Submitted:
24 September 2024
Posted:
25 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
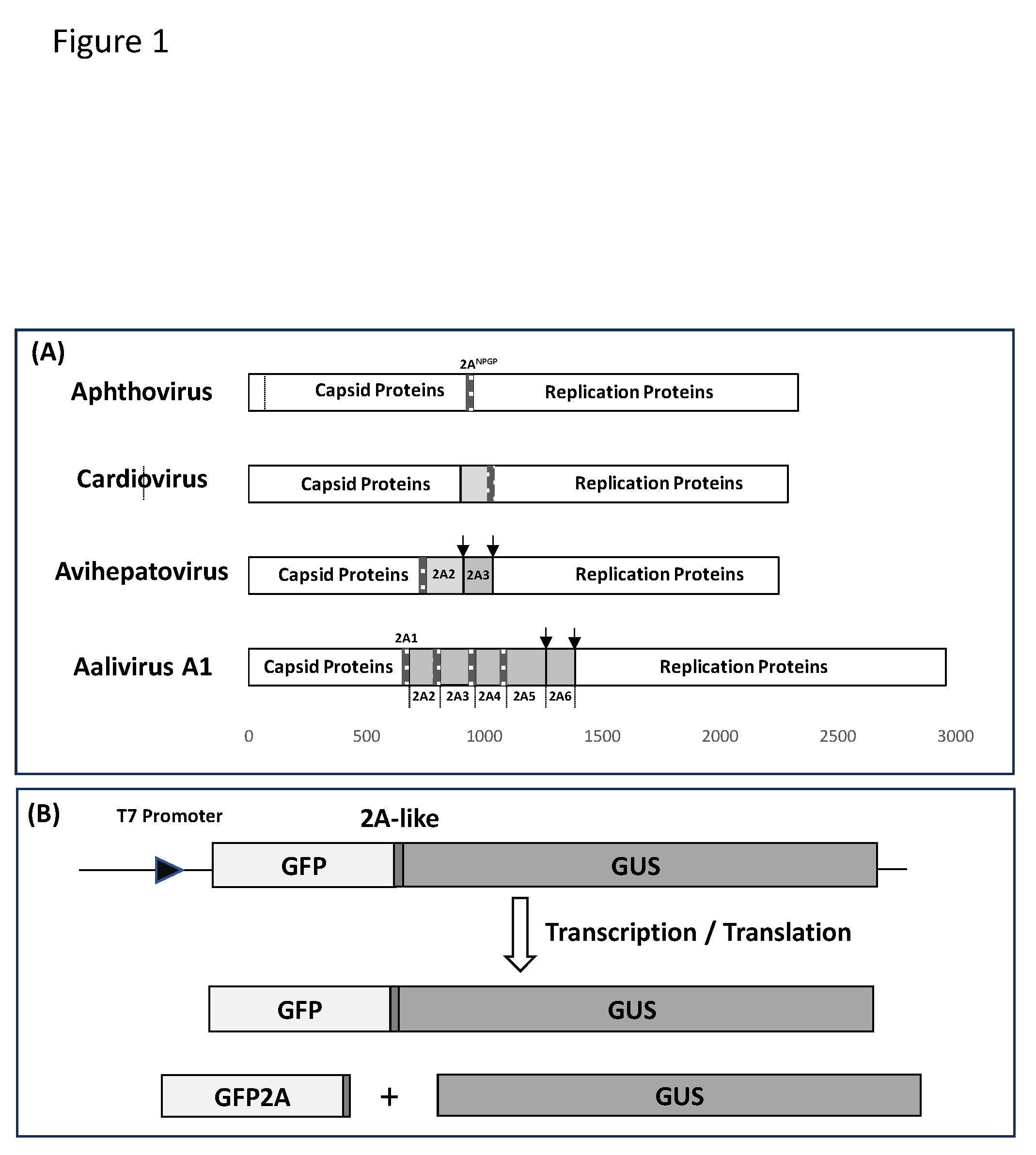
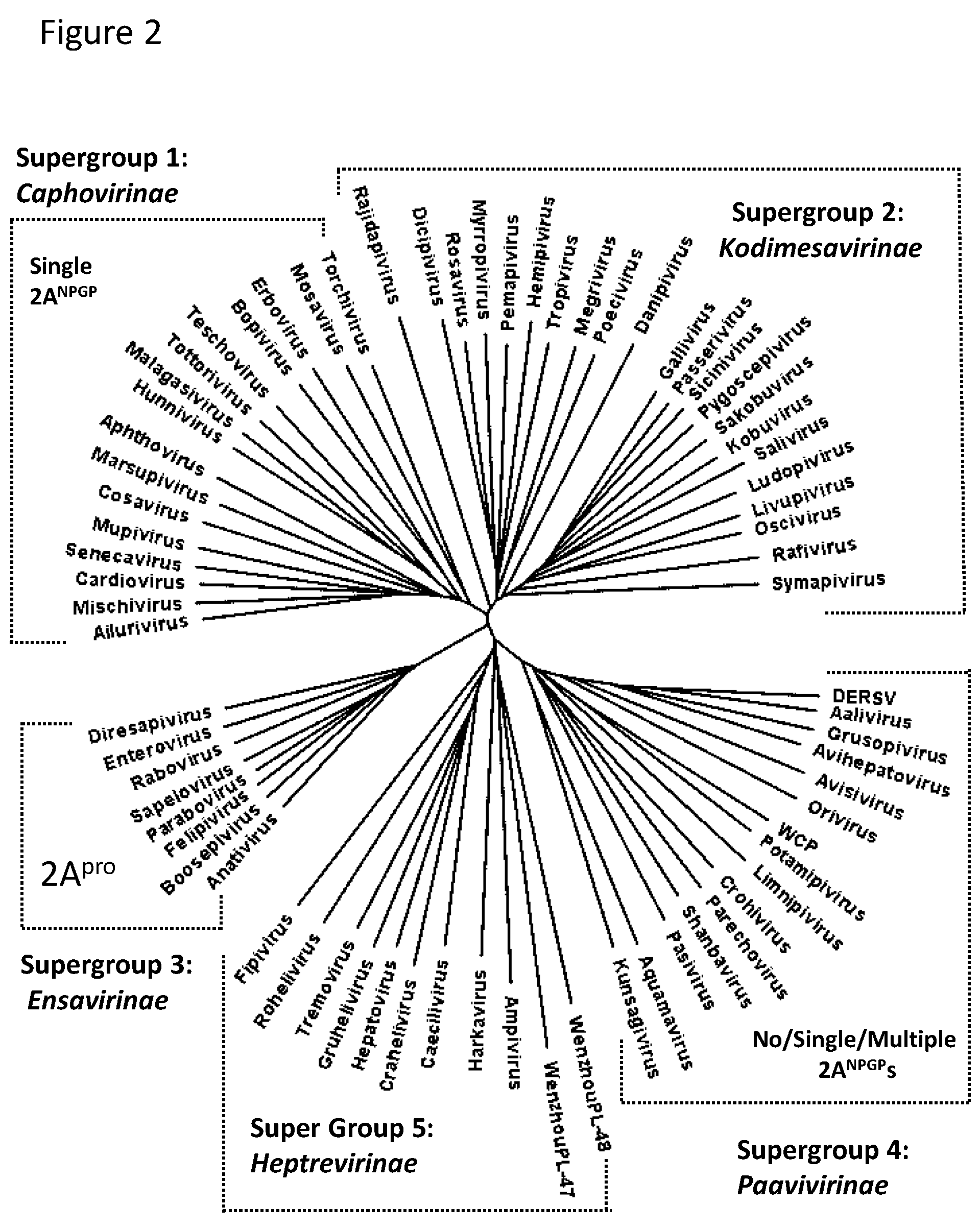
2.1. Bioinformatic Analyses
2.2. Cloning of 2ANPGP Sequences into pSTA1
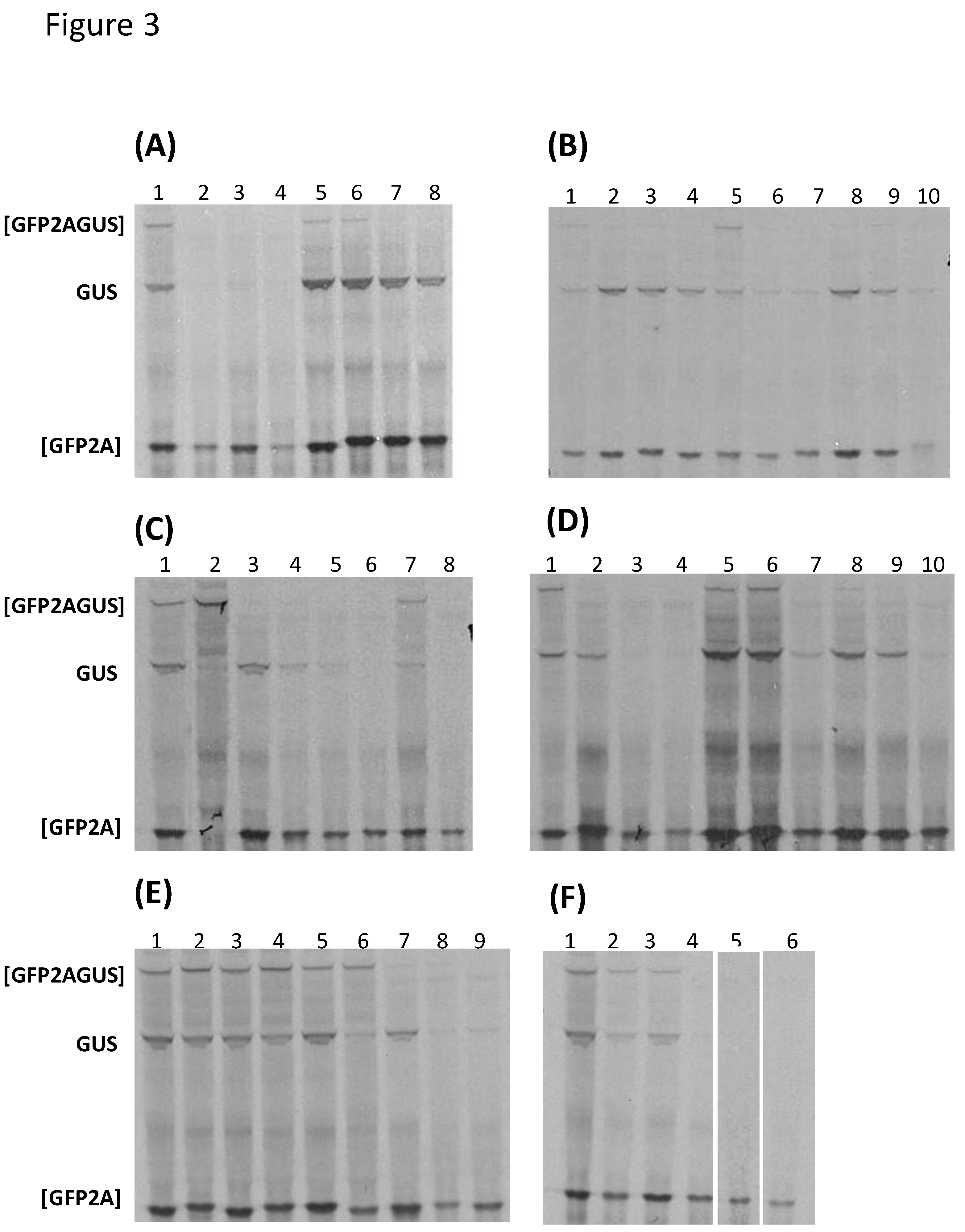
2.3. In Vitro Transcription/Translation
3. Results
3.1. Bioinformatic Analyses
4. Discussion
4.1. XX
4.2. Implications for our Model of Ribosome Skipping
4.3. Biotechnological and Biomedical Applications
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
References
- Martĩnez-Salas, E.; Ryan, M.D. Translation and protein processing. In The Picornaviruses Ehrenfeld, E., Domingo, E., Roos, R.P., Eds.; ASM Press: Washington, DC, USA, 2010; pp. 141–161. [Google Scholar]
- Yu, S.F.; Benton, P.; Bovee, M.; Sessions, J.; Lloyd, R.E. Defective RNA replication by Poliovirus mutants deficient in 2A protease cleavage activity. J Virol 1995, 69, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, H.; Nicklin, M.J.; Murray, M.G.; Anderson, C.W.; Dunn, J.J.; Studier, F.W.; Wimmer, E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell 1986, 45, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-Y.; Liu, Y-N.; Wang, W.; Kao, F.-J.; Kung, S.-H. In vivo dynamics of enterovirus protease revealed by fluorescence resonance emission transfer (FRET) based on a novel FRET pair. Biochem Biophys Res Commun 2007, 353, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Lina, Y.; Zhao, J.; Yu, J.; Chen, Y.; Lin, M.C.; Kung, H.-F.; He, M.-L. Enterovirus 71 disrupts Interferon signaling by reducing the level of Interferon Receptor 1. J Virol 2012, 86, 3767–3776. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Escriou, N.; Chao, S.-F.; Girard, M.; Lemon, S.M.; Wychowski, C. Identification and site-directed mutagenesis of the primary (2A/2B) cleavage site of the Hepatitis A Virus polyprotein: functional impact on the infectivity of HAV RNA transcripts. Virology 1995, 213, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Palmenberg, A.; Neubauer, D.; Skern, T. Genome organization and encoded proteins. In The Picornaviruses Ehrenfeld, E., Domingo, E., Roos, R.P., Eds.; ASM Press: Washington, DC, USA, 2010; pp. 3–17. [Google Scholar]
- Ryan, M.D.; Donnelly, M.L.L.; Lewis, A.; Mehrotra, A.P.; Wilkie, J.; Gani, D. A model for nonstoichiometric, co-translational protein scission in eukaroytic ribosomes. Bioorg Chem 1999, 27, 55–79. [Google Scholar] [CrossRef]
- Donnelly, M.L.L.; Luke, G.A.; Mehotra, A.; Li, X.; Hughes, L.E.; Gani, D.; Ryan, M.D. Analysis of the aphthovirus 2A/2B polyprotein “cleavage” mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal “skip”. J Gen Virol 2001, 82, 1013–1025. [Google Scholar] [CrossRef]
- Luke, G.A.; de Felipe, P.; Lukashev, A.; Kallioinen, S.E.; Bruno, E.A.; Ryan, M.D. Occurrence, function and evolutionary origins of “2A-like” sequences in virus genomes. J Gen Virol 2008, 89, 1036–1042. [Google Scholar] [CrossRef]
- de Lima, J.G.S.; Teixeira, D.G.; Freitas, T.T.; Lima, J.P.M.S. Evolutionary origin of 2A-like sequences in Totiviridae genomes. Virus Res 2019, 259, 1–9. [Google Scholar] [CrossRef]
- de Lima, J.G.S.; Lanza, D.C.F. 2A and 2A-like sequences: Distribution in different virus species and applications in biotechnology. Viruses 2021, 13, 2160. [Google Scholar] [CrossRef]
- Donnelly, M.L.L.; Hughes, L.E.; Luke, G.A.; Mendoza, H.; ten Dam, E.; Gani, D.; Ryan, M.D. The “cleavage” activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring “2A-like” sequences. J Gen Virol 2001, 82, 1027–1041. [Google Scholar] [CrossRef] [PubMed]
- Heras, S.R.; Thomas, M.C.; García-Canadas, M.; de Felipe, P.; García-Perez, J.L.; Ryan, M.D.; Lopez, M.C. L1Tc non-LTR retrotransposons from Trypanosoma cruzi contain functional viral-like self-cleaving 2A sequence in frame with active proteins they encode. Cell Mol Life Sci 2006, 63, 1449–1460. [Google Scholar] [CrossRef] [PubMed]
- Odon, V.; Luke, G.A.; Roulston, C.; de Felipe, P.; Ruan, L.; Escuin-Ordinas, H.; Brown, J.D.; Ryan, M.D.; Sukhodub, A. APE-type non-LTR retrotransposons of multicellular organisms encode virus-like 2A oligopeptide sequences, which mediate translational recoding during protein synthesis. Mol Biol Evol 2013, 30, 1955–1965. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.D.; Ryan, M.D. Ribosome “Skipping”: “Stop-Carry On” or “Stop-Go” Translation. In Recoding: Expansion of Decoding Rules Enriches Gene Expression Atkins, J.F., Gesteland, R.F., Eds.; Springer: New York, NY, USA, 2010; pp. 101–121. [Google Scholar]
- Roulston, C.; Luke, G.A.; de Felipe, P. , Ruan, L.; Cope, J.; Nicholson, J.; Sukhodub, A.; Tilsner, J.; Ryan, M.D. “2A-Like” signal sequences mediating translational recoding: A novel form of dual protein targeting. Traffic 2016, 17, 923–939. [Google Scholar] [CrossRef] [PubMed]
- Minskaia, E.; Nicholson, J.; Ryan, M.D. Optimisation of the foot-and-mouth disease virus 2A co-expression system for biomedical applications. BMC Biotechnology 2013, 13, 67. [Google Scholar] [CrossRef]
- Donnelly, M.L.L.; Gani, D.; Flint, M.; Monaghan, S.; Ryan, M.D. The cleavage activities of aphthovirus and cardiovirus 2A proteins. J Gen Virol 1997, 78, 13–21. [Google Scholar] [CrossRef]
- Loughran, G.; Firth, A.E.; Atkins, J.F. Ribosomal frameshifting into an overlapping gene in the 2B-encoding region of the cardiovirus genome. Proc. Natl Acad. Sci. USA 2011, 108, E1111–E1119. [Google Scholar] [CrossRef]
- Napthine, S.; Ling, R.; Finch, L.K.; Jones, J.D.; Bell, S.; Brierley, I.; Firth, A.E. Protein-directed ribosomal frameshifting temporally regulates gene expression. Nat Commun. 2017, 8, 15582. [Google Scholar] [CrossRef]
- Napthine, S.; Bell, S.; Hill, C.H.; Brierley, I.; Firth, A.E. Characterization of the stimulators of protein-directed ribosomal frameshifting in Theiler's murine encephalomyelitis virus. Nucleic Acids Res 2019, 47, 8207–8223. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Choi, G.K.Y.; Huang, Y.; Teng, J.L.L.; Tsoi, H.-W.; Tse, H.; Yeung, M.L.; Chan, K.-H.; Jin, D.-Y.; Yuen, K.-Y. Natural occurrence and characterization of two internal ribosome entry site elements in a novel virus, canine Picodicistrovirus, in the picornavirus-like superfamily. J Virol 2012, 86, 2797–2808. [Google Scholar] [CrossRef]
- Zell, R. Picornaviridae—the ever-growing virus family. Arch Virol. 2018, 163, 299–317. [Google Scholar] [CrossRef] [PubMed]
- Zell, R.; Knowles, N.J.; Simmonds, P. A proposed division of the family Picornaviridae into subfamilies based on phylogenetic relationships and functional genomic organization. Arch Virol. 2021, 166, 2927–2935. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zeng, Q.; Wang, M.; Cheng, A.; Pan, K.; Zhu, D.; Liu, M.; Jia, R.; Yang, Q.; Wu, Y.; Chen, S.; Zhao, X.; Zhang, S.; Liu, Y.; Yu, Y.; Zhang, L. DHAV-1 2A1 peptide – A newly discovered co-expression tool that mediates the ribosomal “Skipping” function. Front. Microbiol 2018, 9, 2727. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Ou, X.; Zhu, D.; Ma, G.; Cheng, A.; Wang, M.; Chen, S.; Jia, R.; Liu, M.; Sun, K.; Yang, Q.; Wu, Y.; Chen, X. The 2A2 protein of Duck hepatitis A virus type 1 induces apoptosis in primary cell culture. Virus Genes 2016, 52, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Hughes, P.J.; Stanway, G. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J Gen Virol. 2000, 81, 201–207. [Google Scholar] [CrossRef]
- Yang, X.; Cheng, A.; Wang, M.; Jia, R.; Sun, K.; Pan, K.; Yang, Q.; Wu, Y.; Zhu, D.; Chen, S.; Liu, M.; Zhao, X.-X.; Chen, X. Structures and corresponding functions of five types of picornaviral 2A proteins. Front Microbiol. 2017, 8, 1373. [Google Scholar] [CrossRef]
- Wang, X.; Liu, N.; Wang, F.; Ning, K.; Li, Y.; Zhang, D. Genetic characterization of a novel duck-origin picornavirus with six 2A proteins. J Gen Virol. 2014, 95, 1289–1296. [Google Scholar] [CrossRef]
- Su, X.; Shuo, D.; Luo, Y.; Pan, X.; Yan, D.; Li, X.; Lin, W.; Huang, D.; Yang, J.; Yuan, C.; Liu, Q.; Teng, Q.; Li, Z. An emerging duck egg-reducing syndrome caused by a novel picornavirus containing seven putative 2A peptides. Viruses 2022, 14, 932. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Lukashev, A.N. Recombination among picornaviruses. Rev Med Virol 2010, 20, 327–37. [Google Scholar] [CrossRef]
- Dantas, M.D.A.; Cavalcante, G.H.O.; Oliveira, R.A.C.; Lanza, D.C.F. New insights about ORF1 coding regions support the proposition of a new genus comprising arthropod viruses in the family Totiviridae. Virus Res 2016, 211, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Jia, X.; Gao, Y.; Liu, Z.; Zhang, H.; Tan, Q.; Zhang, X.; Zhou, H.; Li, Y.; Wu, D.; Zhang, Q. Cryo-EM reveals a previously unrecognized structural protein of a dsRNA virus implicated in its extracellular transmission. PLOS Pathog 2021. [Google Scholar] [CrossRef] [PubMed]
- Eyler, D.E.; Wehner, K.A.; Green, R. Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1. J Biol Chem. 2013, 288, 29530–8. [Google Scholar] [CrossRef] [PubMed]
- Pisareva, V.P.; Skabkin, M.A.; Hellen, C.U.; Pestova, T.V.; Pisarev, A.V. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011, 30, 1804–1817. [Google Scholar] [CrossRef] [PubMed]
- Yip, M.C.J.; Shao, S. Detecting and rescuing stalled ribosomes. Trends Biochem Sci. 2021, 46, 731–743. [Google Scholar] [CrossRef]
- Powers, K.T.; Szeto, J.A.; Schaffitzel, C. New insights into no-go, non-stop and nonsense-mediated mRNA decay complexes. Curr Opin Struct Biol. 2020, 65, 110–118. [Google Scholar] [CrossRef]
- Harigaya, Y.; Parker, R. No-go decay: a quality control mechanism for RNA in translation. Wiley Interdiscip Rev RNA. 2010, 1, 132–141. [Google Scholar] [CrossRef]
- Chaplin, P.J.; Camon, E.B.; Villarreal-Ramos, B.; Flint, M.; Ryan, M.D.; Collins, R.A. Production of interleukin-12 as a self-processing 2A polypeptide. J Interferon Cytokine Res. 1999, 19, 235–241. [Google Scholar] [CrossRef]
- https://www.st-andrews.ac.uk/ryanlab/page10.htm.
- Ito, K.; Chiba, S. Arrest peptides: cis-acting modulators of translation. Annu Rev Biochem. 2013, 82, 171–202. [Google Scholar] [CrossRef]
- Ramu, H.; Vázquez-Laslop, N.; Klepacki, D.; Dai, Q.; Piccirilli, J.; Micura, R.; Mankin, AS. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol Cell. 2011, 41, 321–330. [Google Scholar] [CrossRef]
- Doronina, V.A.; Wu, C.; de Felipe, P.; Sachs, M.S.; Ryan, M.D.; Brown, J.D. Site-specific release of nascent chains from ribosomes at a sense codon. Mol Cell Biol. 2008, 28, 4227–4239. [Google Scholar] [CrossRef] [PubMed]
- Machida, K.; Mikami, S.; Masutani, M.; Mishima, K.; Kobayashi, T.; Imataka, H. A translation system reconstituted with human factors proves that processing of encephalomyocarditis virus proteins 2A and 2B occurs in the elongation phase of translation without eukaryotic release factors. J Biol Chem. 2014, 289, 31960–31971. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Ripp, S.; Sayler, G.S.; Close, D.M. Expression of a humanized viral 2A-mediated lux operon efficiently generates autonomous bioluminescence in human cells. PLoS One 2014, 9, e96347. [Google Scholar] [CrossRef] [PubMed]
- Geier, M.; Fauland, P.; Vogl, T.; Glieder, A. Compact multi-enzyme pathways in P. pastoris. Chem Commun (Camb). 2015, 51, 1643–1646. [Google Scholar] [CrossRef]
| Genus | Species | 2ANPGP | Amino Acid Sequence | Acc.# |
|---|---|---|---|---|
| Aphthovirus | FMDV O1K | 2A | VAPVKQTLNFDLLKLAGDVESNPGP | GNNYF |
| Aalivirus (SG4) | AalV- A1 | 2A1 | LLTSEGATNSSLLKLAGDVEENPGP | KJ000696 |
| 2A2 | FEMPYDDPEWDRLLQAGDIEQNPGP | |||
| 2A3 | PIPARPDPQWNNLQQAGDVEMNPGP | |||
| 2A4 | EHFNQTGGWVPDLTQCGDVESNPGP | |||
| AalV-B1 | 2A1 | ATTLQVSEYLKDLTIDGDVESNPGP | MH453803 | |
| 2A2 | LKVKKLEGDYVRDLTQEGVEPNPGP | |||
| 2A3 | SVRVTDAGWVRDLTVDGDVESNPGP | |||
| 2A4 | VFKCHDKCWVDDLTNCGDVEPNPGP | |||
| 2A5 | IFKCHEGCWVEDLTVDGDVESNPGP | |||
| DERSV-AH204 | 2A1 | TSTAQATSYVKDLTIDGDVESNPGP | UYL81882 | |
| 2A2 | KTCREVEGSYVKDLTEEGIEPNPGP | |||
| 2A3 | LLKIGNAAWVRDLTEDGDVEENPGP | |||
| 2A4 | VYNCHESCWNRDLTIDGDVELNPGP | |||
| 2A5 | VFKCHEKCWQKDPTQDGDVEQNPGP | |||
| 2A6 | EFKCHEHCWVRDLTMDGDVEENPGP | |||
| Avisivirus (SG4) | AsV-A1 | 2A1 | EVGAYDEVDHRDILMGGDIEENPGP | KC465954 |
| 2A2 | EMGVFDETDHRDILLGGDIEENPGP | |||
| AsV-B1 | 2A1 | PQFEKERSAHEDVLLGGDVESNPGP | KF979333 | |
| 2A2 | SESVQYLEPQIDICVCGDVERNPGP | |||
| Grusopivirus (SG4) | GrV-A1 | 2A1 | FEKHVKPWRSQEDLSKEGIEPNPGP | KY312544 |
| 2A2 | ITDNRYKETDAKWLSRYGVEMNPGP | |||
| 2A3 | VTQDLYAATNQDQLSNQGIESNPGP | |||
| GrV-C | 2A1 | YFEERSPHPTQKELGQFGVETNPGP | MK443503 | |
| 2A2 | ENNSNYSERDAKHLSRYGIEMNPGP | |||
| 2A3 | CVCTRWSPTMQSELGKYGIEKNPGP | |||
| YC-4 | 2A1 | PERQYFSPKAKEELSKYGIEPNPGP | KY312543 | |
| Kunsagivirus (SG4) | Kuv-C1 | 2A1 | IAAASAQGWQRDLTQDGDVESNPGP | KY670597 |
| 2A2 | LGIVISDSVWQRDLPREGVEENPGP | |||
| 2A3 | SYDPLAPSQWCRDLTCEGIEPNPGP | |||
| Limnipivirus (SG4) | A1 | 2A1 | CKEFVRESDNQELLKCGDVESNPGP | JX134222 |
| 2A2 | WDLSTGWFHFFRLLRSGDVEQNPGP | |||
| B1 | 2A1 | MDVVDDYPFKRDLTRDGDVESNPGP | KF306267 | |
| 2A2 | IDLVQAAYSRMRLLLSGDVEQNPGP | |||
| C1 | 2A1 | KLLEQILAYKRDLTACGDVESNPGP | KF874490 | |
| 2A2 | SRWIHARFARLRLLLSGDVEQNPGP | |||
| D1 | 2A1 | EEEVDWGVGRMRLKMSGDVEENPGP | MG600094 | |
| 2A2 | AVHLLVTWMRRRLTLSGDIESNPGP | |||
| 2A3 | DLRAVKSFIESQLMRAGDVERNPGP | |||
| Mosavirus (SG1) | B1 | 2A1 | ESRGTGNCDATTISQCGDVETNPGP | KY855435 |
| 2A2 | YVRRSANRTAADISQDGDVETNPGP | |||
| Parechovirus (SG4) | E | 2A1 | WFDARTGFKTPLMNPCGDVEENPGP | KY645497 |
| 2A2 | QIEKRYGYRFWLLMLCGDVELNPGP | |||
| RtPV | 2A1 | MLDRRMGYRSRILCQCGDVEENPGP | MF352429 | |
| 2A2 | WFNKRSGYRSRLLSQCGDVEENPGP | |||
| Potamipivirus (SG4) | B1 | 2A1 | LMEKTEEAGWLRDLTREGVEENPGP | MK189163 |
| 2A2 | FDDYHQEGGWIRDLTAEGVEPNPGP | |||
| Unassigned (SG4) | WCP | 2A1 | MKEDEAGGWKEDLTEDGDVESNPGP | MG600066 |
| 2A2 | EQAIPETTWRRDLTQSGDVESNPGP | |||
| 2A3 | PGAIPASVWVHDLTTDGDVESNPGP | |||
| Unassigned | WP-LV 48 | 2A1 | GPSCYDRNNHCNILLSGDIEENPGP | NC_032820 |
| 2A2 | VFNASYLDCFISLLSCGDIESNPGP | |||
| 2A3 | PIQGLTQRFESTLLLGGDIEENPGP |
| Genus | Species | 2ANPGP | Reverse Primer Sequence (5’-3’) |
|---|---|---|---|
| Aalivirus | AalV- A1 | 2A1 | GCGCGCGGGCCCTGGATTCTCTTCCACATCTCCAGCTAACTTTAACAGAC TTGAATTTGTGGCTCCCTCTGATGTGAGCAATCTAGACCCGGACTTGTAT |
| 2A2 | GCGCGCGGGCCCAGGATTCTGTTCTATGTCTCCAGCCTGGAGCAGCCTGT CCCATTCTGGGTCATCATATGGCATTTCGAATCTAGACCCGGACTTGTAT |
||
| 2A3 | GCGCGCGGGCCCTGGATTCATCTCAACATCACCAGCTTGCTGCAAATTAT TCCATTGTGGGTCAGGCCTGGCTGGAATTGGTCTAGACCCGGACTTGTAT |
||
| 2A4 | GCGCGCGGGCCCGGGATTGGACTCTACATCACCACACTGCGTCAGATCGG GGACCCATCCCCCTGTCTGGTTGAAGTGCTCTCTAGACCCGGACTTGTAT |
||
| AalV-B1 | 2A1 | GCGCGCGGGCCCAGGATTTGATTCAACATCTCCGTCAATGGTTAAATCTT TCAGATACTCAGACACTTGCAAAGTAGTTGCTCTAGACCCGGACTTGTAT |
|
| 2A2 | GCGCGCGGGCCCAGGGTTAGGTTCCACACCCTCTTGAGTTAAATCTCTAA CATAATCTCCCTCAAGTTTCTTAACTTTCAATCTAGACCCGGACTTGTAT |
||
| 2A3 | GCGCGCGGGCCCAGGGTTTGATTCCACATCTCCATCAACTGTGAGGTCTC TCACCCACCCAGCATCTGTTACTCTAACCGATCTAGACCCGGACTTGTAT |
||
| 2A4 | GCGCGCGGGCCCTGGATTTGGCTCAACATCCCCACAATTCGTCAGGTCGT CAACCCAACATTTATCGTGGCACTTAAAAACTCTAGACCCGGACTTGTAT |
||
| 2A5 | GCGCGCGGGCCCAGGGTTCGACTCCACATCACCATCAACAGTTAGATCCT CAACCCAACAGCCCTCATGACACTTAAAAATTCTAGACCCGGACTTGTAT |
||
| Avisivirus | AsV-A1 | 2A1 | GCGCGCGGGCCCAGGGTTTTCTTCAATGTCACCCCCCATGAGAATGTCTC TGTGGTCCACTTCATCATAAGCTCCAACTTCTCTAGACCCGGACTTGTAT |
| 2A2 | GCGCGCGGGCCCTGGATTCTCTTCAATGTCACCTCCAAGTAGTATGTCTC TGTGGTCAGTCTCATCAAAGACTCCCATCTCTCTAGACCCGGACTTGTAT |
||
| AsV-B1 | 2A1 | GCGCGCGGGCCCAGGGTTTGATTCTACATCTCCACCTAGCAGAACATCCT CATGGGCTGAGCGCTCCTTTTCAAACTGTGGTCTAGACCCGGACTTGTAT |
|
| 2A2 | GCGCGCGGGCCCGGGATTCCTCTCTACATCACCACAAACACAGATATCAA TCTGGGGCTCCAAATATTGAACAGACTCACTTCTAGACCCGGACTTGTAT |
||
| Grusopivirus | GrV-A1 | 2A1 |
GGGCCCAGGGTTTGGTTCAATTCCCTCCTTAGATAGATCTTCTTGTGATT TCCAAGGTTTCCCATGTTTTTCAAATCTAGACCCGGACTTGTATAGTTC |
| 2A2 |
GGGCCCTGGGTTCATTTCCACTCCATATCGGCTCAACCATTTAGCGTCGG TTTCCTTATAACGATTGTCCGTAATTCTAGACCCGGACTTGTATAGTTC |
||
| 2A3 |
GGGCCCAGGATTTGATTCAATGCCTTGATTTGATAACTGATCTTGATTAG TAGCAGCATAAAGATCCTGAGTGACTCTAGACCCGGACTTGTATAGTTC |
||
| GrV-C | 2A1 | GCGCGCGGGCCCAGGATTAGTTTCTACTCCAAATTGCCCCAATTCCTTCT GAGTTGGATGTGGAGATCTTTCTTCAAAATATCTAGACCCGGACTTGTAT |
|
| 2A2 | GCGCGCGGGCCCAGGATTCATCTCTATGCCATATCGTGATAAGTGTTTGG CATCTCTCTCAGAATAATTTGAGTTGTTCTCTCTAGACCCGGACTTGTAT |
||
| 2A3 | GCGCGCGGGCCCTGGATTCTTCTCAATTCCATACTTACCTAATTCAGACT GCATGGTGGGACTCCACCTAGTGCAAACACATCTAGACCCGGACTTGTAT |
||
| YC-4 | 2A1 | GCGCGCGGGCCCAGGATTAGGCTCGATACCATATTTAGACAGTTCTTCCT TCGCCTTTGGAGAGAAATATTGACGTTCTGGTCTAGACCCGGACTTGTAT |
|
| Kunsagivirus | Kuv-C | 2A1 | GCGCGCGGGCCCAGGATTGCTCTCAACATCACCATCTTGAGTAAGGTCTC TTTGCCAGCCCTGTGCACTAGCCGCGGCAATTCTAGACCCGGACTTGTAT |
| 2A2 | GCGCGCGGGCCCTGGATTTTCCTCAACACCTTCGCGGGGTAGATCCCGCT GCCACACAGAGTCGGAGATGACAATACCTAATCTAGACCCGGACTTGTAT |
||
| 2A3 | GCGCGCGGGCCCTGGATTAGGCTCGATACCCTCACAAGTCAAATCCCTAC ACCACTGGCTGGGGGCCAGAGGGTCGTAGCTTCTAGACCCGGACTTGTAT |
||
| Limnipivirus | A1 | 2A1 | GCGCGCGGGCCCTGGGTTAGACTCCACATCTCCACACTTGAGTAGCTCCT GGTTGTCTGATTCTCTTACAAATTCTTTGCATCTAGACCCGGACTTGTAT |
| 2A2 | GCGCGCGGGCCCAGGGTTCTGTTCAACATCTCCGCTCCTCAACAACCGGA AAAAGTGAAACCATCCTGTTGAAAGGTCCCATCTAGACCCGGACTTGTAT |
||
| B1 | 2A1 | GCGCGCGGGCCCTGGGTTGCTCTCAACATCTCCATCACGTGTTAAGTCAC GTTTGAAAGGGTAATCATCAACGACATCCATTCTAGACCCGGACTTGTAT |
|
| 2A2 | GCGCGCGGGCCCGGGATTTTGTTCAACGTCCCCCGATAACAACAACCTCA TGCGTGAGTAGGCAGCTTGCACCAAGTCGATTCTAGACCCGGACTTGTAT |
||
| C1 | 2A1 | GCGCGCGGGCCCAGGGTTGGACTCCACATCGCCACAAGCAGTCAAATCTC GCTTGTATGCCAGAATTTGTTCAAGCAGTTTTCTAGACCCGGACTTGTAT |
|
| 2A2 | GCGCGCGGGCCCAGGGTTGGACTCCACATCGCCACAAGCAGTCAAATCTC GCTTGTATGCCAGAATTTGTTCAAGCAGTTTTCTAGACCCGGACTTGTAT |
||
| D1 | 2A1 | GCGCGCGGGCCCTGGGTTCTCCTCAACATCACCAGACATCTTCAGCCGCA TCCTGCCCACGCCCCAGTCGACTTCCTCCTCTCTAGACCCGGACTTGTAT |
|
| 2A2 | GCGCGCGGGCCCTGGGTTGGATTCAATGTCTCCAGAAAGCGTCAATCGTC TGCGCATCCAAGTAACCAGTAAATGAACAGCTCTAGACCCGGACTTGTAT |
||
| 2A3 | GCGCGCGGGCCCTGGGTTTCTCTCCACGTCACCAGCGCGCATCAATTGAC TTTCAATGAATGACTTCACTGCTCTTAAATCTCTAGACCCGGACTTGTAT |
||
| Mosavirus | B1 | 2A1 | GCGCGCGGGCCCAGGATTGGTTTCAACATCCCCGCACTGACTGATAGTCG TCGCATCACAGTTTCCTGTGCCACGAGATTCTCTAGACCCGGACTTGTAT |
| 2A2 | GCGCGCGGGCCCGGGGTTGGTCTCAACATCTCCATCCTGACTGATATCAG CGGCAGTACGGTTTGCGGACCGCCTGACGTATCTAGACCCGGACTTGTAT |
||
| Parechovirus | E | 2A1 | GCGCGCGGGCCCTGGGTTTTCTTCCACATCACCACAGGGGTTCATTAGGG GTGTTTTAAACCCCGTGCGTGCATCAAACCATCTAGACCCGGACTTGTAT |
| 2A2 | GCGCGCGGGCCCAGGATTTAACTCAACATCTCCACAGAGCATTAGCAACC AGAAACGATAGCCATATCGCTTCTCTATCTGTCTAGACCCGGACTTGTAT |
||
| RtPV | 2A1 | GCGCGCGGGCCCTGGATTTTCCTCGACATCTCCACATTGACAGAGGATTC TGCTCCGATAGCCCATTCTCCTGTCAAGCATTCTAGACCCGGACTTGTAT |
|
| 2A2 | GCGCGCGGGCCCAGGATTTTCTTCTACATCACCACATTGAGACAACAATC TTGACCTGTATCCTGATCTTTTGTTGAACCATCTAGACCCGGACTTGTAT |
||
| Potamipivirus | A | 2A1 | GCGCGCGGGCCCTGGATTTGGTTCAACTCCTTCTTGTGTCAGATCTCTGA TCCACATCTTGTCCGTGAGTATTTCGCCAATTCTAGACCCGGACTTGTAT |
| B | 2A1 | GCGCGCGGGCCCGGGGTTCTCCTCAACTCCCTCTCTTGTCAAATCTCTTA GCCATCCTGCTTCTTCTGTTTTCTCCATCAATCTAGACCCGGACTTGTAT |
|
| 2A2 | GCGCGCGGGCCCCGGGTTGGGCTCCACACCCTCAGCAGTGAGGTCCCGTA TCCAACCACCTTCCTGGTGGTAATCATCAAATCTAGACCCGGACTTGTAT |
||
| Unassigned | WCP | 2A1 | CGCGCGGGGCCCAGGGTTACTCTCCACATCACCGTCCTCAGTGAGGTCTT CTTTCCACCCACCAGCTTCATCCTCCTTCATTCTAGACCCGGACTTGTAT |
| 2A2 | GCGCGCGGGCCCTGGATTGGATTCCACATCACCAGATTGTGTGAGATCTC GACGCCATGTGGTTTCAGGAATTGCTTGCTCTCTAGACCCGGACTTGTAT |
||
| 2A3 | GCGCGCGGGCCCAGGATTGGATTCAACATCACCATCTGTTGTGAGGTCAT GAACCCAGACACTTGCTGGTATGGCACCAGGTCTAGACCCGGACTTGTAT |
||
| Unassigned | WP-LV 48 | 2A1 | GCGCGCGGGCCCTGGATTCTCTTCAATATCTCCTGAAAGTAAGATGTTGC AATGATTATTCCTGTCGTAGCAAGATGGACCTCTAGACCCGGACTTGTAT |
| 2A2 | GCGCGCGGGCCCTGGATTTGACTCGATATCCCCACAAGATAATAAGCTGA TGAAACAATCTAAATAACTGGCATTAAAAACTCTAGACCCGGACTTGTAT |
||
| 2A3 | GCGCGCGGGCCCTGGATTTTCTTCAATATCGCCCCCCAAAAGAAGAGTTG ACTCAAAACGTTGTGTAAGACCTTGTATTGGTCTAGACCCGGACTTGTAT |
| Genus | Species | 2ANPGP | [GFP2AGUS] | [GFP2A] | GUS |
|---|---|---|---|---|---|
| Aphthovirus | FMDV O1K | 2A | + | ++++ | ++ |
| Aalivirus (SG4) | AalV- A1 | 2A1 | + | +++++ | +++ |
| 2A2 | + | +++++ | +++ | ||
| 2A3 | - | +++++ | +++ | ||
| 2A4 | - | +++++ | +++ | ||
| AalV-B1 | 2A1 | - | ++++ | ++ | |
| 2A2 | - | ++++ | ++ | ||
| 2A3 | - | ++++ | ++ | ||
| 2A4 | + | ++++ | ++ | ||
| 2A5 | - | ++++ | + | ||
| Unassigned | DERSV | 2A1-6 | ND | ND | ND |
| Avisivirus (SG4) | AsV-A1 | 2A1 | - | ++++ | + |
| 2A2 | - | ++++ | ++ | ||
| AsV-B1 | 2A1 | - | ++++ | ++ | |
| 2A2 | - | ++ | + | ||
| Grusopivirus (SG4) | GrV-A1 | 2A1 | + | ++++ | ++ |
| 2A2 | + | ++++ | ++ | ||
| 2A3 | + | ++++ | ++ | ||
| GrV-C | 2A1 | + | ++++ | ++ | |
| 2A2 | + | ++++ | (+) | ||
| 2A3 | (+) | ++++ | ++ | ||
| YC-4 | 2A1 | - | ++++ | (+) | |
| Kunsagivirus (SG4) | Kuv-C1 | 2A1 | - | ++++ | - |
| 2A2 | ND | ND | ND | ||
| 2A3 | ND | ND | ND | ||
| Limnipivirus (SG4) | A1 | 2A1 | - | ++++ | ++ |
| 2A2 | - | ++++ | - | ||
| B1 | 2A1 | - | +++ | - | |
| 2A2 | ++ | +++ | ++++ | ||
| C1 | 2A1 | ++ | ++++ | ++++ | |
| 2A2 | - | ++++ | + | ||
| D1 | 2A1 | - | ++++ | ++ | |
| 2A2 | - | ++++ | ++ | ||
| 2A3 | - | ++++ | + | ||
| Mosavirus (SG1) | B1 | 2A1 | + | ++++ | ++ |
| 2A2 | + | ++++ | ++ | ||
| Parechovirus (SG4) | E | 2A1 | + | - | - |
| 2A2 | - | ++++ | + | ||
| RtPV | 2A1 | - | ++++ | + | |
| 2A2 | - | ++++ | + | ||
| Potamipivirus (SG4) | B1 | 2A1 | (+) | ++++ | + |
| 2A2 | - | +++ | - | ||
| Unassigned (SG4?) | WCP | 2A1 | - | +++ | - |
| 2A2 | - | +++ | - | ||
| 2A3 | - | +++ | - | ||
| Unassigned (SG5?) | WP-LV 48 | 2A1 | - | +++ | - |
| 2A2 | - | ++++ | - | ||
| 2A3 | - | +++ | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





