Introduction
Clostridium difficile is a toxin producing gram positive anaerobic bacteria. It is the leading cause of hospital/healthcare-acquired diarrhea all over the world and antibiotic-associated diarrhea as well [
1,
2]. C. Difficile is spore forming bacteria. Its spores can enter the host through inhalation and begin to germinate in the gut [
3]. Gut microflora is highly susceptible to antibiotics. Once disrupted by the antibiotics, C. difficile begins to colonize the colon. Once colonized it can cause mild diarrhea or more serious manifestations such as pseudomonas colitis or toxic megacolon which is the major cause of increasing mortality and morbidity associated with C. difficile infection [
4,
5]. C difficile is rapidly spreading all over the world. Due to its high prevalence and severe complications, the diagnosis and management are becoming a bit of a challenge. C. Difficile has an enzyme called Glutamate dehydrogenase which plays an important role in the metabolism of glutamate, an amino acid. The NAD-specific GDH enzyme in C. difficile causes the oxidative deamination of glutamate to create ammonia and a-ketoglutarate [
6]. C. difficile also produces toxins which are toxin A and toxin B. It accounts for the major virulence factor causing CDI. The latex agglutination test was first thought to detect the C. difficile toxins but later it was found that it detects the enzyme GDH [
7]. The diagnostic tests today are based on the finding of toxin A and toxin B from the stool samples of the patient suffering from Clostridium difficile Infection (CDI)[
8].
Toxigenic C. difficile can be definitively diagnosed only by microbiologic testing. Nowadays laboratories struggle to produce tests that are reliable as well as fast and affordable. The use of a low-sensitivity test can compromise the efficacy of control measures if infected individuals are kept without the necessary isolation and are not properly treated [
9,
10].
The commercially available enzyme immunoassay method to detect the enzyme GDH involves two steps [
3]. The first step involves the detection of GDH. The GDH enzyme is found in all strains of C.Difficile regardless of the strain’s toxigenicity. When this is positive the second step is used to detect the toxins which are responsible for the actual symptoms of the C. Difficile infection [
11].
This meta-analysis aims to critically review and compare the diagnostic modalities utilized for the detection of GDH in clinical samples, evaluating their sensitivity, specificity, cost-effectiveness, and practical utility in routine clinical practice. By comprehensively assessing the strengths and limitations of these diagnostic approaches, this research endeavors to provide insights into optimizing CDI diagnosis and guiding evidence-based decision-making in healthcare settings. Eventually, this enhances the accuracy and efficiency of GDH detection which can facilitate early intervention, improve patient outcomes, and reduce the transmission of C. difficile within healthcare facilities.
Methodology
Data Collection
For the collection of the data, a search was done by two individuals using PubMed, Google Scholar, and Cochrane Library databases for all relevant literature. Full - Text Articles written only in English were considered.
The medical subject headings (MeSH) and keywords “_”, “_”, “_”, and “_” were used. References, reviews, and meta-analyses were scanned for additional articles.
Inclusion and Exclusion Criteria
Screening of titles and abstracts was carried out, wherein duplicates and citations were filtered out of consideration. Reviewing references of papers found to be relevant was carried out further for underlying additional articles, following which papers with detailed and reliable patient information and results with a strong statistical base were selected.
We searched for papers showing more accurate diagnoses, wherein the modality considered was Glutamate Dehydrogenase enzyme for Clostridium difficile.
The inclusion criteria were as follows: (1) studies that provided information about the accurate diagnosis of C. difficile using GDH; (2) studies published in English; (3) Studies comparing GDH to the Gold Standard (PCR/Culture essay) as a diagnostic modality for cases of C. difficile.
The exclusion criteria were: (1) articles that were not full text, (2) unpublished articles, and (3) articles in other languages.
Assessment of Study Quality
Using the QualSyst tool, two writers independently assessed the caliber of each included study. This test consists of 10 questions, each with a score between 0 and 2, with 20 being the maximum possible overall score. Two authors independently rated each article based on the above criteria. The weighted Cohen’s Kappa coefficient (K) was used to determine the interobserver agreement for the study selection. For evaluating the bias risk of the studies, we also employed the Cochrane tool. No assumptions were made about any missing or unclear information. Neither the collection nor the review of data were funded.
Statistical Analysis
The statistical software packages RevMan (Review Manager, version 5.3), SPSS (Statistical Package for the Social Sciences, version 20), Google Sheets, and Excel in Stata 14 were used to perform the statistical analyses. The data was obtained and entered into analytic software [21]. Fixed- or random-effects models were used to estimate Sensitivity, Specificity, positive predictive value (PPV), diagnostic odds ratios (DOR), and relative risk (RR) with 95 percent confidence intervals to examine critical clinical outcomes (CIs). Diagnosis accuracy and Younden Index were calculated for each result. Individual study sensitivity and specificity were plotted on Forest plots and in the receiver operating characteristic (ROC) curve. The forest plot and Fagan’s Nomogram were used to illustrate the sensitivity and specificity of different papers.
Bias Study
The risk of bias was evaluated by using QUADAS-2 analysis. This tool includes 4 domains as Patient selection, Index test, Reference standard, Flow of the patients, and Timing of the Index tests.
Results
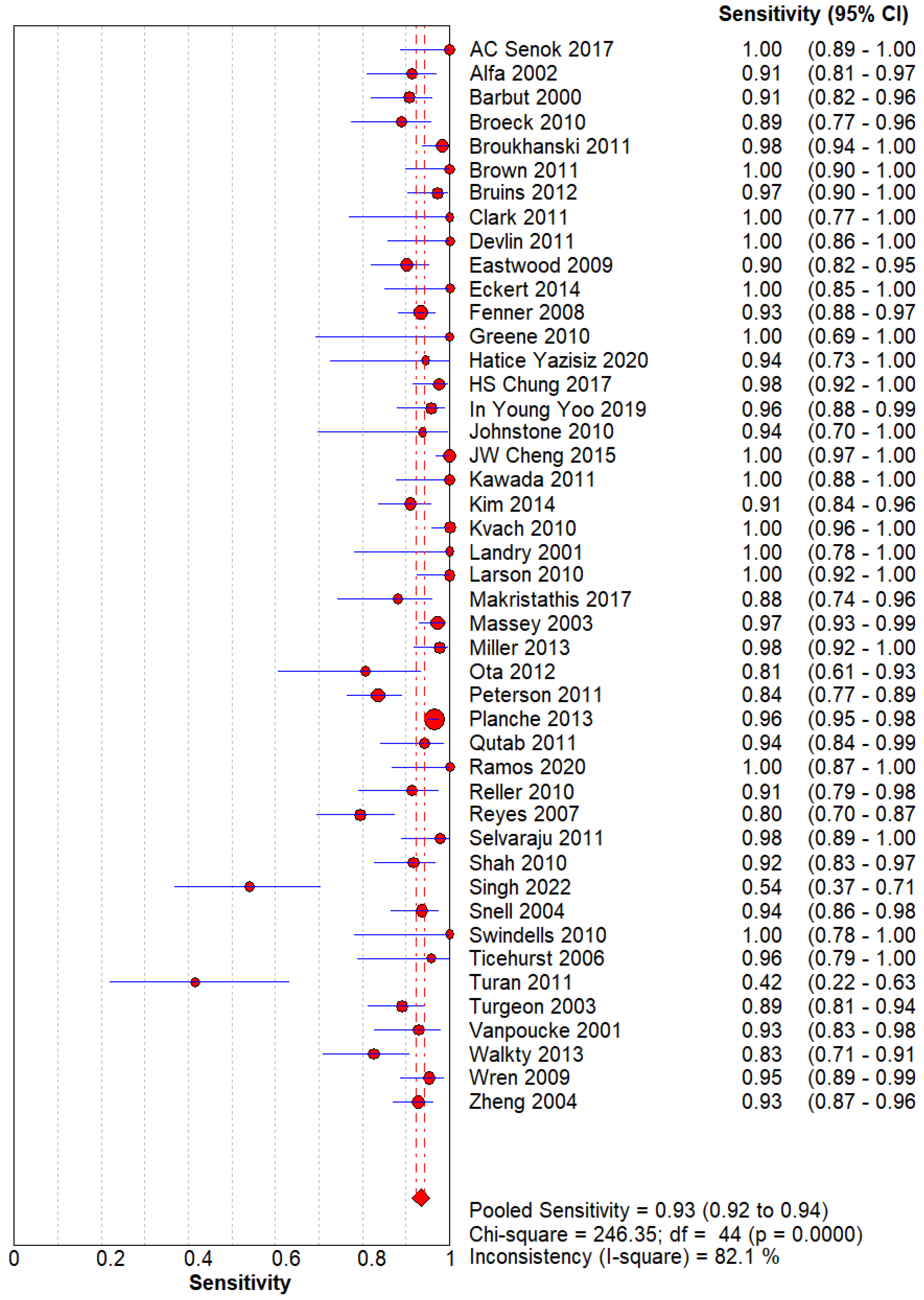
Figure 1.
Sensitivity of GDH for C. difficle. The pooled sensitivity is 0.93(0.92-0.94) with a high heterogeneity across research.
Figure 1.
Sensitivity of GDH for C. difficle. The pooled sensitivity is 0.93(0.92-0.94) with a high heterogeneity across research.
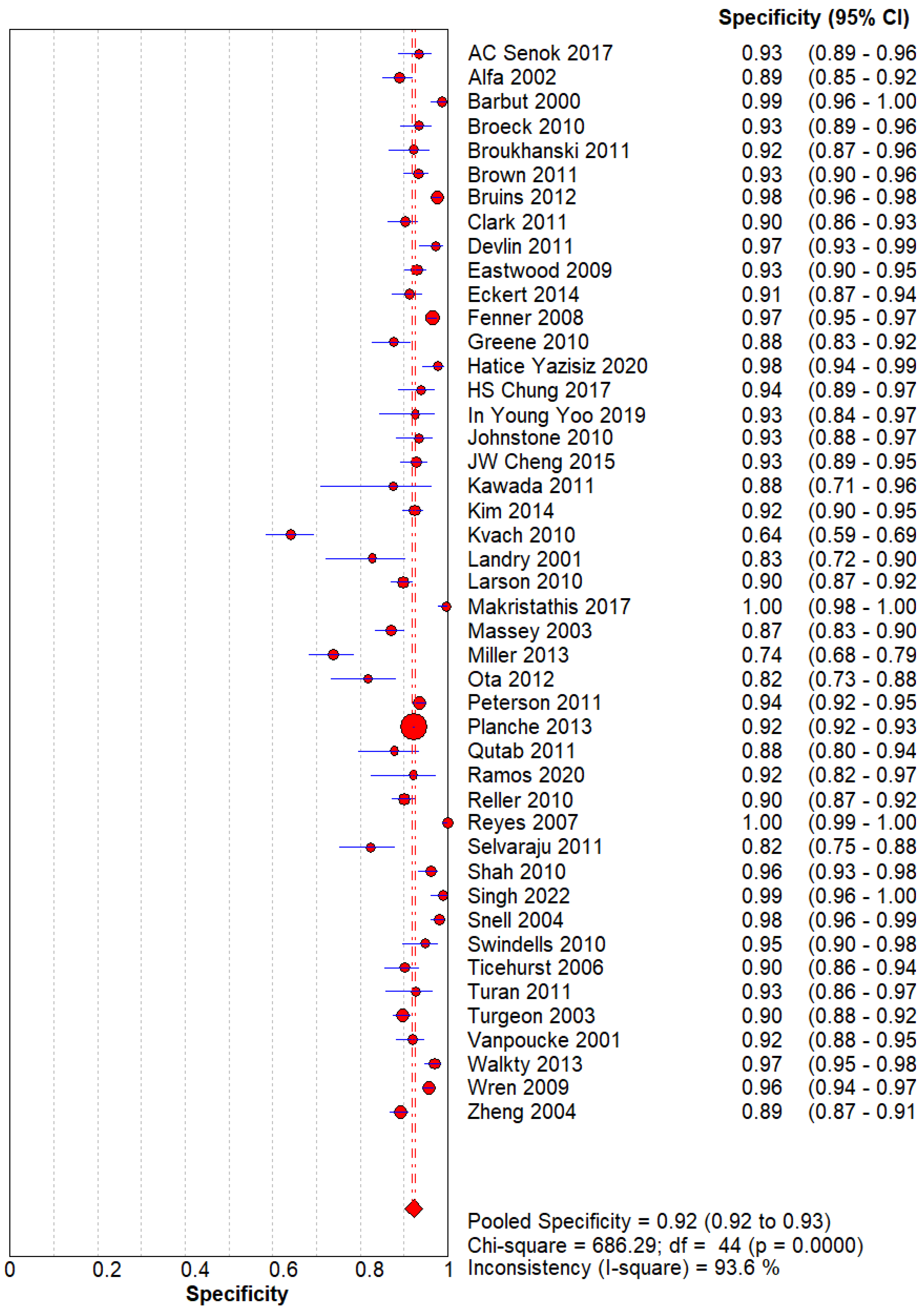
Figure 2.
– Specificity of GDH for C. difficle. The pooled specificity is 0.92(0.92-0.93).
Figure 2.
– Specificity of GDH for C. difficle. The pooled specificity is 0.92(0.92-0.93).
The Sensitivity and Specificity are evident that GDH can be used as a screening and diagnostic modality for Clostridium difficile.
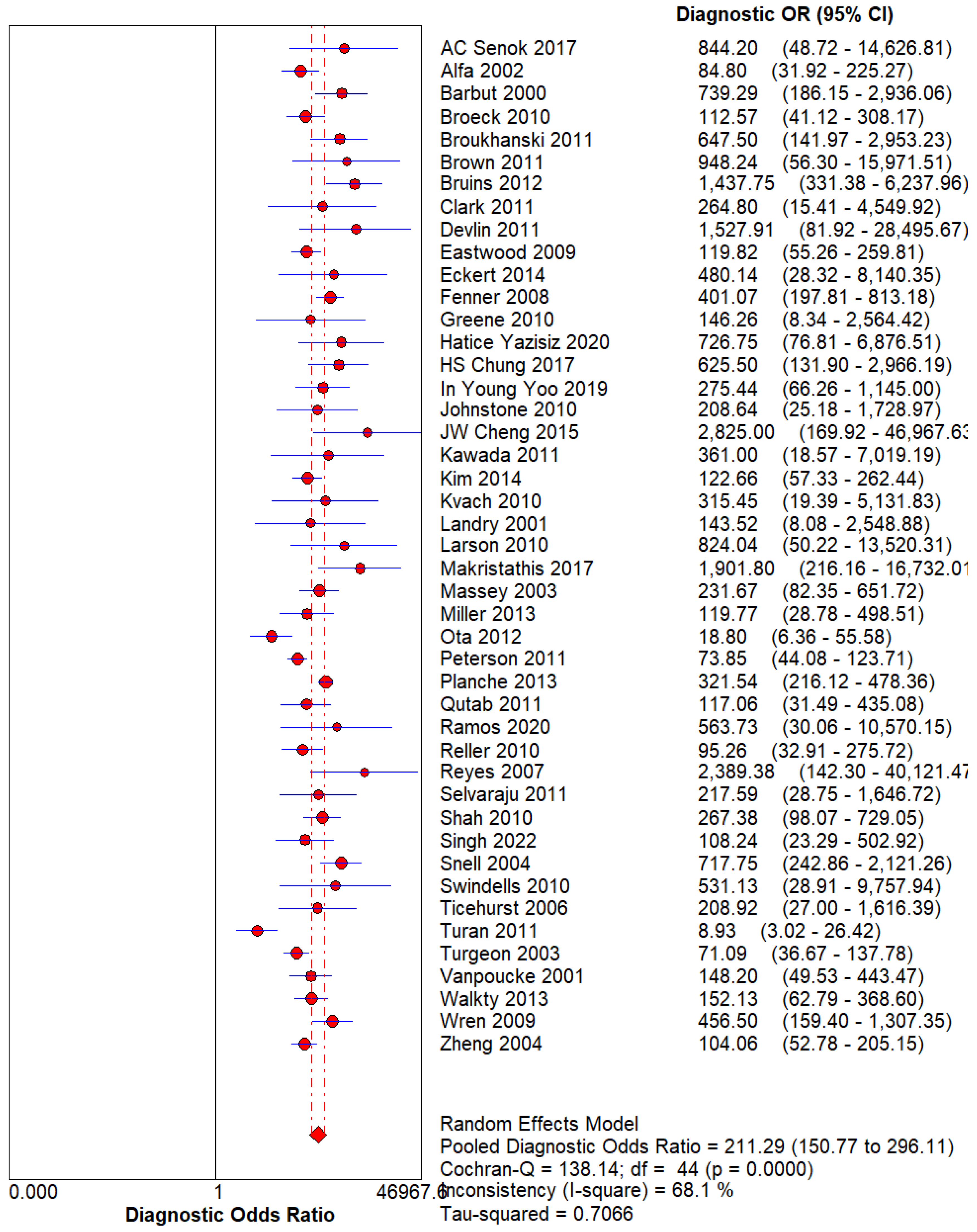
Figure 3.
– Diagnostic Odds Ratio of 18-FDG PET. Using the Random effect model for Heterogeneity higher than 50%, The DOR for 18-FDG PET can be calculated at 30.98(21.18 – 45.31).
Figure 3.
– Diagnostic Odds Ratio of 18-FDG PET. Using the Random effect model for Heterogeneity higher than 50%, The DOR for 18-FDG PET can be calculated at 30.98(21.18 – 45.31).
Figure 4.
– SROC curve. With an AUC (Area Under the Curve) of 0.9794, GDH demonstrates its brilliant diagnostic prowess. Index Q* at 0.9361 is exceptional.
Figure 4.
– SROC curve. With an AUC (Area Under the Curve) of 0.9794, GDH demonstrates its brilliant diagnostic prowess. Index Q* at 0.9361 is exceptional.
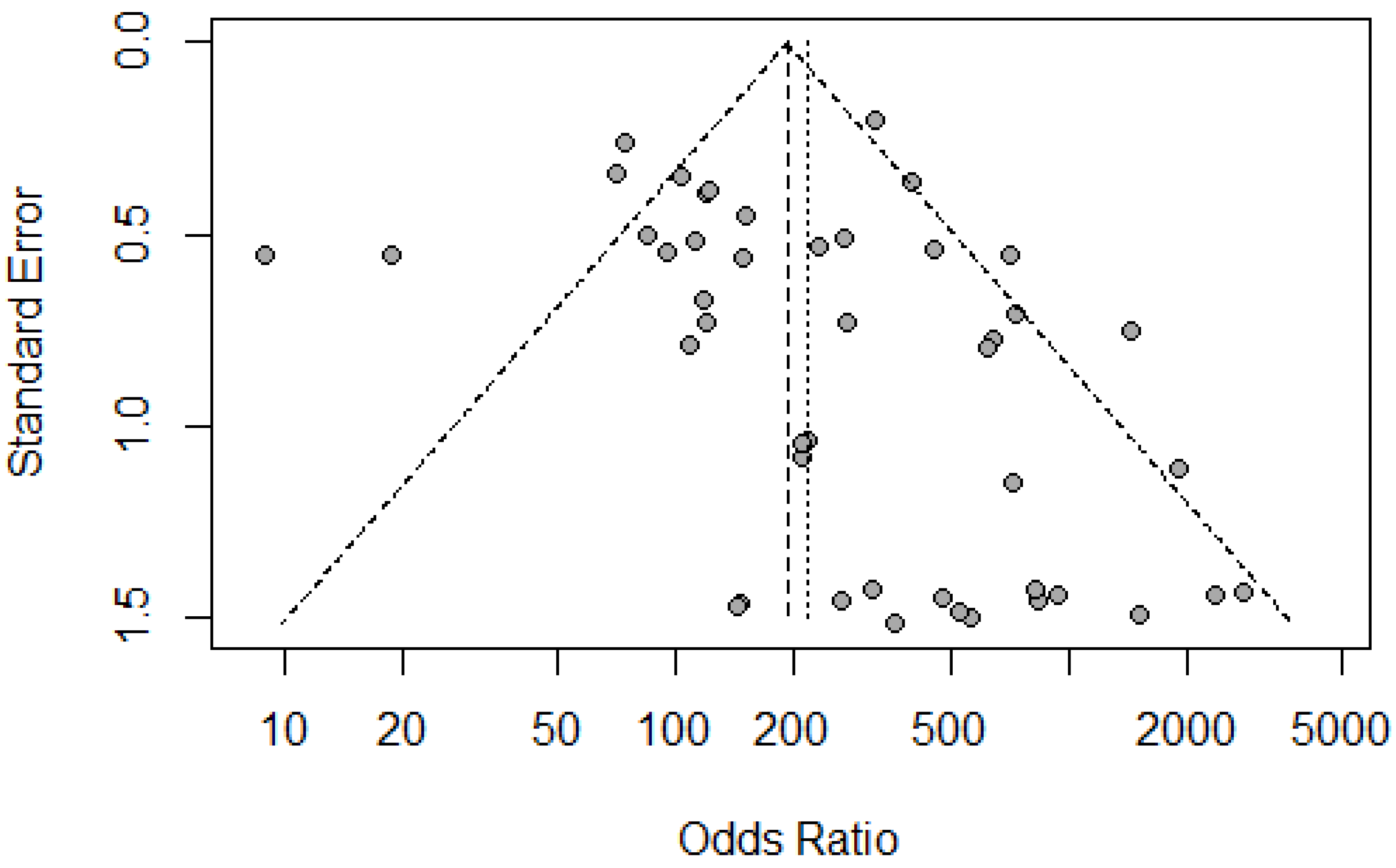
Figure 5.
– Funnel Plot for Bias. Individual dots represent different research papers which are plotted on a graph of the Odds ratio vs. the Standard Error.
Figure 5.
– Funnel Plot for Bias. Individual dots represent different research papers which are plotted on a graph of the Odds ratio vs. the Standard Error.
Discussion:
This meta-analysis is aimed at assessing the diagnostic accuracy of glutamate dehydrogenase (GDH) for Clostridium difficile infection (CDI). Our findings demonstrate that GDH testing is a highly effective diagnostic tool, with pooled sensitivity and specificity values of 93% and 92%, respectively. These results are consistent with the previous studies which describe the importance of GDH in the diagnosis of Clostridium Difficile Infection [
12,
13,
14,
15].
The high sensitivity of GDH (93%) highlights its capability to detect CDI which is important for the diagnosis in clinical settings because missing an infection could lead to severe outcomes. This sensitivity supports the findings from a recent meta-analysis that reported GDH sensitivity exceeding 90% when compared to traditional diagnostic methods such as cell culture neutralization (CCNA) or toxigenic culture [
11]. Such high sensitivity reduces the risk of false negatives, ensuring that patients with CDI are identified and treated as early as possible [
16].
Conversely, the specificity of GDH (92%) highlights its reliability in identifying non-infected individuals, thus reducing the incidence of false positives. This balance of high sensitivity and specificity tells that GDH is a valuable tool for both screening and definitive diagnosis of CDI. The diagnostic odds ratio (DOR) for glutamate dehydrogenase (GDH) in our meta-analysis was calculated at 30.98, which indicates a strong association between positive GDH test results and the presence of Clostridium difficile infection (CDI). The DOR value suggests that GDH is highly effective in identifying CDI cases, which is consistent with its excellent area under the curve (AUC) of 0.9794. An AUC nearing 1.0 indicates that GDH is highly efficient in differentiating between infected and non-infected patients. This states its potential utility in clinical settings.
There are currently different techniques available for diagnosing CDI based on various assay principles. These techniques include tissue culture neutralization (TCN), immunochromatographic test devices, enzyme immunoassay (EIA), nucleic acid amplification test (NAAT) for toxin gene detection, and the rapid detection of GDH or CD toxin A/B by toxigenic CD culture [
13,
15,
17]. Compared to the identification of CD toxin A/B in stool samples, which is specific but has low sensitivity and a significant percentage of false negatives [
18], GDH assay has a high negative predictive value and sensitivity [
19]. Because of its high accuracy in diagnosing CDI patients, outcomes can be clinically improved because of its faster diagnosis and management. The healthcare cost can also be reduced.
However, some limitations need to be considered. Variability in GDH test performance across different studies may be affected by factors such as assay quality, patient demographics, and study design [
20]. To address these limitations, future research should focus on further validating GDH testing in diverse clinical environments and exploring its association with other diagnostic methods. Large-scale studies should aim to improve GDH testing protocols and address any limitations observed in the current meta-analysis. GDH testing implementation can offer practical advantages such as its being non-invasive and its efficiency in detecting CDI [
16]. However, variability in test performance across different environments must be managed to ensure consistent and accurate results.
In summary, GDH testing has shown to be a promising diagnostic tool for detecting CDI, with high sensitivity and specificity supporting its inclusion in diagnostic protocols. The excellent AUC and high DOR further emphasize its role in improving diagnostic accuracy. Continued research and clinical validation are necessary to fully establish GDH’s place in the diagnostic algorithm and to ensure its effective implementation across various healthcare settings.
Conclusion
In conclusion, this meta-analysis reinforces the diagnostic value of glutamate dehydrogenase (GDH) testing for Clostridium difficile infection (CDI), demonstrating a high pooled sensitivity of 93% and specificity of 92%. These findings align with previous research, underscoring GDH’s ability to accurately identify CDI cases and reduce the risk of false negatives, thereby facilitating timely treatment and improving clinical outcomes. The strong diagnostic odds ratio and excellent area under the curve further attest to GDH’s efficacy in differentiating between infected and non-infected individuals.
While the advantages of GDH testing are evident, including its non-invasive nature and potential to lower healthcare costs, variability in performance across different studies necessitates caution. Future research should focus on validating GDH testing in diverse clinical settings, refining protocols, and exploring its integration with other diagnostic methods to ensure consistent accuracy. Ultimately, GDH testing holds significant promise as a critical component in CDI diagnostic protocols, and ongoing investigation will be essential to fully realize its potential across healthcare environments.
References
- Girinathan BP, Braun SE, Govind R. Clostridium difficile glutamate dehydrogenase is a secreted enzyme that confers resistance to H2O2. Microbiology. 2014;160(Pt 1):47-55. [CrossRef]
- Czepiel J, Dróżdż M, Pituch H, et al. Clostridium difficile infection: review. Eur J Clin Microbiol Infect Dis. 2019;38(7):1211-1221. [CrossRef]
- Girinathan BP, Braun S, Sirigireddy AR, Lopez JE, Govind R. Importance of Glutamate Dehydrogenase (GDH) in Clostridium difficile Colonization In Vivo. PLOS ONE. 2016;11(7):e0160107. [CrossRef]
- Bartlett JG, Moon N, Chang TW, Taylor N, Onderdonk AB. Role of Clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology. 1978;75(5):778-782. [CrossRef]
- Larson HE, Price AB, Honour P, Borriello SP. CLOSTRIDIUM DIFFICILE AND THE ÆTIOLOGY OF PSEUDOMEMBRANOUS COLITIS. The Lancet. 1978;311(8073):1063-1066. [CrossRef]
- Anderson BM, Anderson CD, Vantassell RL, Lyerly DM, Wilkins TD. Purification and Characterization of Clostridium difficile Glutamate Dehydrogenase. Arch Biochem Biophys. 1993;300(1):483-488. [CrossRef]
- Lyerly DM, Barroso LA, Wilkins TD. Identification of the latex test-reactive protein of Clostridium difficile as glutamate dehydrogenase. J Clin Microbiol. 1991;29(11):2639-2642. [CrossRef]
- McCollum DL, Rodriguez JM. Detection, Treatment, and Prevention of Clostridium difficile Infection. Clin Gastroenterol Hepatol. 2012;10(6):581-592. [CrossRef]
- Sydnor ERM, Lenhart A, Trollinger B, et al. Antimicrobial Prescribing Practices in Response to Different Clostridium difficile Diagnostic Methodologies. Infect Control Hosp Epidemiol. 2011;32(11):1133-1136. [CrossRef]
- Silva ROS, Neves MS, Ribeiro MG, Palhares MS, Albuquerque Maranhão R de P, Faria Lobato FC. Evaluation of Three Enzyme Immunoassays for Diagnosis of Clostridium difficile–Associated Diarrhea in Foals. J Equine Vet Sci. 2014;34(8):1032-1035. [CrossRef]
- Shetty N, Wren MWD, Coen PG. The role of glutamate dehydrogenase for the detection of Clostridium difficile in faecal samples: a meta-analysis. J Hosp Infect. 2011;77(1):1-6. [CrossRef]
- Eastwood K, Else P, Charlett A, Wilcox M. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol. 2009;47(10):3211-3217. [CrossRef]
- Kim H, Kim WH, Kim M, Jeong SH, Lee K. Evaluation of a rapid membrane enzyme immunoassay for the simultaneous detection of glutamate dehydrogenase and toxin for the diagnosis of Clostridium difficile infection. Ann Lab Med. 2014;34(3):235-239. [CrossRef]
- Sharp SE, Ruden LO, Pohl JC, Hatcher PA, Jayne LM, Ivie WM. Evaluation of the C.Diff Quik Chek Complete Assay, a new glutamate dehydrogenase and A/B toxin combination lateral flow assay for use in rapid, simple diagnosis of clostridium difficile disease. J Clin Microbiol. 2010;48(6):2082-2086. [CrossRef]
- Kawada M, Annaka M, Kato H, et al. Evaluation of a simultaneous detection kit for the glutamate dehydrogenase antigen and toxin A/B in feces for diagnosis of Clostridium difficile infection. J Infect Chemother Off J Jpn Soc Chemother. 2011;17(6):807-811. [CrossRef]
- Cheng JW, Xiao M, Kudinha T, et al. The Role of Glutamate Dehydrogenase (GDH) Testing Assay in the Diagnosis of Clostridium difficile Infections: A High Sensitive Screening Test and an Essential Step in the Proposed Laboratory Diagnosis Workflow for Developing Countries like China. PLoS ONE. 2015;10(12):e0144604. [CrossRef]
- Vanpoucke H, De Baere T, Claeys G, Vaneechoutte M, Verschraegen G. Evaluation of six commercial assays for the rapid detection of Clostridium difficile toxin and/or antigen in stool specimens. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2001;7(2):55-64. [CrossRef]
- Kvach EJ, Ferguson D, Riska PF, Landry ML. Comparison of BD GeneOhm Cdiff real-time PCR assay with a two-step algorithm and a toxin A/B enzyme-linked immunosorbent assay for diagnosis of toxigenic Clostridium difficile infection. J Clin Microbiol. 2010;48(1):109-114. [CrossRef]
- Wren MWD, Kinson R, Sivapalan M, Shemko M, Shetty NR. Detection of Clostridium difficile infection: a suggested laboratory diagnostic algorithm. Br J Biomed Sci. 2009;66(4):175-179. [CrossRef]
- Planche TD, Davies KA, Coen PG, et al. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. Lancet Infect Dis. 2013;13(11):936-945. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).