Submitted:
24 September 2024
Posted:
25 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
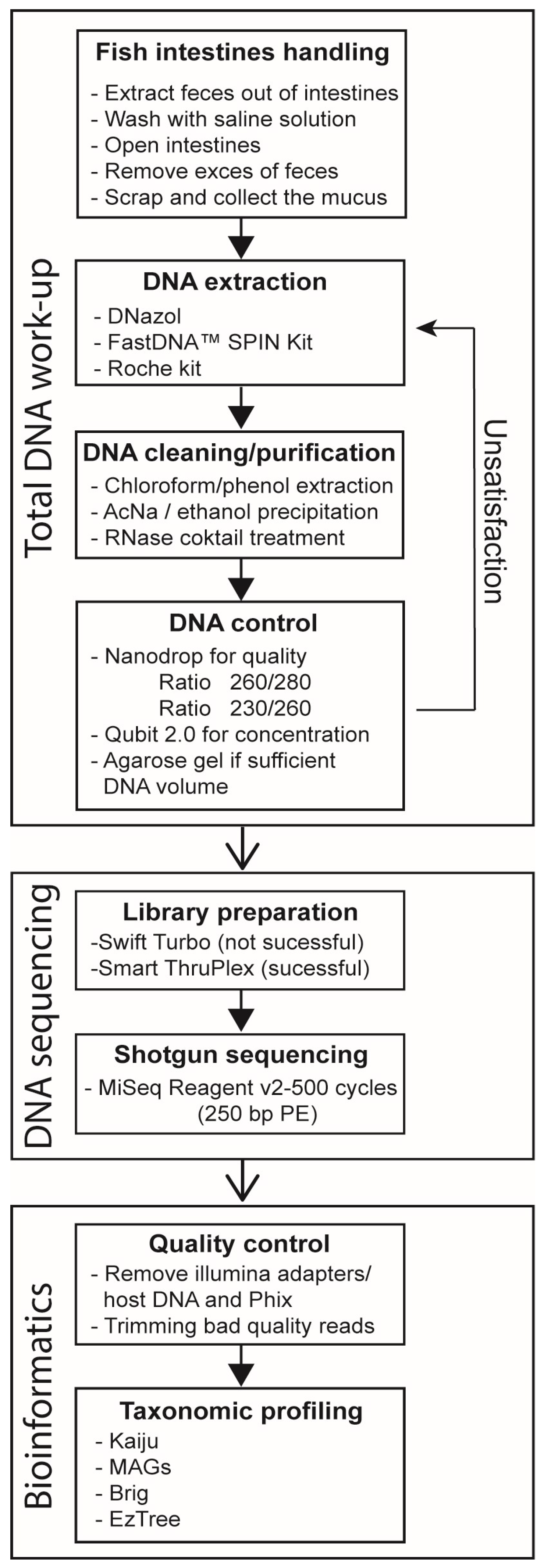
2. Materials and Methods
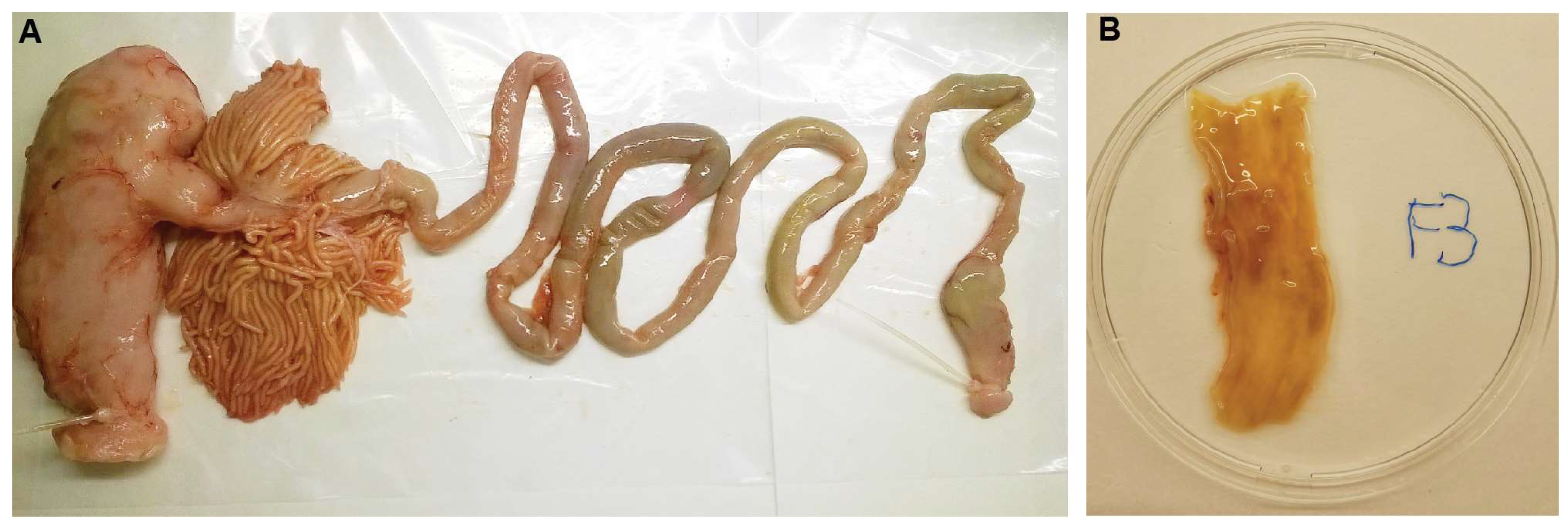
2.1. Sampling of Fish and Intestinal Mucus
2.2. Isolation of DNA and Shotgun Sequencing
2.3. Quality Control of DNA Sequence Reads
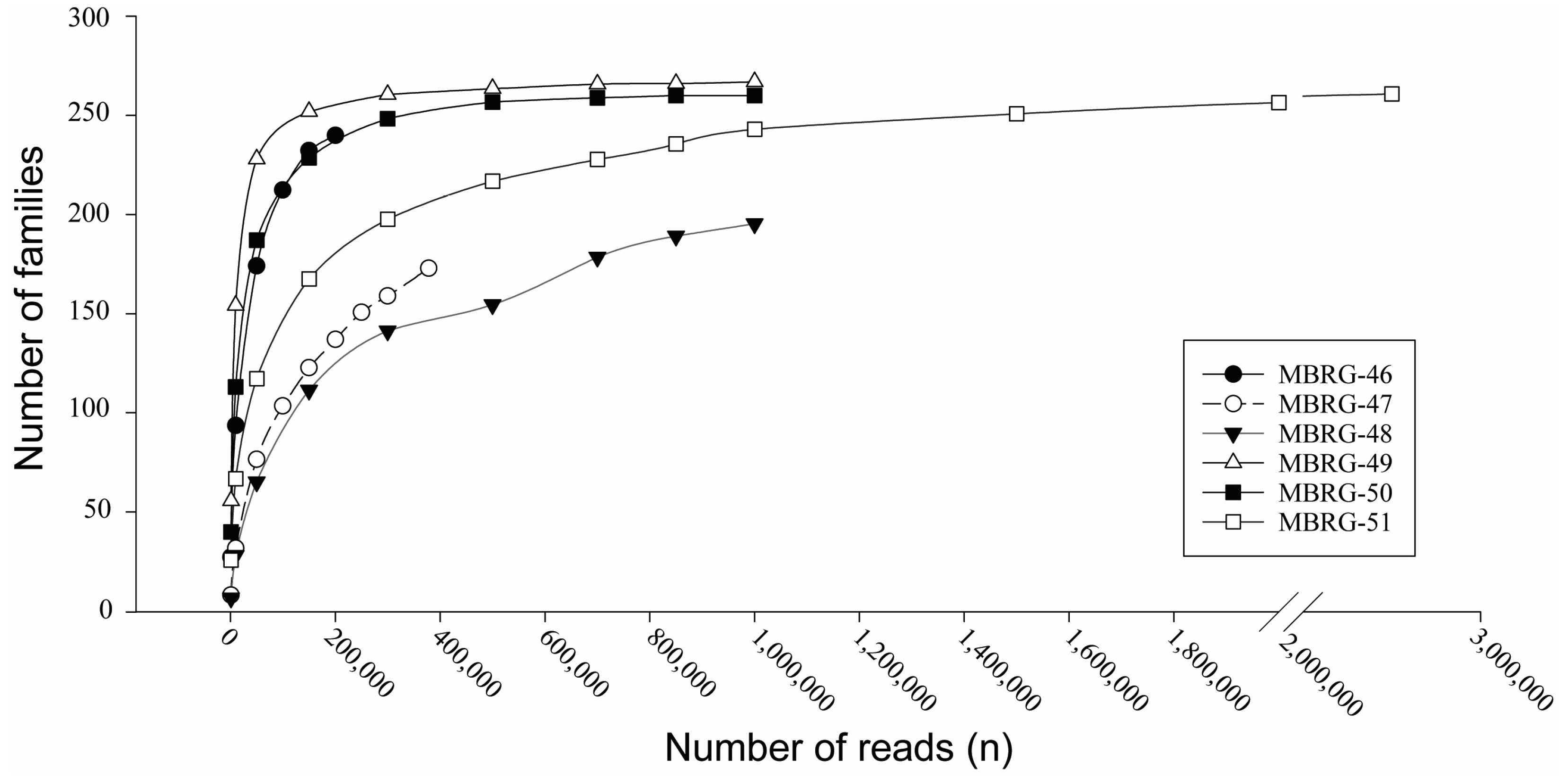
2.4. Rarefaction Curves
2.5. Taxonomic Profiling and Metagenome-Assembled Genomes (MAGs)
2.6. Phylogenomic Analyses of the MAGs
3. Results
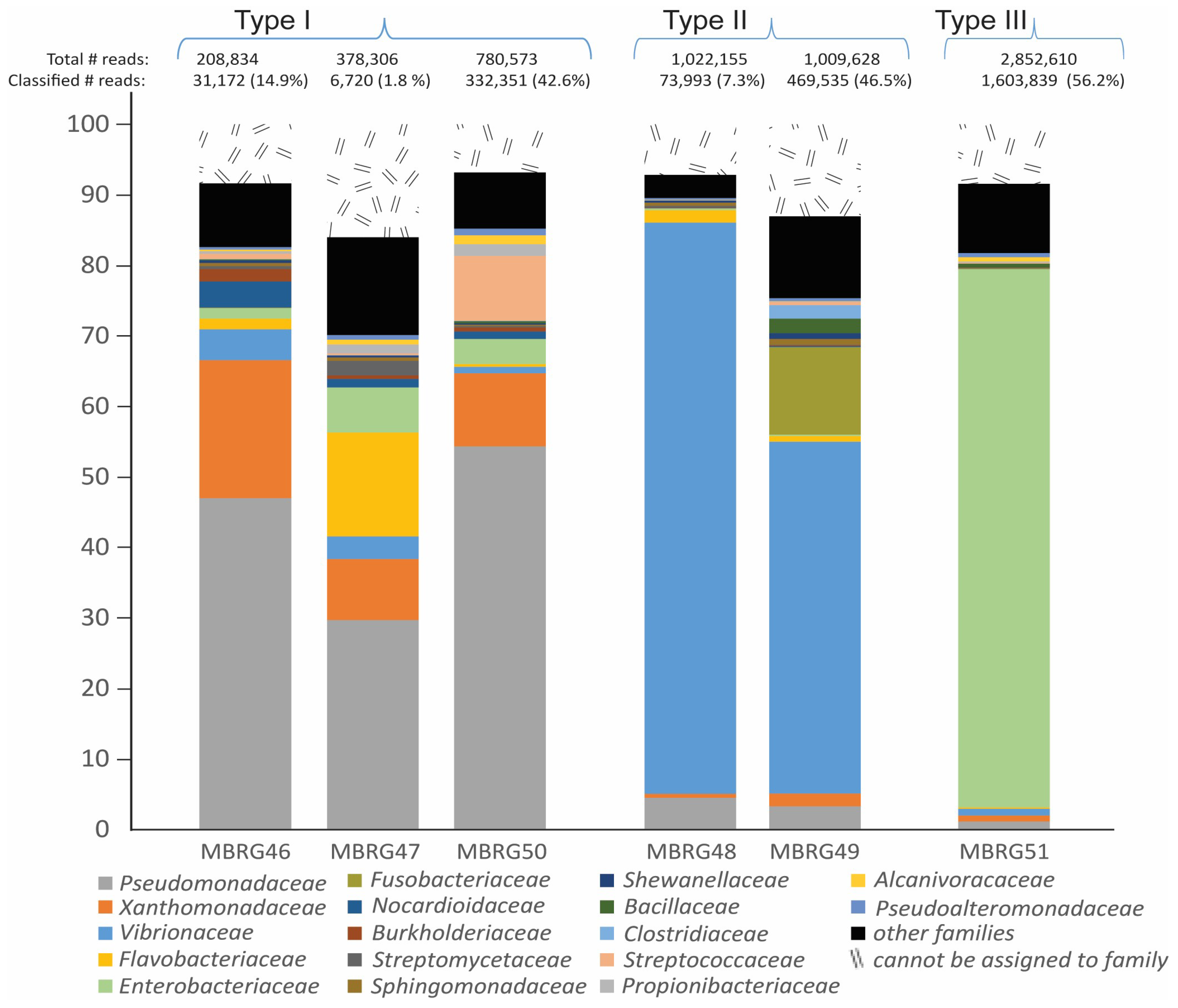
3.1. DNA Sequencing Revealed Three Different Taxonomic Profile Types among Six Mucosal Samples from Migrating Northeast Arctic Cod
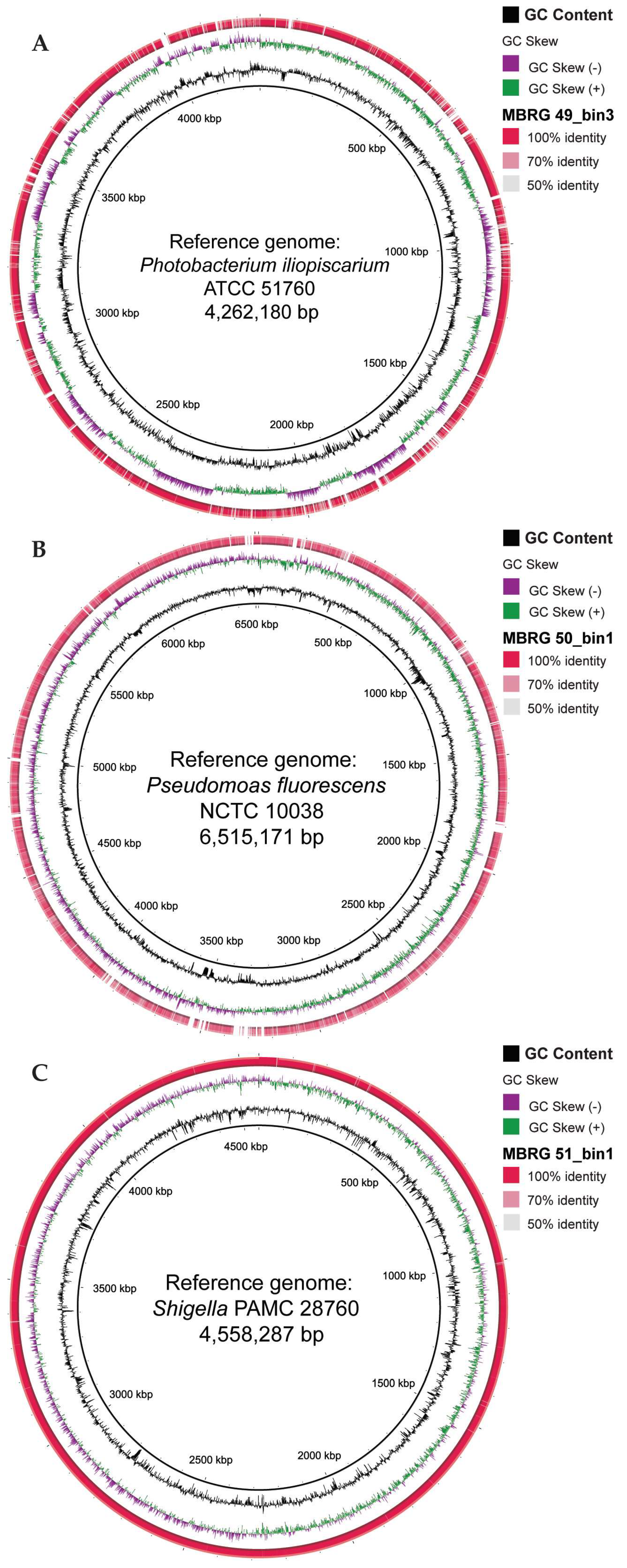
3.2. Binning of Assembled Contigs Produce High-Completeness bins/MAGs
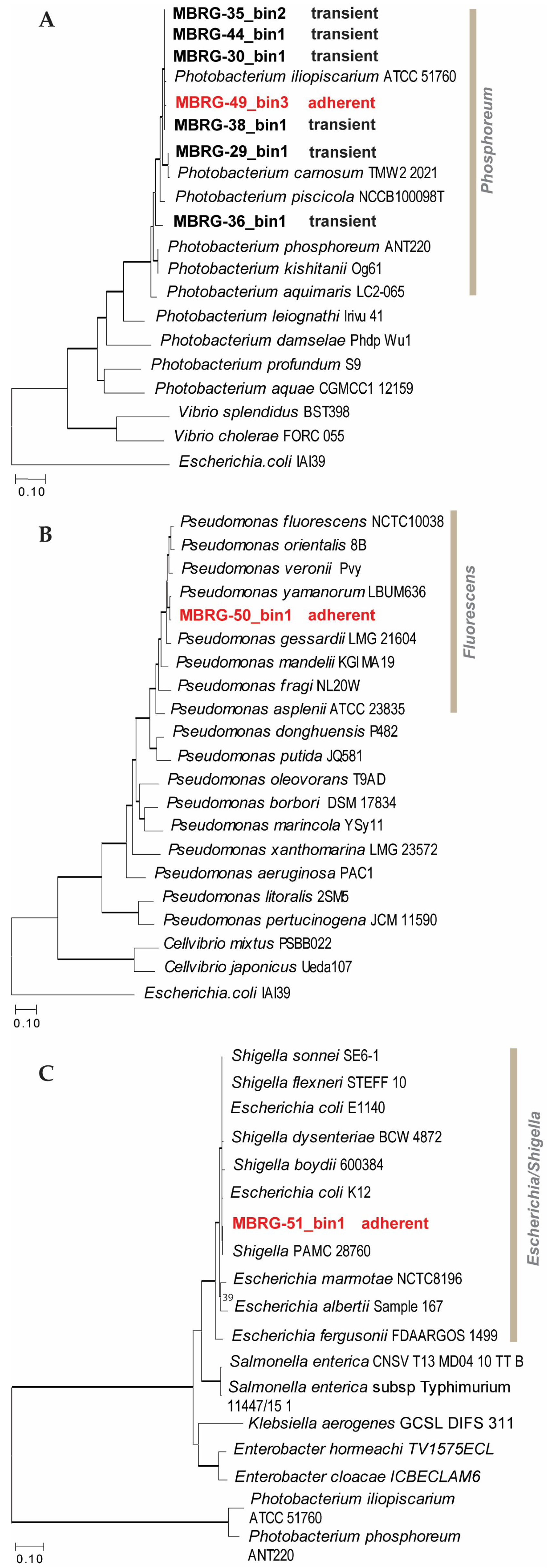
3.3. EzTree Robustly Places High-Completeness Bins on Maximum Likelihood Trees
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Esser, D.; Lange, J.; Marinos, G.; Sieber, M.; Best, L.; Prasse, D.; et al. Functions of the microbiota for the physiology of animal metaorganisms. Journal of innate immunity. 2019, 11, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Runge, S.; Rosshart, S.P. The mammalian metaorganism: A holistic view on how microbes of all Kingdoms and niches shape local and systemic immunity. Frontiers in Immunology. 2021, 12, 702378. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, J.; Moeller, A.H. The effects of temperature on animal gut microbiomes. Frontiers in microbiology. 2020, 11, 384. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS biology. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Tuddenham, S.; Sears, C.L. The intestinal microbiome and health. Current opinion in infectious diseases. 2015, 28, 464. [Google Scholar] [CrossRef]
- Oren A, Arahal DR, Rosselló-Móra R, Sutcliffe IC, Moore ER: Emendation of Rules 5b, 8, 15 and 22 of the International Code of Nomenclature of Prokaryotes to include the rank of phylum. In.: Microbiology Society; 2021. [CrossRef]
- Finlayson-Trick, E.C.; Getz, L.J.; Slaine, P.D.; Thornbury, M.; Lamoureux, E.; Cook, J.; et al. Taxonomic differences of gut microbiomes drive cellulolytic enzymatic potential within hind-gut fermenting mammals. PloS one. 2017, 12, e0189404. [Google Scholar] [CrossRef]
- Kim, P.S.; Shin, N.-R.; Lee, J.-B.; Kim, M.-S.; Whon, T.W.; Hyun, D.-W.; et al. Host habitat is the major determinant of the gut microbiome of fish. Microbiome. 2021, 9, 166. [Google Scholar] [CrossRef]
- Consortium, T.H.M.P. Structure, function and diversity of the healthy human microbiome. nature. 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Reddy, D.N. Role of the normal gut microbiota. World journal of gastroenterology: WJG. 2015, 21, 8787. [Google Scholar] [CrossRef]
- Jiang, H.-Y.; Ma, J.-E.; Li, J.; Zhang, X.-J.; Li, L.-M.; He, N.; et al. Diets alter the gut microbiome of crocodile lizards. Frontiers in Microbiology. 2017, 8, 2073. [Google Scholar] [CrossRef]
- Korry, B.J.; Belenky, P. Trophic level and proteobacteria abundance drive antibiotic resistance levels in fish from coastal New England. Animal Microbiome. 2023, 5, 16. [Google Scholar] [CrossRef]
- Johny, T.K.; Puthusseri, R.M.; Bhat, S.G. Metagenomic landscape of taxonomy, metabolic potential and resistome of Sardinella longiceps gut microbiome. Archives of Microbiology. 2022, 204, 87. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Limenitakis, J.P.; Fuhrer, T.; Geuking, M.B.; Lawson, M.A.; Wyss, M.; et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nature communications. 2015, 6, 8292. [Google Scholar] [CrossRef] [PubMed]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochemical journal. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Herath, M.; Hosie, S.; Bornstein, J.C.; Franks, A.E.; Hill-Yardin, E.L. The role of the gastrointestinal mucus system in intestinal homeostasis: implications for neurological disorders. Frontiers in cellular and infection microbiology. 2020, 10, 248. [Google Scholar] [CrossRef]
- Gustafsson, J.K.; Johansson, M.E. The role of goblet cells and mucus in intestinal homeostasis. Nature reviews Gastroenterology & hepatology. 2022, 19, 785–803. [Google Scholar] [CrossRef]
- Cornuault, J.K.; Byatt, G.; Paquet, M.-E.; De Koninck, P.; Moineau, S. Zebrafish: a big fish in the study of the gut microbiota. Current Opinion in Biotechnology. 2022, 73, 308–313. [Google Scholar] [CrossRef]
- Star, B.; Haverkamp, T.H.; Jentoft, S.; Jakobsen, K.S. Next generation sequencing shows high variation of the intestinal microbial species composition in Atlantic cod caught at a single location. BMC microbiology. 2013, 13, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Riiser, E.S.; Haverkamp, T.H.; Varadharajan, S.; Borgan, Ø.; Jakobsen, K.S.; Jentoft, S.; et al. Metagenomic shotgun analyses reveal complex patterns of intra-and interspecific variation in the intestinal microbiomes of codfishes. Applied and Environmental Microbiology. 2020, 86, e02788-19. [Google Scholar] [CrossRef]
- Le Doujet, T.; De Santi, C.; Klemetsen, T.; Hjerde, E.; Willassen, N.-P.; Haugen, P. Closely-related Photobacterium strains comprise the majority of bacteria in the gut of migrating Atlantic cod (Gadus morhua). Microbiome. 2019, 7, 1–12. [Google Scholar] [CrossRef]
- Chomczynski, P.; Mackey, K.; Drews, R. Wilfinger WDNAzol®: a reagent for the rapid isolation of genomic DNA. Biotechniques. 1997, 22, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Wingett SW, Andrews S. FastQ Screen: A tool for multi-genome mapping and qualit y control [version 1; peer review: 3 approved, 1 approved with. 2018. [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Andrews, S. A quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 2 February 2023).
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nature communications. 2016, 7, 11257. [Google Scholar] [CrossRef]
- Klemetsen, T.; Raknes, I.A.; Fu, J.; Agafonov, A.; Balasundaram, S.V.; Tartari, G.; et al. The MAR databases: development and implementation of databases specific for marine metagenomics. Nucleic acids research. 2018, 46, D692-D9. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: a new versatile metagenomic assembler. Genome research. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Wu, Y.-W.; Tang, Y.-H.; Tringe, S.G.; Simmons, B.A.; Singer, S.W. MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome. 2014, 2, 1–18. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome research. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Bushnell B: BBMap: a fast, accurate, splice-aware aligner. In.: Lawrence Berkeley National Lab.(LBNL), Berkeley, CA (United States); 2014.
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC genomics. 2011, 12, 1–10. [Google Scholar] [CrossRef]
- Wu, Y.-W. ezTree: an automated pipeline for identifying phylogenetic marker genes and inferring evolutionary relationships among uncultivated prokaryotic draft genomes. BMC genomics. 2018, 19, 7–16. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Molecular biology and evolution. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Magnuson, J.T.; Monticelli, G.; Schlenk, D.; Bisesi Jr, J.H.; Pampanin, D.M. Connecting gut microbiome changes with fish health conditions in juvenile Atlantic cod (Gadus morhua) exposed to dispersed crude oil. Environmental Research. 2023, 234, 116516. [Google Scholar] [CrossRef]
- Riiser, E.S.; Haverkamp, T.H.; Borgan, Ø.; Jakobsen, K.S.; Jentoft, S.; Star, B. A single Vibrionales 16S rRNA oligotype dominates the intestinal microbiome in two geographically separated Atlantic cod populations. Frontiers in Microbiology. 2018, 1561. [Google Scholar] [CrossRef] [PubMed]
- Riiser, E.S.; Haverkamp, T.H.; Varadharajan, S.; Borgan, Ø.; Jakobsen, K.S.; Jentoft, S.; et al. Switching on the light: using metagenomic shotgun sequencing to characterize the intestinal microbiome of Atlantic cod. Environmental Microbiology. 2019, 21, 2576–2594. [Google Scholar] [CrossRef] [PubMed]
- Ringø, E.; Sperstad, S.; Myklebust, R.; Refstie, S.; Krogdahl, Å. Characterisation of the microbiota associated with intestine of Atlantic cod (Gadus morhua L. ): the effect of fish meal, standard soybean meal and a bioprocessed soybean meal. Aquaculture. 2006, 261, 829–841. [Google Scholar] [CrossRef]
- Dhanasiri, A.K.; Brunvold, L.; Brinchmann, M.F.; Korsnes, K.; Bergh, Ø.; Kiron, V. Changes in the intestinal microbiota of wild Atlantic cod Gadus morhua L. upon captive rearing. Microbial ecology. 2011, 61, 20–30. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; von Wright, A.; Vilpponen-Salmela, T.; Ben-Amor, K.; Akkermans, A.D.; de Vos, W.M. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Applied and environmental microbiology. 2002, 68, 3401–3407. [Google Scholar] [CrossRef] [PubMed]
- Vaga, S.; Lee, S.; Ji, B.; Andreasson, A.; Talley, N.J.; Agréus, L.; et al. Compositional and functional differences of the mucosal microbiota along the intestine of healthy individuals. Scientific reports. 2020, 10, 14977. [Google Scholar] [CrossRef]
- Luan, Y.; Li, M.; Zhou, W.; Yao, Y.; Yang, Y.; Zhang, Z.; et al. The fish microbiota: Research progress and potential applications. Engineering. 2023. [Google Scholar] [CrossRef]
- Wenne, R.; Bernaś, R.; Kijewska, A.; Poćwierz-Kotus, A.; Strand, J.; Petereit, C.; et al. SNP genotyping reveals substructuring in weakly differentiated populations of Atlantic cod (Gadus morhua) from diverse environments in the Baltic Sea. Scientific Reports. 2020, 10, 9738. [Google Scholar] [CrossRef]
- Barney, B.T.; Munkholm, C.; Walt, D.R.; Palumbi, S.R. Highly localized divergence within supergenes in Atlantic cod (Gadus morhua) within the Gulf of Maine. BMC genomics. 2017, 18, 1–14. [Google Scholar] [CrossRef]
- Loeng, H. Features of the physical oceanographic conditions of the Barents Sea. Polar research. 1991, 10, 5–18. [Google Scholar] [CrossRef]
- Sakshaug, E.; Johnsen, G.; Kovacs, K. Ecosystem Barents Sea. Tapir Acad. Press; 2009.
- Ringø, E.; Zhou, Z.; Vecino, J.G.; Wadsworth, S.; Romero, J.; Krogdahl, Å.; et al. Effect of dietary components on the gut microbiota of aquatic animals. A never-ending story? Aquaculture nutrition. 2016, 22, 219–282. [Google Scholar] [CrossRef]
- Johannesen, E.; Høines, Å.S.; Dolgov, A.V.; Fossheim, M. Demersal fish assemblages and spatial diversity patterns in the Arctic-Atlantic transition zone in the Barents Sea. PLoS One. 2012, 7, e34924. [Google Scholar] [CrossRef] [PubMed]
- Bogstad, B.; Gjøsæter, H.; Haug, T.; Lindstrøm, U. A review of the battle for food in the Barents Sea: cod vs. marine mammals. Frontiers in Ecology and Evolution. 2015, 3, 29. [Google Scholar] [CrossRef]
- Scales, B.S.; Dickson, R.P.; LiPuma, J.J.; Huffnagle, G.B. Microbiology, genomics, and clinical significance of the Pseudomonas fluorescens species complex, an unappreciated colonizer of humans. Clinical microbiology reviews. 2014, 27, 927–948. [Google Scholar] [CrossRef]
- Ringø, E.; Strøm, E.; Tabachek, J.A. Intestinal microflora of salmonids: a review. Aquaculture Research. 1995, 26, 773–789. [Google Scholar] [CrossRef]
- Gołaś, I.; Szmyt, M.; Potorski, J.; Łopata, M.; Gotkowska-Płachta, A.; Glińska-Lewczuk, K. Distribution of Pseudomonas fluorescens and Aeromonas hydrophila bacteria in a recirculating aquaculture system during farming of european grayling (Thymallus thymallus L.) Broodstock. Water. 2019, 11, 376. [Google Scholar] [CrossRef]
- Shakila, R.; Saravanakumar, R.; Vyla, S.; Jeyasekaran, G.; Jasmine, G. Antagonistic activity of the gut microflora isolated from farmed tiger shrimp (Penaeus monodon). 2007.
- González-Palacios, C.; Fregeneda-Grandes, J.M.; Aller-Gancedo, J.M. Biocontrol of saprolegniosis in rainbow trout (Oncorhynchus mykiss Walbaum) using two bacterial isolates (LE89 and LE141) of Pseudomonas fluorescens. Journal of fish diseases. 2019, 42, 269–275. [Google Scholar] [CrossRef]
- Korkea-Aho, T.; Papadopoulou, A.; Heikkinen, J.; von Wright, A.; Adams, A.; Austin, B.; et al. Pseudomonas M162 confers protection against rainbow trout fry syndrome by stimulating immunity. Journal of applied microbiology. 2012, 113, 24–35. [Google Scholar] [CrossRef]
- Eissa, N.; Abou El-Gheit, N.; Shaheen, A.A. Protective effect of Pseudomonas fluorescens as a probiotic in controlling fish pathogens. American Journal of BioScience. 2014, 2, 175–181. [Google Scholar] [CrossRef]
- Egerton, S.; Culloty, S.; Whooley, J.; Stanton, C.; Ross, R.P. The gut microbiota of marine fish. Frontiers in Microbiology. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Hilgarth, M.; Fuertes, S.; Ehrmann, M.; Vogel, R.F. Photobacterium carnosum sp. nov., isolated from spoiled modified atmosphere packaged poultry meat. Systematic and applied microbiology. 2017. [CrossRef]
- Fuertes-Perez, S.; Hauschild, P.; Hilgarth, M.; Vogel, R.F. Biodiversity of Photobacterium spp. isolated from meats. Frontiers in Microbiology. 2019, 10, 480482. [Google Scholar] [CrossRef]
- Figge, M.J.; Cleenwerck, I.; van Uijen, A.; De Vos, P.; Huys, G.; Robertson, L. Photobacterium piscicola sp. nov., isolated from marine fish and spoiled packed cod. Systematic and applied microbiology. 2014, 37, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Itoi, S.; Okamura, T.; Koyama, Y.; Sugita, H. Chitinolytic bacteria in the intestinal tract of Japanese coastal fishes. Canadian journal of microbiology. 2006, 52, 1158–1163. [Google Scholar] [CrossRef] [PubMed]
- Ringø, E.; Zhou, Z.; Olsen, R.; Song, S. Use of chitin and krill in aquaculture–the effect on gut microbiota and the immune system: a review. Aquaculture Nutrition. 2012, 18, 117–131. [Google Scholar] [CrossRef]
- Caipang, C.M.A.; Brinchmann, M.F.; Kiron, V. Antagonistic activity of bacterial isolates from intestinal microbiota of Atlantic cod, Gadus morhua, and an investigation of their immunomodulatory capabilities. Aquaculture research. 2010, 41, 249–256. [Google Scholar] [CrossRef]
- van den Beld, M.; Reubsaet, F. Differentiation between Shigella, enteroinvasive Escherichia coli (EIEC) and noninvasive Escherichia coli. European journal of clinical microbiology & infectious diseases. 2012, 31, 899–904. [Google Scholar] [CrossRef]
- Carlos, C.; Pires, M.M.; Stoppe, N.C.; Hachich, E.M.; Sato, M.I.; Gomes, T.A.; et al. Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC microbiology. 2010, 10, 1–10. [Google Scholar] [CrossRef]
- Sun, F.; Wang, Y.; Wang, C.; Zhang, L.; Tu, K.; Zheng, Z. Insights into the intestinal microbiota of several aquatic organisms and association with the surrounding environment. Aquaculture. 2019, 507, 196–202. [Google Scholar] [CrossRef]
- Wu, P.; Liu, Y.; Li, C.; Xiao, Y.; Wang, T.; Lin, L.; et al. The Composition of Intestinal Microbiota From Collichthys lucidus and Its Interaction With Microbiota From Waters Along the Pearl River Estuary in China. Frontiers in Environmental Science. 2021, 9, 675856. [Google Scholar] [CrossRef]
- European Nucleotide Archive (ENA). 2018. https://www.ebi.ac.uk/ena/data/view/%3CACCESSION (accessed on 2 February 2023).
|
Fish ID |
DNA quality |
DNA Concentration in ng/µL1 |
Available DNA for sequencing |
|||
|---|---|---|---|---|---|---|
| 260/280 | 230/260 | No RNase treatment | Treated with RNase cocktail2 | Volume (µL) | Concentration (ng) | |
| MBRG46 | 1.8 | 1.83 | 2.56 | 0.29 | 15 | 4.35 |
| MBRG47 | 2 | 1.6 | 2.09 | 0.45 | 20 | 9 |
| MBRG48 | 1.86 | 1.84 | 17 | 10 | 19 | 19 |
| MBRG49 | 1.95 | 1.72 | 19.4 | 0.70 | 10| | 7 |
| MBRG50 | 1.91 | 1.73 | 1.6 | 0.11 | 12 | 1.34 |
| MBRG51 | 1.89 | 1.4 | 0.261 | 0.14 | 19 | 2.6 |
| Fish ID 1 | Raw data | Final dataset | ||
|---|---|---|---|---|
| # reads | Cod DNA (%) | # reads 2 | Average length (bp) | |
| MBRG46 | 9 805 968 | 96.5 | 327 852 | 116 |
| MBRG47* | 12 534 644 | 95.4 | 541 404 | 54 |
| MBRG48* | 21 894 396 | 94.7 | 1 496 529 | 57 |
| MBRG49 | 8 376 018 | 76.4 | 1 964 746 | 106 |
| MBRG50 | 10 955 154 | 86.2 | 1 491 889 | 118 |
| MBRG51* | 16 305 536 | 65.5 | 5 543 182 | 101 |
| MAXBIN | CHECKM | SENDSKETCH | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample ID | Bin ID | Rel Abund1 (%) |
Contigs (n) | Comp2 (%) | Genome size (bp) |
GC content (%) | Comp2 (%) | Cont3 (%) | Bacteria IDs | KWID (%) | KID (%) |
| MBRG49 |
1 |
52.5 |
88 |
96.3 |
1 796 209 |
28.5 |
100 |
0 |
Photobacterium iliopiscarium |
1.2 |
0.3 |
| 2 | 34.7 | 274 | 90.7 | 1 902 394 | 30.7 | 98.6 | 0 | Photobacterium iliopiscarium | 12.7 | 3.6 | |
| 3 | 12.8 | 1 396 | 98.1 | 4 168 186 | 41.7 | 95 | 3.2 | Photobacterium iliopiscarium | 80.8 | 50.4 | |
|
MBRG50 |
2 |
36.4 |
653 |
99.1 |
7 662 027 |
60.8 |
96.5 |
6.5 |
Pseudomonas fluorescens |
78.1 |
59.2 |
|
MBRG51 |
3 |
85.3 |
126 |
99.1 |
4 535 891 |
50.6 |
99.4 |
0.2 |
Shigella sp. |
99.9 |
49.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





