phys. Chem. C. 2024, 128, 7627–7638. [Google Scholar] [CrossRef]
- Hou, J.Z.; Xiong, W.; Shao, X.Y.; Long, L.; Chang, Y.; Chen, G.H.; Wang, L.; Wang, Z.C.; Huang, Y.Z. Liposomal Resveratrol Alleviates Platelet Storage Lesion via Antioxidation and the Physical Buffering Effect. ACS Appl. Mater. 2023, 15, 45658–45667. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, Y.C.; Gu, D.Y.; Liu, C.; Wang, Y.; Tang, S.S.; Yin, Y.X.; Tian, J. Fermentation of Robinia Pseudoacacia Flower for Improving the Antioxidation: Optimized Conditions, Active Composition, Mechanism, and Biotransformation Process. PREP Biochem Biotech. 2023, 53, 11–13. [Google Scholar] [CrossRef]
- Li, Z.J.; Xue, Y.B.; Li, M.X.; Guo, Q.B.; Sang, Y.X.; Wang, C.L.; Luo, C. The Antioxidation of Different Fractions of Dill (Anethum graveolens) and Their Influences on Cytokines in Macrophages RAW264.7. J. Oleo Sci. 2018, 67, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.B.; Liu, X.L.; Cheng, R.Q.; Jia, H.X.; Wang, X.Y.; Kong, L.N.; Li, Y.J.; Cui, T.T.; Qu, H.D.; Feng, C. Effect of β-sitosterol on The Side Effects and Pharmacological Effects of Aspirin-induced Gastric Mucosal Injury. Chin. J. Ex. Trad. Med. Formulae. 2016, 22, 148–152. [Google Scholar] [CrossRef]
- Long, B.y. Effect of Oleic Acid on Lipopolysaccharide-induced Inflammatory Response of Mouse Macrophages and Its Mechanism. D. Univ. South Chin. 2019. [Google Scholar] [CrossRef]
- Sun, J.L.; Yan, J.F.; Li, J.; Wang, W.R.; Yu, S.B.; Zhang, H.Y.; Huang, F.; Niu, L.N.; Jiao, K. Conditional Deletion of Adrb2 in Mesenchymal Stem Cells Attenuates Osteoarthritis-like Defects in Temporomandibular Joint. Bone 2020, 33, 115229. [Google Scholar] [CrossRef] [PubMed]
- Kunzmann, A.T.; Murray, L.J.; Cardwell, C.R.; McShane, C.M.; McMenamin, U.C.; Cantwell, M.M. PTGS2 (Cyclooxygenase-2) Expression and Survival among Colorectal Cancer Patients: A Systematic Review. Cancer Epidemiol Biomarkers Prev. 2013, 22, 1490–1497. [Google Scholar] [CrossRef]
- Mousavi, S.E.; Saberi, P.; Ghasemkhani, N.; Fakhraei, N.; Mokhtari, R.; Dehpour, A.R. Licofelone Attenuates LPS-induced Depressive-like Behavior in Mice: A Possible Role for Nitric Oxide. Pharm. Sci. 2018, 21, 184–194. [Google Scholar] [CrossRef]
- Hellmann, J.; Tang, Y.; Zhang, M.J.; Hai, T.; Bhatnagar, A.; Srivastava, S.; Spite, M. Atf3 Negatively Regulates Ptgs2/Cox2 Expression During Acute Inflammation. Prostag Oth. Lipid. M. 2015, 116–117, 49–56. [Google Scholar] [CrossRef]
- Ricciotti, E.; FitzGerald, G.A. Prostaglandins and Inflammation. Ioscler Thromb Vasc. Biol. 2011, 5, 986–1000. [Google Scholar] [CrossRef]
- Neto, A.B.L.; Vasconcelos, N.B.R.; Santos, T.R.D.; Duarte, L.E.C.; Assunção, M.L.; Sales-Marques, C.; Silva Ferreira, H. Prevalence of IGFBP3, NOS3 and TCF7L2 Polymorphisms and Their Association with Hypertension: A Population-based Study with Brazilian Women of African Descent. BMC Res. Notes 2021, 17, 186. [Google Scholar] [CrossRef]
- Sadati, S.M.; Radfar, M.; Kakavand Hamidi, A.; Abdollahi, M.; Qorbani, M.; Nasli Esfahani, E.; Amoli, M.M. Association Between the Polymorphism of Glu298Asp in Exon 7 of the eNOS Gene with Foot Ulcer and Oxidative Stress in Adult Patients with Type 2 Diabetes. Can. J. Diabetes 2018, 42, 18–22. [Google Scholar] [CrossRef]
- Björkholm, C.; Monteggia, L.M. BDNF - A Key Transducer of Antidepressant Effects. Neuropharmacology 2016, 102, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Ming, Y.; Zhou, X.N.; Liu, G.; Abudupataer, M.; Zhu, S.C.; Xiang, B.T.; Yin, X.J.; Lai, H.; Sun, Y.X.; Wang, C.S.; Li, J.; Zhu, K. PM2.5 Exposure Exacerbates Mice Thoracic Aortic Aneurysm and Dissection by Inducing Smooth Muscle Cell Apoptosis via the MAPK Pathway. Chemosphere. 2023, 313, 137500. [Google Scholar] [CrossRef] [PubMed]
- Jin, W. Regulation of BDNF-TrkB Signaling and Potential Therapeutic Strategies for Parkinson's Disease. Clin. Med. Insights-Pe. 2020, 17, 257. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Zhu, X.Y.; Si, J.R. Angelica Polysaccharide Ameliorates Memory Impairment in Alzheimer's Disease Rat Through Activating BDNF/TrkB/CREB Pathway. Exp. Biol. Med (Maywood) 2020, 245, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nava-Villalba, M.; Nuñez-Anita, R.E.; Bontempo, A.; Aceves, C. Activation of Peroxisome Proliferator-activated Receptor Gamma is Crucial for Antitumoral Effects of 6-iodolactone. Mol. Cancer Res. 2015, 7, 168. [Google Scholar] [CrossRef]
- Wang, B.; Song, Z.J.; Qiu, L.; Ling, J.P. Experimental Study on Yiqi Huayu Zhiwei Recipe Relieving Chronic Atrophic Gastritis by Regulating PI3K/AKT Signaling Pathway. J. Zhejiang Uni. Trad. Chin. Med. 2023, 47, 1123–1128. [Google Scholar] [CrossRef]
- Sun, J.P.; Shi, L.; Wang, F.; Qin, J.; Ke, B. Modified Linggui Zhugan Decoction Ameliorates Glycolipid Metabolism and Inflammation via PI3K-Akt/mTOR-S6K1/AMPK-PGC-1 α Signaling Pathways in Obese Type 2 Diabetic Rats. Chin. J. Integr. Med. 2022, 28, 52–59. [Google Scholar] [CrossRef]
- Dobrovolskaia, M.A.; Vogel, S.N. Toll Receptors, CD14, and Macrophage Activation and Deactivation by LPS. Microbes Infect. 2002, 4, 903–914. [Google Scholar] [CrossRef]
- Maldonado, R.F.; Sá-Correia, L.; Valvano, M.A. Lipopolysaccharide Modification in Gram-negative Bacteria During Chronic Infection. FEMS Microbiol. Rev. 2016, 40, 480–493. [Google Scholar] [CrossRef]
- Olivry, T.; Foster, A.P.; Mueller, R.S.; McEwan, N.A.; Chesney, C.; Williams, H.C. Interventions for Atopic Dermatitis in Dogs: A Systematic Review of Randomized Controlled Trials. Vet. Dermatol. 2010, 21, 4–22. [Google Scholar] [CrossRef]
| Molecular ID | Molecule name | OB (%) | DL |
|---|---|---|---|
| MOL001641 MOL001735 MOL001749 MOL002032 MOL002879 MOL003180 MOL003344 MOL000359 MOL000449 MOL004746 MOL000057 MOL005736 MOL005840 MOL005842 MOL005846 MOL000612 MOL000675 |
Methyl linoleate Dinatin ZINC03860434 DNOP Diop Widdrene β-amyrin acetate Sitosterol Stigmasterol (E,7S,11R)-3,7,11,15-tetramethylhexadec-2-en-1-ol DIBP Cyperene PANA Pectolinarigenin Pectolinarin (-)-Alpha-cedrene Oleic acid |
41.93 30.97 43.59 40.59 43.59 53.81 42.06 36.91 43.83 49.63 51.87 50.35 41.17 47.62 43.08 55.56 33.13 |
0.17 0.27 0.35 0.4 0.39 0.12 0.74 0.75 0.76 0.13 0.13 0.11 0.13 0.3 0.65 0.1 0.14 |
| MOL000635 | Vanillin | 51.99 | - |
| MOL000676 | DBP | 64.54 | 0.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





