1. Introduction
The development of automated computer-aided diagnosis (CAD) systems is a prominent area of research [
12]. Numerous endeavors have focused on creating CAD systems for a variety of health applications. Research efforts include studies using mammography images to develop systems for detecting and classifying breast cancer [
13,
14,
15], and projects using computed tomography (CT) scans for lung cancer detection and classification [
16,
17,
18]. Additionally, CAD systems targeting other types of cancer, such as prostate cancer, often utilize MRI technology for diagnostic and classification purposes [
19,
20,
21].
Moreover, CAD systems find use in diverse medical fields beyond oncology. For instance, they are instrumental in grading diabetic retinopathy using optical coherence tomography (OCT) images [
22,
23,
24], and in detecting ulcers where capsule endoscopy [
25,
26] and even smartphone images [
27] are employed.
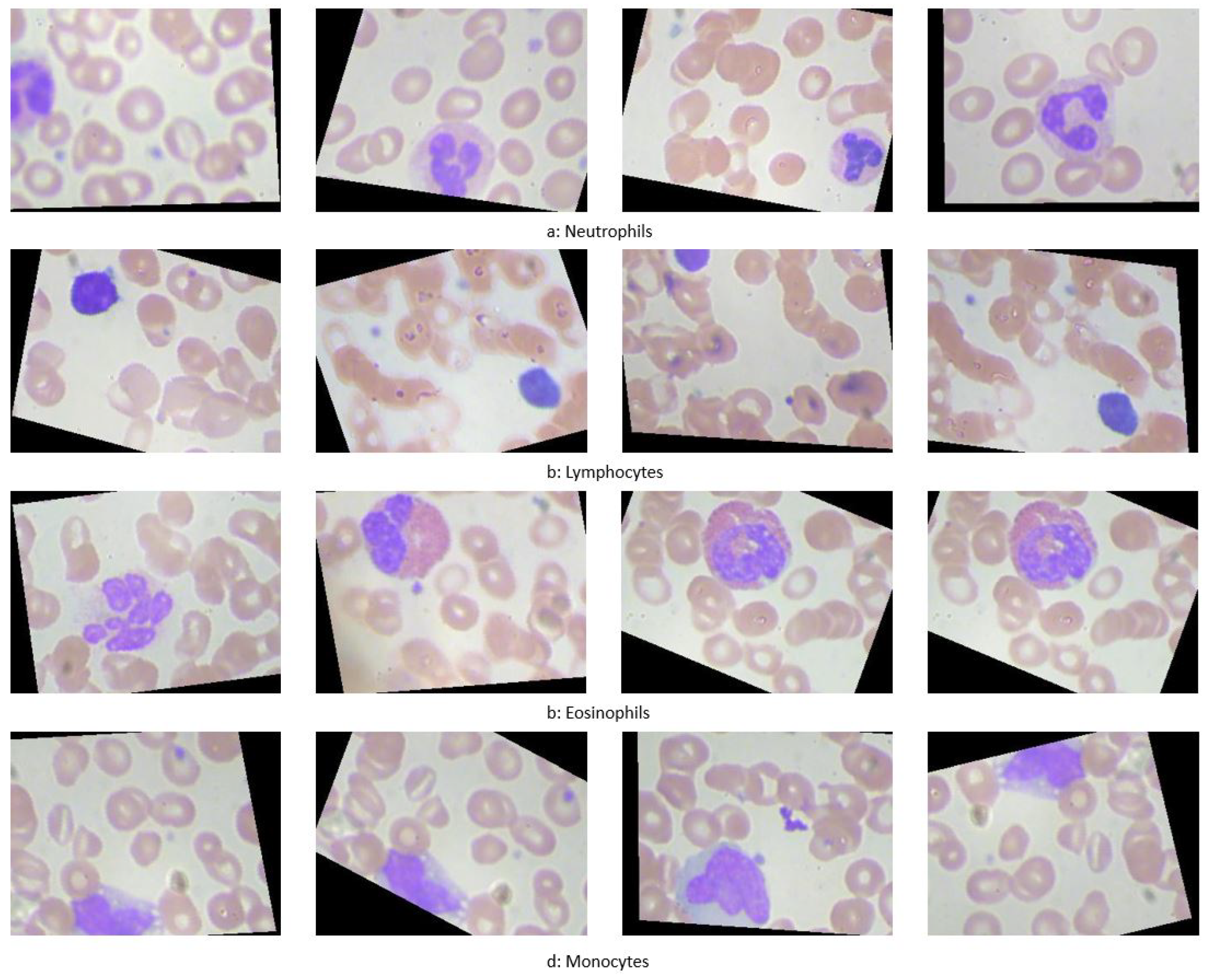
This research introduces a CAD system aimed at classifying and localizing white blood cells (WBCs) using microscopic imaging. Such imaging is crucial for providing insights into a patient’s hematological health, with differential blood counts revealing conditions like infections, leukemia, and specific cancers. Traditionally, such counts are performed manually by technicians using microscopes to assess the prevalence of different WBC types in smear slides, a process that is both time-consuming and labor-intensive [
28]. As illustrated in
Figure 1, different types of WBCs include Neutrophils, Lymphocytes, Eosinophils, and Monocytes.
2. Literature Review
2.1. Overview of White Blood Cells
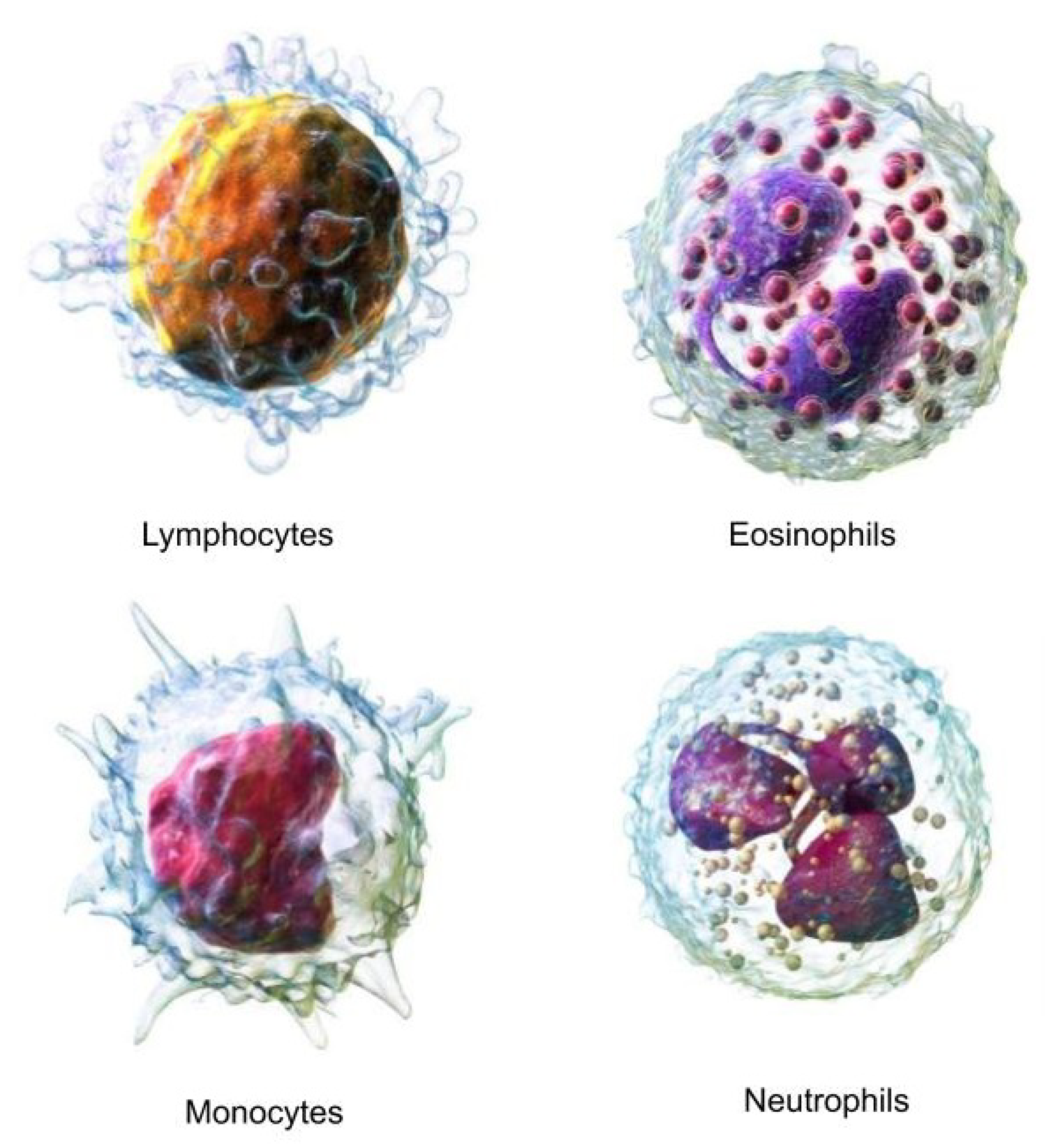
White blood cells (WBCs) are essential components of the immune system, grouped into four main categories: Neutrophils, Lymphocytes, Eosinophils, and Monocytes.[
28] These cells are instrumental in the body’s defenses against infections, diseases, and foreign intrusions.
Neutrophils act as a primary defense mechanism against infections, constituting a significant portion of the WBC count. Lymphocytes are critical to the adaptive immune response, whereas Eosinophils combat parasites and allergens. Monocytes are crucial for phagocytosis.
Figure 2 provides a 3D visualization of these cells.
2.2. Disorders of White Blood Cells
White blood cell (WBC) disorders can be primarily categorized into two types: proliferative disorders and leukopenias. Proliferative disorders are characterized by an increase in the number of WBCs, while leukopenias are marked by a decrease in WBC counts.
2.2.1. Proliferative Disorders
Proliferative disorders lead to an abnormal increase in white blood cells, with the most common condition being leukocytosis. Leukocytosis can be triggered by various factors including:
Bacterial or viral infections, which typically induce a rapid increase in WBCs as a part of the immune response.
Inflammation and allergic reactions that stimulate the production of white blood cells.
Cancer, particularly hematologic malignancies like leukemia, which directly result in the overproduction of abnormal white cells.
Other contributing factors may include smoking, certain medications, and genetic predispositions.[
56]
2.2.2. Leukopenias
On the other end of the spectrum, leukopenias are characterized by a reduced WBC count, significantly impacting the body’s ability to fight infections. Key disorders under this category include:
Autoimmune Neutropenia: This condition arises when antibodies produced by the body’s immune system mistakenly target and destroy neutrophils. It is often associated with autoimmune diseases such as Crohn’s disease and rheumatoid arthritis.[
57]
Severe Congenital Neutropenia: Caused by genetic mutations, this rare disorder features marked susceptibility to recurrent bacterial infections due to dramatically reduced neutrophil counts.[
58]
Cyclic Neutropenia: This genetic disorder causes fluctuations in neutrophil levels, typically cycling every three weeks, leading to periodic susceptibilities to infections.[
59]
Chronic Granulomatous Disease: Affects multiple types of white blood cells including neutrophils and monocytes. Patients with this inherited disorder suffer from frequent bacterial and fungal infections due to the cells’ inability to effectively kill certain types of bacteria and fungi.[
60]
Leukocyte Adhesion Deficiency (LAD) Syndrome: This rare genetic condition prevents white blood cells from migrating to the site of infection, resulting in severe, recurrent bacterial infections.[
61]
2.3. Advancements in WBC Classification and Localization
Research efforts have significantly advanced the classification and localization of WBCs, employing both traditional machine learning and modern deep learning techniques to enhance the accuracy and efficiency of these processes.
Traditional Machine Learning Methods
Ongun
et al. [
30] developed a multi-step system for WBC classification, involving segmentation using active contours and morphological operations, followed by feature extraction focusing on shape, texture, and color. Classification was performed using methods including K-Nearest Neighbor (KNN) and Support Vector Machine (SVM), with SVM achieving an accuracy of 91.03%.
Another approach by Sinha and Ramakrishnan [
34] utilized K-means for segmentation and extracted features for classification via Neural Networks (NN), KNN, and SVM, reaching a 94.1% accuracy on a modest dataset.
Hu
et al. [
35] proposed a segmentation method using wavelet transforms, applying different processing on high and low frequency components to achieve enhanced segmentation.
Segmentation Innovations
Shahin
et al. [
68] introduced a technique utilizing neutrosophic similarity to segment WBCs, adapting the approach for various color spaces. Sadeghian
et al. [
69] combined several image processing steps to segment nuclei and cytoplasm, employing techniques such as GVF snake algorithms and Zack thresholding, resulting in high accuracy for nucleus segmentation.
A notable study by Hegde
et al. [
72] combined color channel manipulations with TissueQuant algorithms for segmenting and classifying nuclei using shape and texture features, achieving an accuracy of 96.64% on a sample set of 117 images.
Innovative Neural Architectures
Research has increasingly incorporated deep learning for enhanced accuracy in WBC classification. Shahin
et al. [
73] explored the efficacy of pre-trained networks like OverfeatNet, AlexNet, and VGG, alongside novel architectures, with deep learning models surpassing traditional methods in accuracy.
Kutlu
et al. [
74] implemented a CAD system employing Single Shot Detector (SSD) for detection and various pre-trained networks for classification, achieving remarkable success with an accuracy rate up to 97%.
Tiwari
et al. [
77] introduced a simpler Double Convolution Layer Neural Network (DCLNN) architecture, which provided competitive results with an 88% accuracy on a large dataset.
Emerging Technologies
Baydilli
et al. [
78] utilized capsule networks, a novel approach offering robust feature extraction through dynamic routing between capsules, showcasing the potential for visual inspection of reconstructed features.
Tran
et al. [
80] employed the SegNet architecture for segmenting both WBCs and red blood cells, demonstrating the utility of encoder-decoder structures in medical image segmentation with a mean intersection over union of 79%.
These studies reflect the dynamic evolution of techniques from traditional machine learning to sophisticated deep learning models, each contributing uniquely to the field of medical image analysis for WBC classification and localization.
3. Methods
This Section discusses the methodologies employed in the experiments, which are categorized into three primary areas:
Region of Interest (ROI) extraction: Various techniques for identifying and defining the ROIs that contain WBCs in the images are examined. These methods are utilized to evaluate model performance using both entire images and cropped images based on the extracted ROI.
Customized deep learning models: Two CNN architectures specifically developed for classifying white blood cell (WBC) data are detailed.
3.1. Region of Interest Extraction
Two methodologies are implemented to extract the Region of Interest (ROI) in this study: a color-based method and a deep learning-based method.
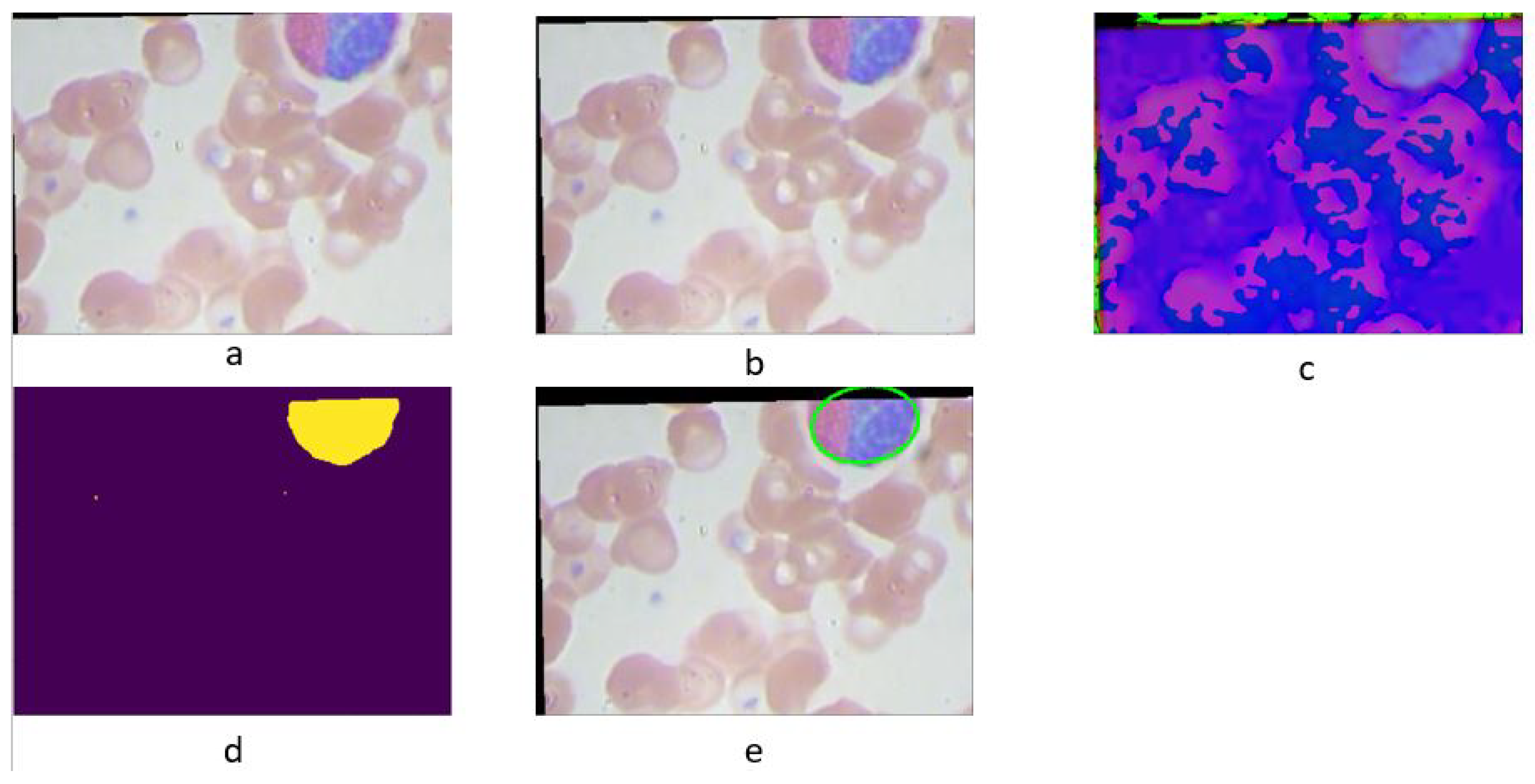
3.1.1. Color-Based ROI Extraction
The color-based method exploits the consistent color range of white blood cells (WBCs) across the dataset as shown in
Figure 3. The ROI extraction algorithm operates through the following steps:
Apply a Gaussian blur (size ) to reduce image noise.
Convert the image to the HSV color space.
Binarize the image within a predefined color range (typically RGB (80, 60, 140) to (255, 255, 255)) that encompasses all WBCs.
Identify the largest contour in the binary image using a contour tracing algorithm.
The contour tracing algorithm, designed to outline the largest object by its external borders, is described below:
|
Algorithm 1: Contour Tracing Algorithm |
 |
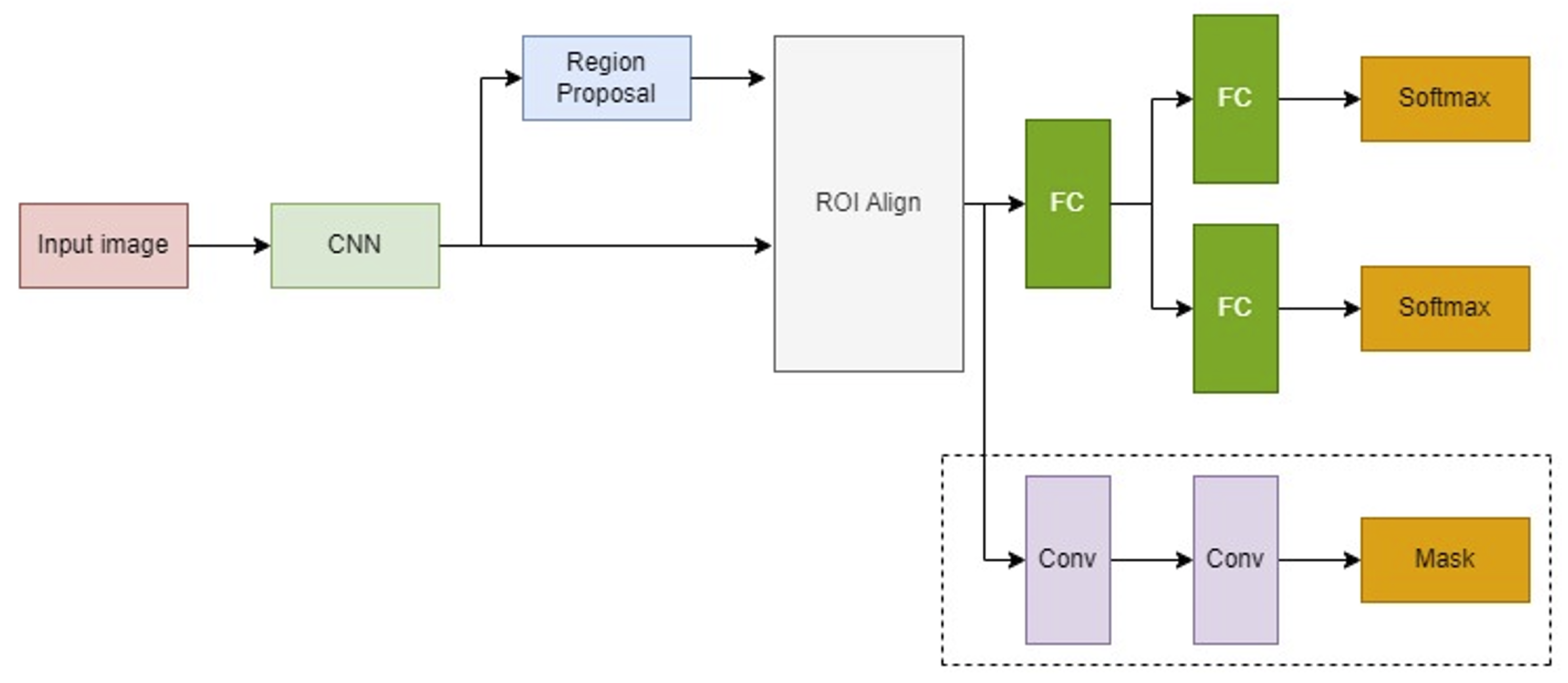
3.1.2. Deep Learning-Based ROI Extraction
The deep learning approach utilizes a Mask R-CNN model, which builds upon the Faster R-CNN by adding a branch for binary mask prediction. This enables precise pixel-wise object segmentation. Mask R-CNN incorporates the ROI Align technique to finely map the predicted masks to the original image coordinates through bilinear interpolation. The architecture of the Mask R-CNN and its operational scheme are illustrated in
Figure 4.
3.2. Customized Deep Learning Models
Two non-pretrained, customized deep learning models are developed, distinct from the standard pretrained models.
3.2.1. Customized Model 1
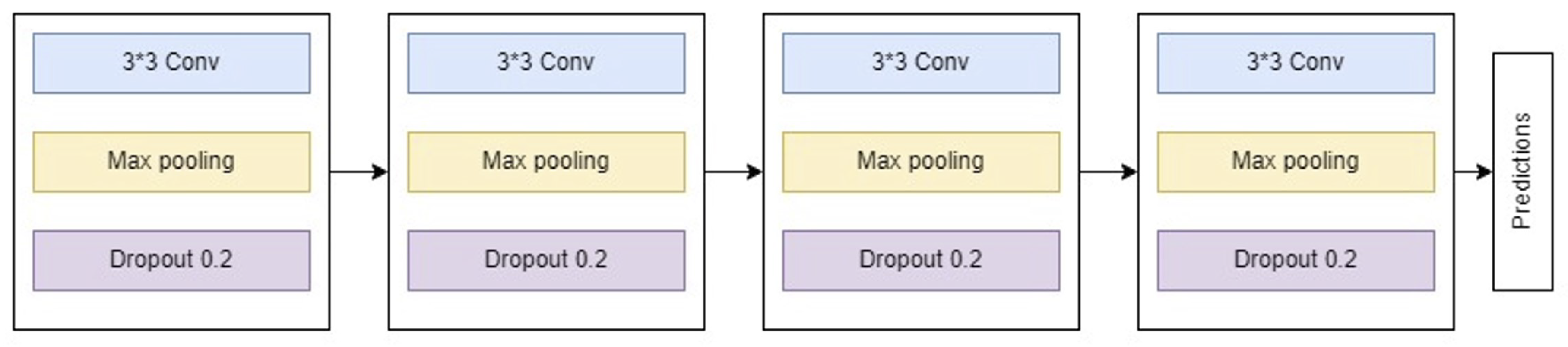
The first model, a traditional convolutional neural network (CNN), consists of four principal blocks:
Block 1: Includes a convolutional layer producing 32 feature maps, followed by a max pooling layer and a dropout layer with a dropout rate of 0.2 to mitigate overfitting.
Block 2: Similar to Block 1 but outputs 64 feature maps.
Block 3: Extends feature map generation to 128 and follows the same structure as previous blocks.
Block 4: Incorporates a flatten layer followed by three dense layers with sizes 64, 128, and 64, respectively. It employs ReLU activation and concludes with a softmax output layer with four nodes.
Figure 5 depicts the architecture of this customized CNN model.
3.2.2. Customized Model 2
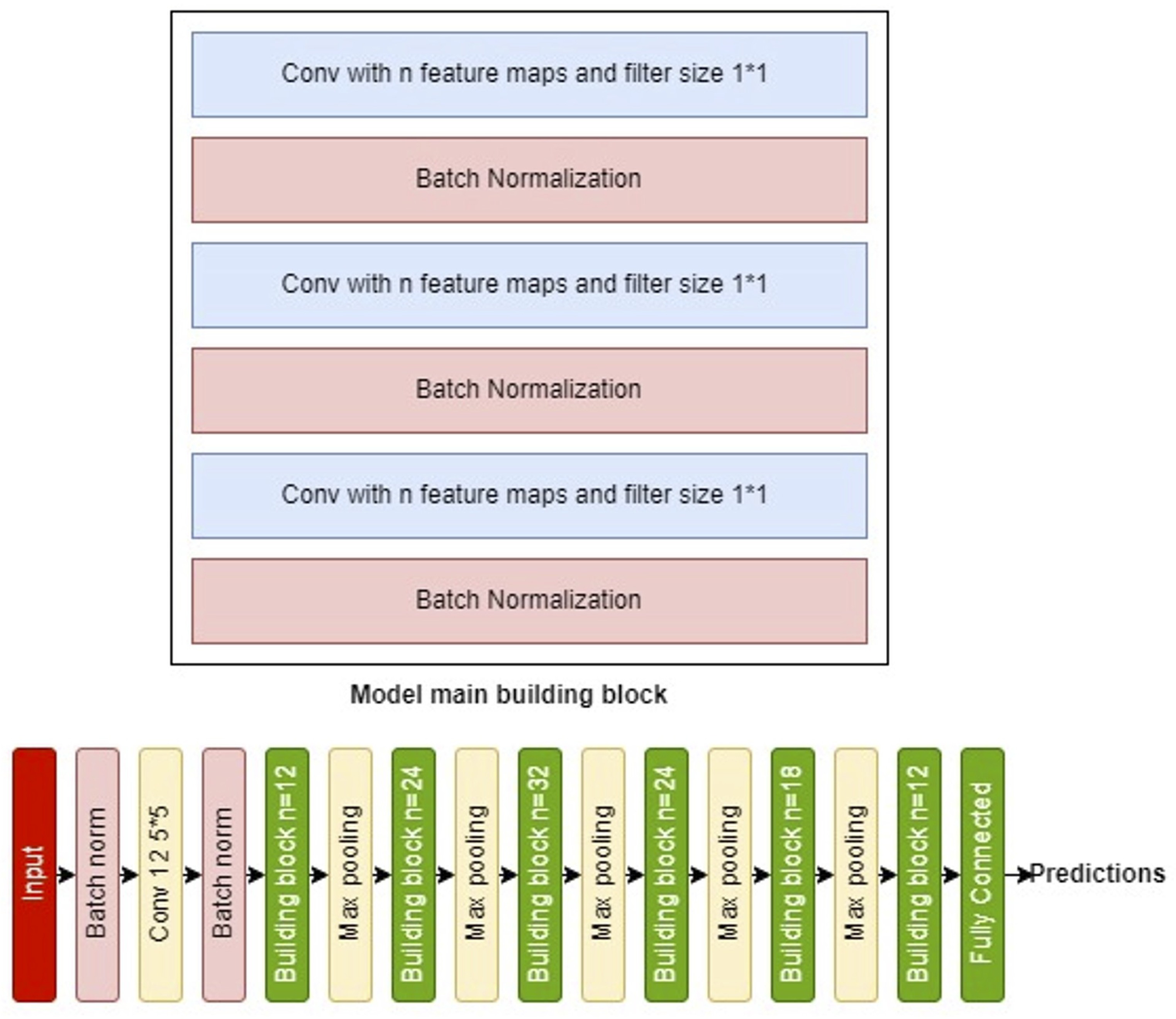
The second model is a deeper architecture, each building block comprising six layers:
Layer 1: Features n convolutional filters for channel-wise pooling and dimensionality reduction.
Layer 2: A batch normalization layer to facilitate higher learning rates and faster convergence, acting also as a regularizer.
Layer 3 and 4: Replicate the structure of the first two layers.
Layer 5: Includes n convolutional filters.
Layer 6: Another batch normalization layer.
The ’same’ convolution mode maintains the input shape, with the output being a concatenation of the features from Layer 4 and Layer 6.
The full model begins with three introductory layers—a batch normalization layer, a
convolution layer with 12 filters, another batch normalization—followed by five repetitions of the described building block and
max pooling, using progressively adjusted filter counts (12, 24, 32, 24, 18, and finally 12). A dense layer with sigmoid activation finalizes the model.
Figure 6 displays both the building block and the complete model architecture.
4. Results
4.1. Dataset Description
The dataset utilized in this study comprises 12,444 microscopic images of blood cells, categorized into four classes of white blood cells (WBCs): eosinophils, lymphocytes, monocytes, and neutrophils. The images were obtained from the publicly available dataset hosted on Kaggle, which can be accessed at
this link. The data is split into training (75%) and testing (25%) sets. The distribution across the four classes in both datasets is shown in
Table 1. All images are 24-bit in depth with a resolution of
pixels.
Figure 1 illustrates samples from the four WBC classes.
4.2. ROI Extraction Experiments
Two distinct methodologies were employed to extract Regions of Interest (ROIs) from microscopic images of White Blood Cells (WBCs): a color-based approach and a deep learning-based method using Mask R-CNN.
4.2.1. Color-Based ROI Extraction
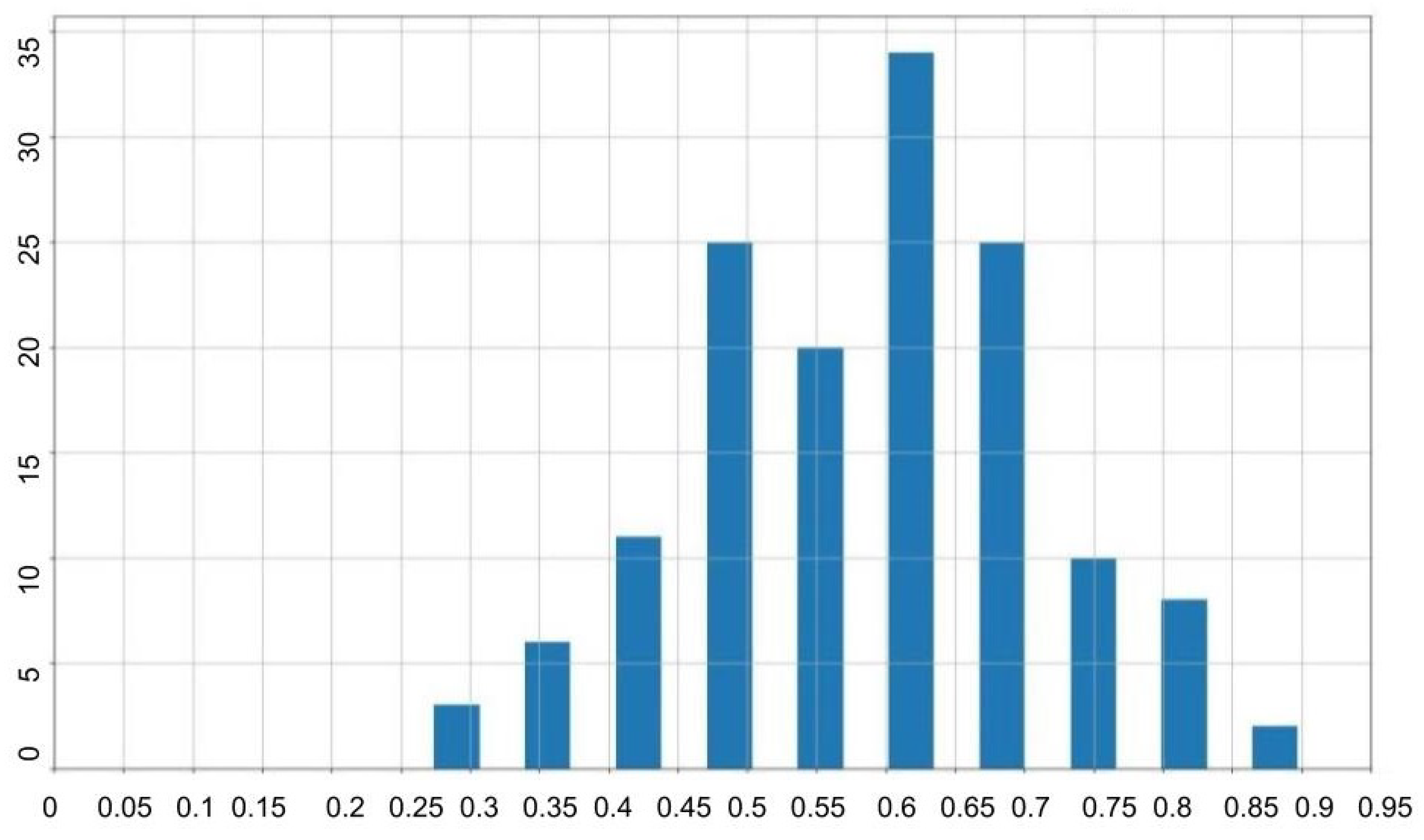
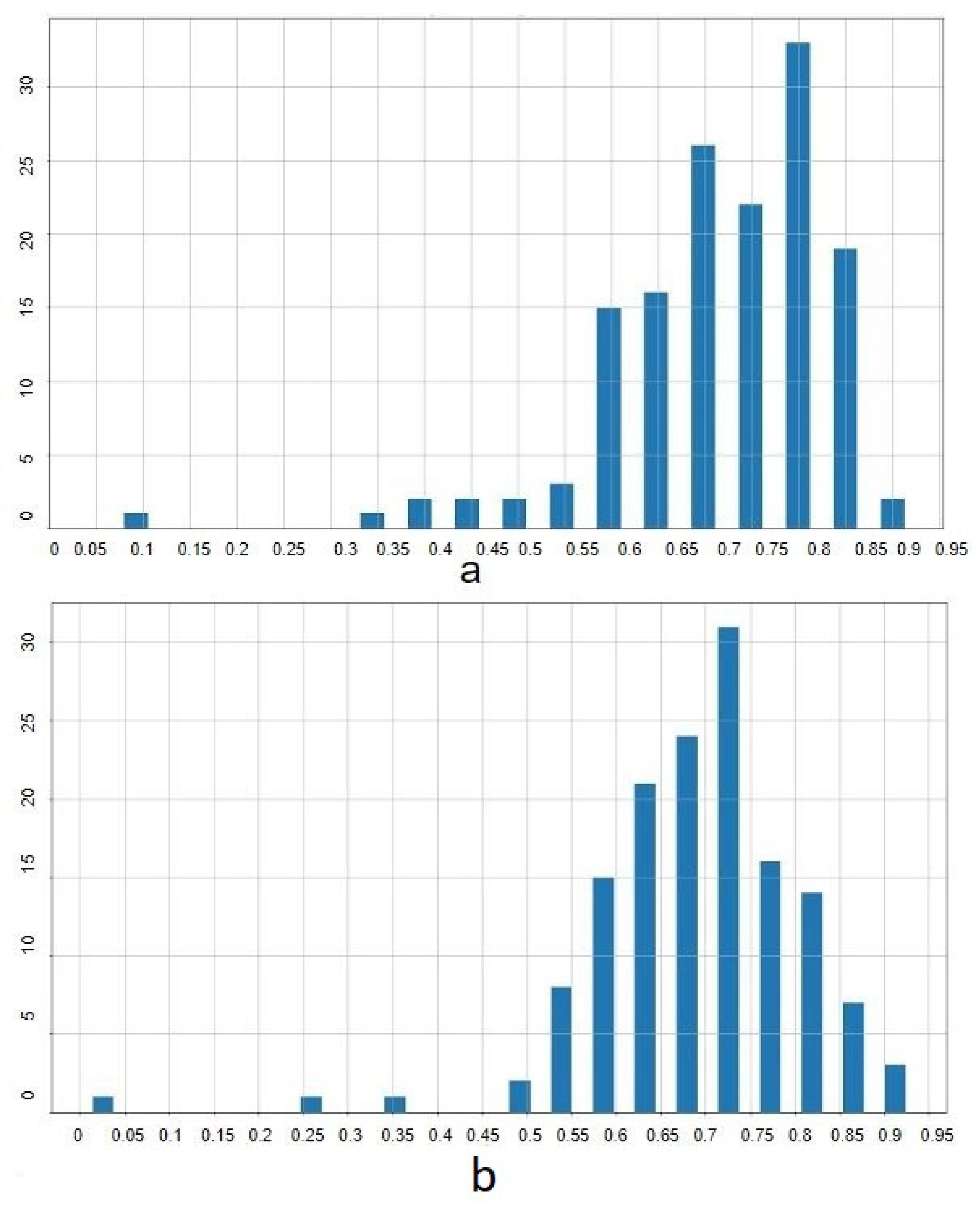
The color-based technique computes bounding boxes by identifying the extremal coordinates that enclose the WBCs based on color thresholds. The effectiveness of this approach was quantified using the Intersection over Union (IoU) metric, which compared the algorithm-generated bounding boxes to the ground truth.
Results indicated an average IoU of 0.59, suggesting moderate accuracy in delineating WBC regions. The IoU values’ distribution is illustrated in
Figure 7, and examples of the algorithm outputs versus the ground truth bounding boxes are shown in
Figure 8, highlighting the method’s reliance on accurate color differentiation.
Table 2 provides a summary of the statistical measures of IoU for the color-based ROI extraction, including mean, minimum, maximum, standard deviation, and median values.
4.2.2. Mask R-CNN ROI Extraction
Mask R-CNN was applied using pretrained networks, specifically VGG16 and ResNet50, described in Sub
Section 3.1.2. The experiment was conducted over 50 epochs with a batch size of 32, using the Adam optimizer. The dataset was partitioned into 220 training images and 144 testing images.
This approach yielded superior results, achieving mean IoU scores of 0.71 with VGG16 and 0.69 with ResNet50. Detailed IoU statistics are provided in
Table 3, and the distribution is depicted in
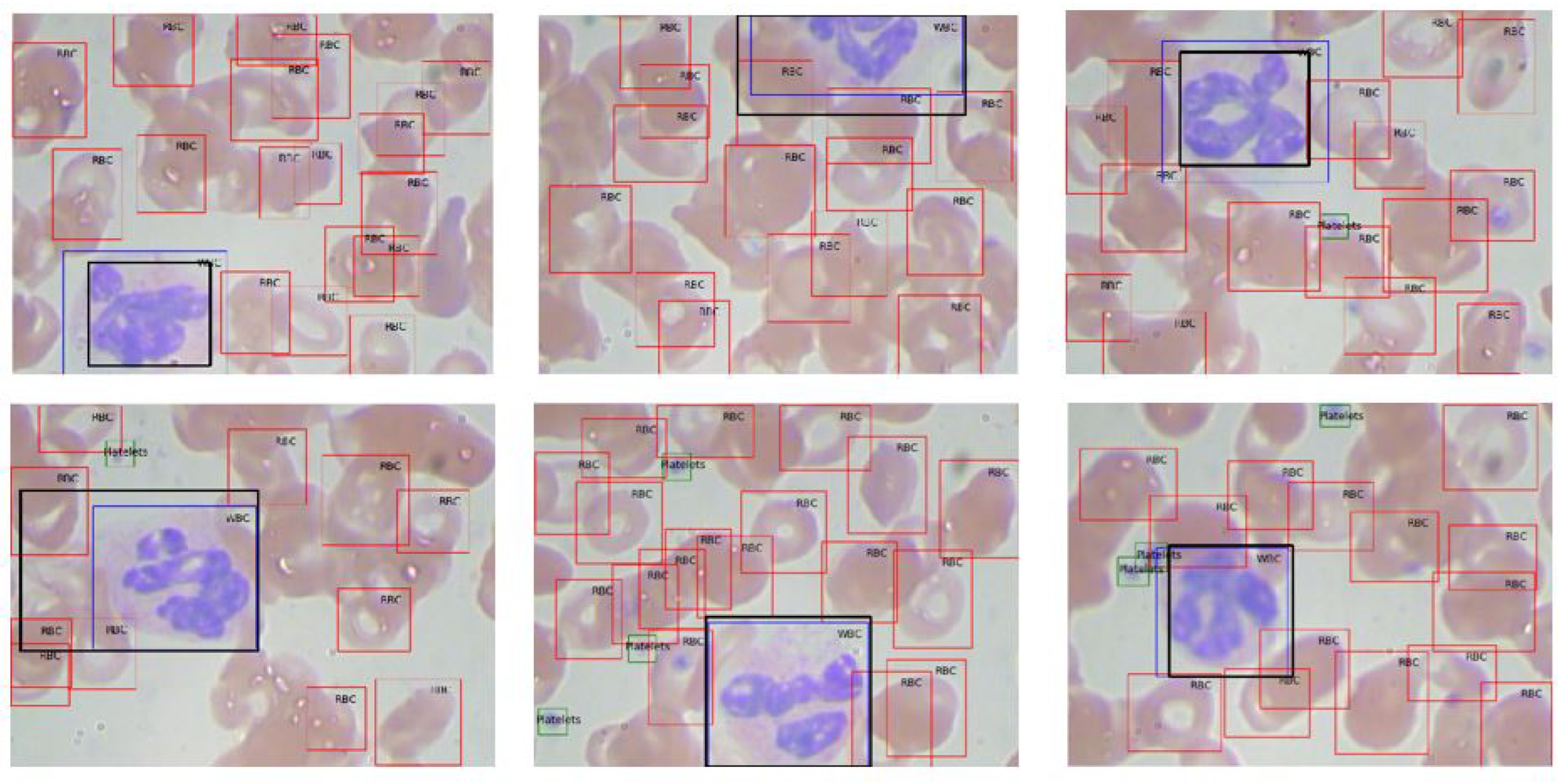
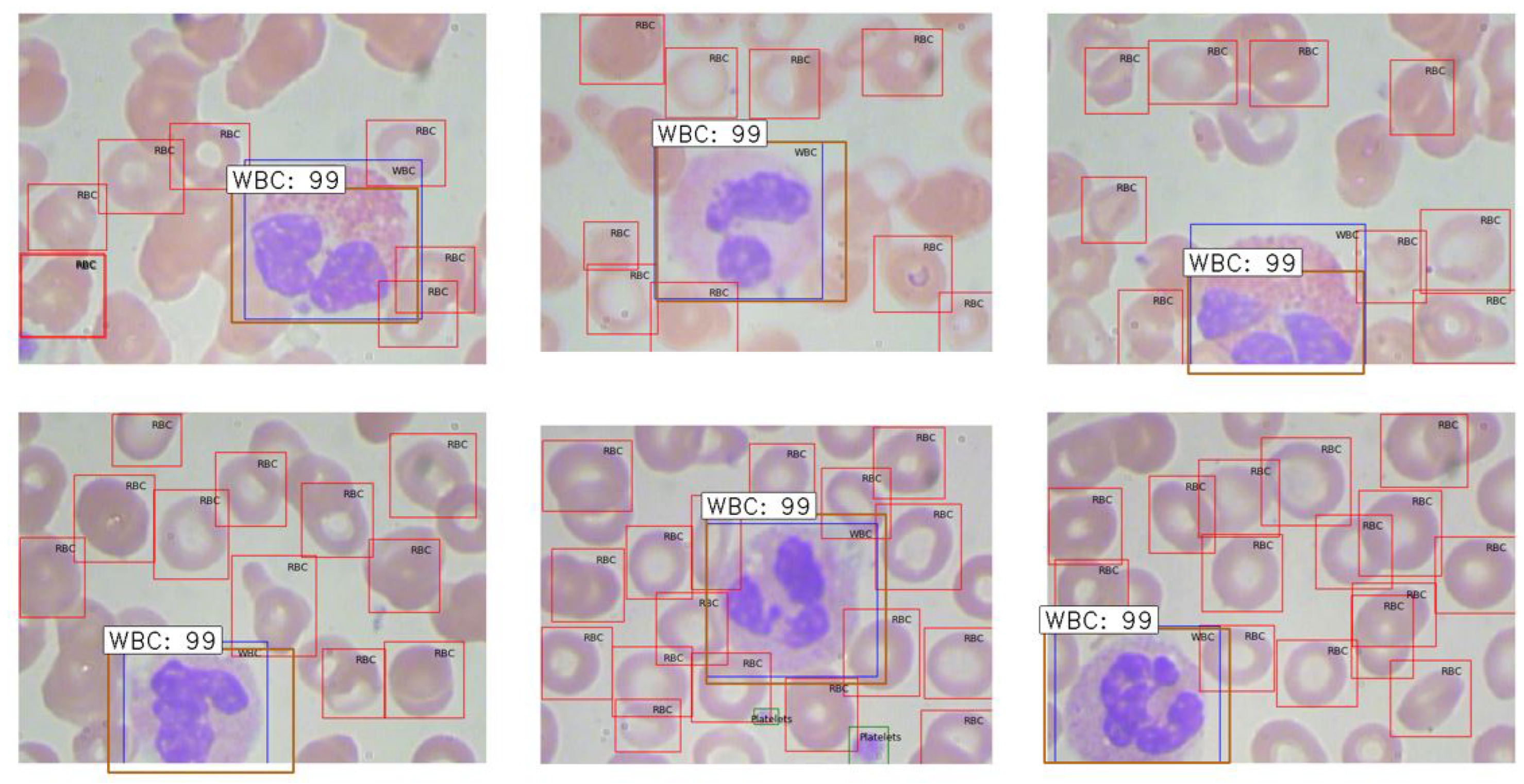
Figure 9. Examples of extracted ROIs using Mask R-CNN are shown in
Figure 10, demonstrating the method’s efficacy in leveraging higher-level features to enhance extraction accuracy.
4.3. Classification Experiments
Extensive classification experiments were conducted using various CNN architectures, including VGG16, ResNet50, and custom models, on both whole images and extracted ROIs. The performance was quantitatively assessed based on classification accuracy, and improvements were noted when models utilized extracted ROIs. This section explores the classification performance of four different WBC types using various CNN architectures as detailed in the following subsections. The results from whole images and regions of interest (ROIs) are compared, with metrics like accuracy and confusion matrices being key evaluative factors.
4.3.1. VGG Classification
Moving to a more complex architecture, VGG16, pre-trained on ImageNet and adapted here by modifying the final layers, provided a noticeable improvement. The model reached an accuracy of 48.4% after 20 training epochs. Although better than LeNet, VGG16 still leaves room for performance enhancement, potentially through more advanced models or further tuning.
4.3.2. ResNet Classification
The ResNet50 model, known for its deep architecture and ability to prevent gradient vanishing, was also pre-trained on ImageNet. With adjustments similar to VGG16, ResNet50 significantly improved the classification accuracy to 74.6%. This model’s deeper layers and residual connections likely helped capture more complex patterns in the WBC images.
4.3.3. Inception Classification
Lastly, the Inception network, utilizing multiple filter sizes at each layer, was tested. Despite its sophisticated architecture, it only achieved 43% accuracy, underscoring some challenges it faced, perhaps due to the distinctive nature of medical imagery compared to ImageNet. Interestingly, it failed to classify any images as Neutrophil, indicating potential issues with feature representation at different scales.
4.3.4. Classification Using the First Customized Model with Whole Images
The initial customized model achieved an 80% accuracy, significantly outperforming the pretrained networks. This model was specifically designed for WBC image analysis, emphasizing simplicity and efficiency in learning from the dataset’s unique characteristics. The performance details are illustrated in the confusion matrix shown in
Table 4.
4.3.5. Classification Using the Second Customized Model with Whole Images
This more complex customized model recorded the highest accuracy of 89.5%, demonstrating its advanced capability to discern intricate features from WBC images. For detailed performance metrics, see
Table 5.
4.4. Impact of ROI Extraction on Classification Outcomes
This section explores how extracting regions of interest (ROIs) that contain white blood cells (WBCs) prior to classification can enhance feature relevance and diminish background noise that might otherwise impair classification accuracy.
The superior performance of the Mask R-CNN algorithm using the VGG pretrained network in extracting ROIs, as discussed in
3.1.2, led to its selection for pre-classification ROI extraction. The subsequent subsections discuss how this step has improved the performance of both pretrained and custom architectures.
4.4.1. Impact of ROI Extraction on Architectural Performances
Implementing ROI extraction has significantly enhanced classification accuracies across different architectures:
VGG16 Results: With the VGG16 architecture, accuracy improved from 48.8% using whole images to 64% with ROIs. Notable improvements were seen in the LYMPHOCYTE and EOSINOPHIL classes, although the NEUTROPHIL class accuracy was notably lower due to misclassifications as EOSINOPHIL.
ResNet50 Results: ROI extraction with ResNet50 led to a substantial gain in performance, increasing accuracy from 74.6% to 84%. This model provided the highest accuracies for LYMPHOCYTE and EOSINOPHIL classes. The most common error involved misclassifying NEUTROPHIL images as MONOCYTE.
Inception Architecture Results: The Inception network also saw improvements with ROI extraction, where classification accuracy rose from 43% to 55%. However, the system struggled with distinguishing between NEUTROPHIL and EOSINOPHIL classes.
These results underscore the effectiveness of ROI extraction in enhancing feature relevancy, thereby boosting the overall classification performance of various deep learning architectures.
4.4.2. Enhancements from ROI Extraction in Customized Architectures
First Customized Architecture: The ROI extraction had a substantial impact on our first customized architecture, boosting accuracy from 81% to 88%. The primary issue observed was misclassifications of MONOCYTE as NEUTROPHIL. Detailed confusion matrix is presented in
Table 6.
Second Customized Architecture: Similarly, the second customized architecture demonstrated the highest improvement, with accuracy improving from 89% to 92%. Misclassifications primarily occurred with LYMPHOCYTE being classified as MONOCYTE. This model’s results, which are summarized in
Table 7, confirm its superiority and suitability for further experiments involving noise resilience in image classification.
4.5. Noise Robustness Testing
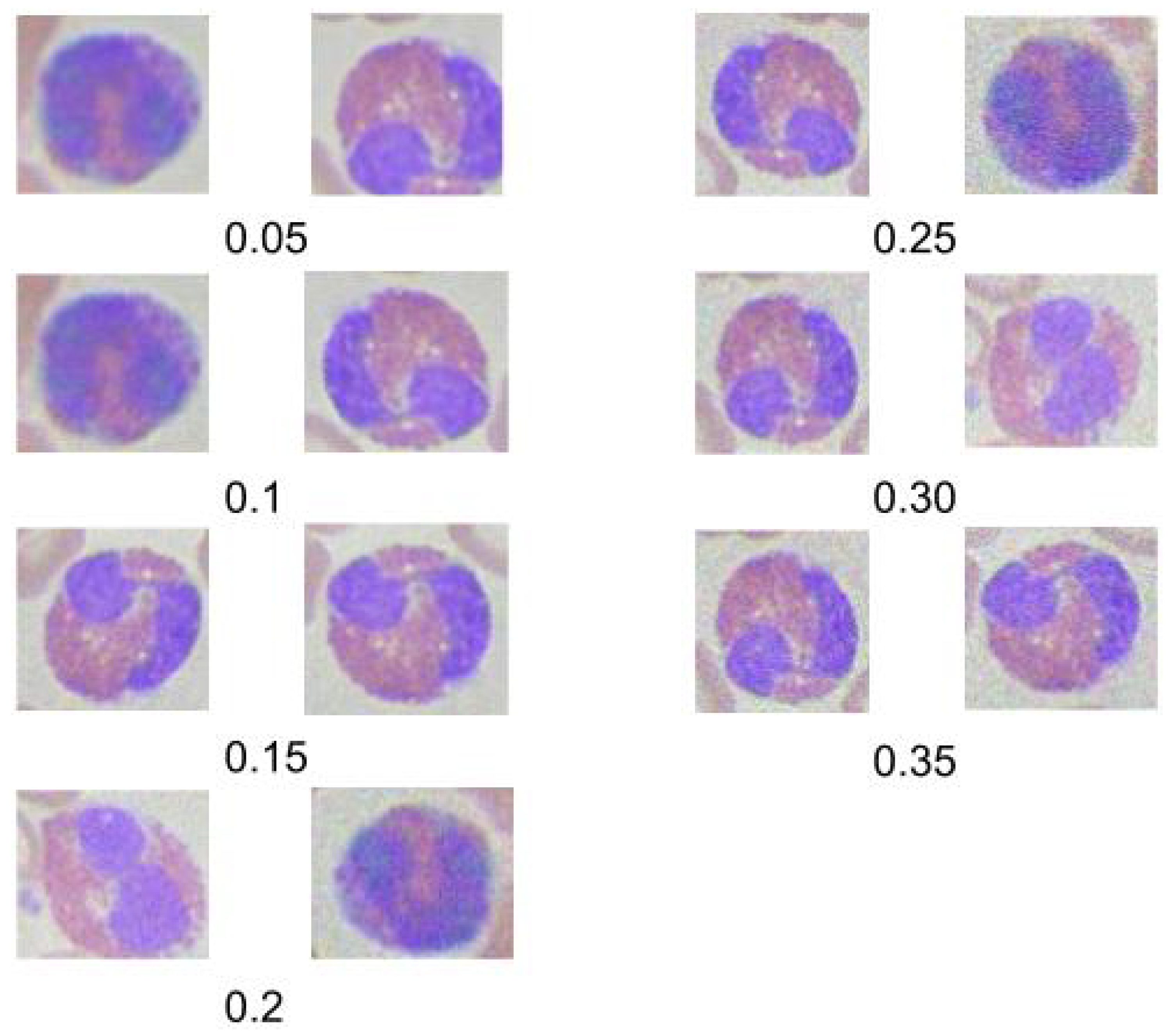
In this section, we explore the impact of introducing Gaussian noise into our testing dataset to evaluate the robustness of the second customized model, which previously demonstrated the highest accuracy. The study focuses on Gaussian noise due to its prevalence in real-world medical imaging scenarios, where electronic and sensor noise often distorts images.
Figure 11 displays examples of images at different
values, illustrating how Gaussian noise affects image clarity. The effect of the noise on classification accuracy is quantified and presented below:
4.5.1. Gaussian Noise
Gaussian noise, characterized by its statistical properties of having a mean of zero and a defined variance, closely simulates the electronic noise encountered in medical imaging devices. To simulate this effect, we added varying levels of Gaussian noise to the test images and assessed the model’s performance.
The Gaussian noise was generated and added to the images using the following equation:
where
I represents the original image,
G is the generated Gaussian noise, and
is the noise factor controlling the signal-to-noise ratio (SNR). We experimented with
values ranging from 0.05 to 0.35 to cover a broad spectrum of common noise intensities.
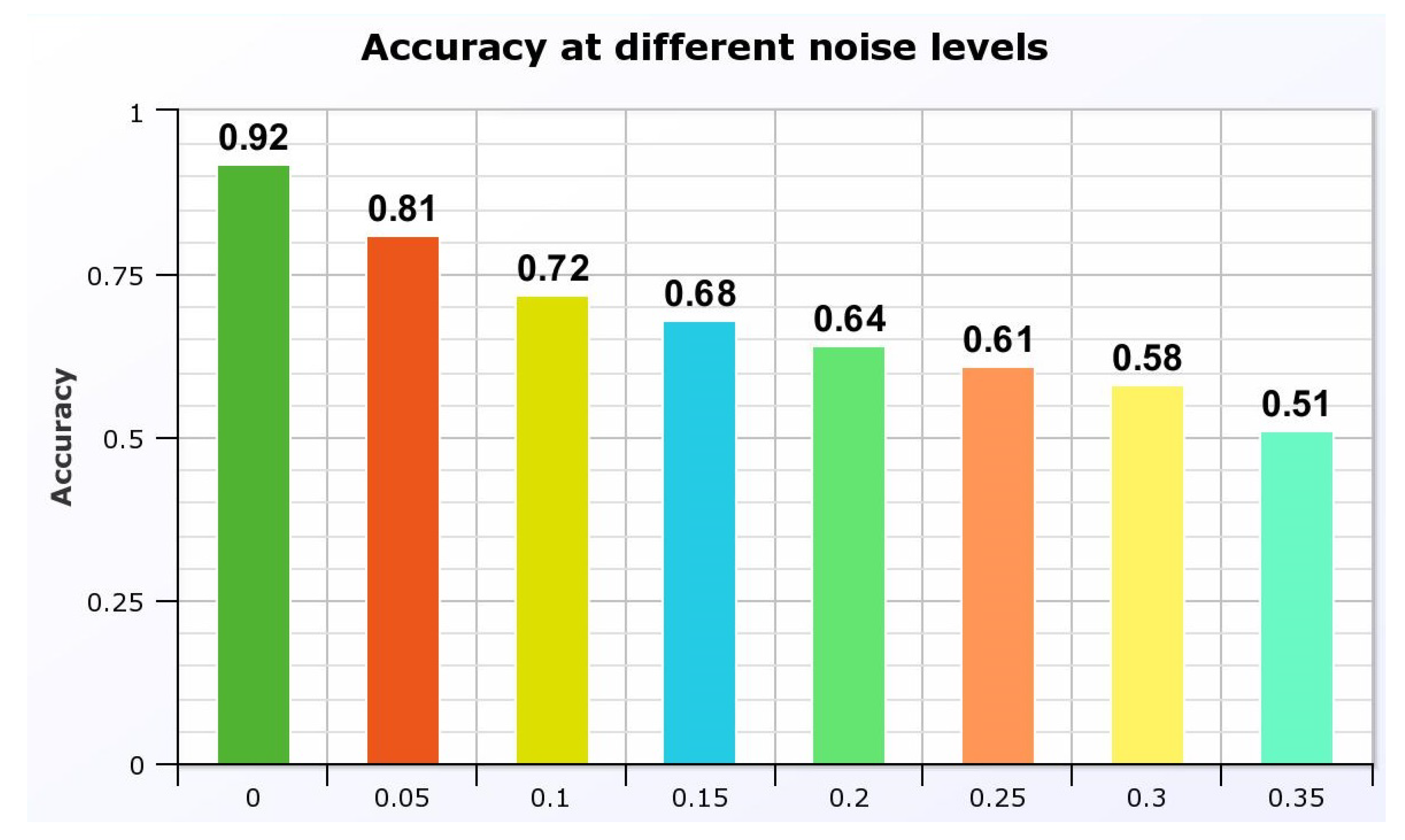
The impact on the model’s accuracy as shown in
Figure 12 was significant, demonstrating a decline as the noise level increased. At
, the model maintained an accuracy of 81%, which dropped to 51% at
. These results are critical as they highlight the need for further refining the model to handle common noise levels effectively, ensuring reliability in practical medical settings.
This experiment underlines the importance of noise handling capabilities in automated diagnostic systems, particularly in environments where electronic interference is prevalent. Future work will aim to enhance the model’s noise resilience, potentially incorporating noise reduction techniques at the preprocessing stage.
5. Discussion
The results demonstrated significant improvements in classification accuracy when ROIs were used, underscoring the importance of focused image analysis in medical diagnostics. Custom models, tailored specifically for the task, outperformed pre-trained models, likely due to better feature alignment with the target domain. Furthermore, the resilience of these models to image noise highlights their potential for deployment in varied clinical settings.
5.1. Region of Interest Extraction
Two methodologies were explored for extracting the region of interest (ROI) from microscopic images of blood cells: a color-based technique and the application of Mask R-CNN using a VGG pretrained architecture. The intersection over union (IoU) metric was used to evaluate both methods, with Mask R-CNN demonstrating superior performance, achieving a mean IoU of 0.71 compared to 0.59 for the color-based method. Detailed performance metrics are presented in
Table 8.
5.2. Classification Experiments
Both pretrained and custom models were assessed using whole images and extracted ROIs. Custom models, especially when utilizing extracted ROIs, consistently outperformed pretrained models. The greatest accuracy improvements were noted when models were applied to extracted ROIs, suggesting that ROI-focused features enhance model learning capabilities. The comparative results of different architectures are shown in
Table 9.
5.2.1. Comparative Analysis
The proposed models were benchmarked against other studies, reflecting not only higher or comparable accuracy but also robustness given the large dataset used. Notably, while some studies achieved slightly higher accuracies, they often relied on substantially smaller datasets, limiting their generalizability. The comparative analysis is summarized in
Table 10.
Overall, the study not only advances the understanding of effective techniques in medical image analysis but also sets a foundation for future research to explore novel architectures and fusion techniques that could offer even better performance and robustness.
Author Contributions
All authors contributed equally to the study’s conception. All authors have read and agreed to the published version of the manuscript.The corresponding author is the main contributor to the coding.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dekhil, O. Computational Techniques in Medical Image Analysis Application for White Blood Cells Classification. PhD Thesis, University of Louisville, Louisville, KY, USA, 2020. Available online: https://doi.org/10.18297/etd/3424.
- Janowczyk, A.; Madabhushi, A. A Comprehensive Review of Deep Learning Applications in Digital Pathology. IEEE Transactions on Medical Imaging 2020. [Google Scholar]
- Zhang, R.; Isola, P.; Efros, A. Enhancing Deep Learning Models with Noise-Robust Training. Journal of Machine Learning Research 2021. [Google Scholar]
- Wang, J.; Li, X.; Hu, Y. Enhancing Noise Robustness in Medical Image Segmentation Using Deep Neural Networks. IEEE Transactions on Medical Imaging 2024. [Google Scholar]
- Rahman, M.A.; Haque, R.; Alam, T. Deep Learning-Based White Blood Cell Classification: A Comparative Study of State-of-the-Art Models. IEEE Access 2023, 11, 11567–11578. [Google Scholar]
- Kumar, S.; Singh, P.; Verma, A.K. Recent Advances in Hematological Image Analysis Using Deep Learning Techniques. Computers in Biology and Medicine 2023, 153, 106319. [Google Scholar]
- Zhao, M.; Feng, Y.; Lin, Z. Exploring Transformer-Based Architectures for Medical Image Analysis. Journal of Medical Imaging 2023, 10, 3–035501. [Google Scholar]
- Daskalov, G.M.; Löffler, E.; Williamson, J.F. Monte Carlo-aided dosimetry of a new high dose-rate brachytherapy source. Med. Phys. 1998, 25, 2200–2208. [Google Scholar] [CrossRef]
- Bielajew, A.F.; Rogers, D.W.O. Electron Step-Size Artefacts and PRESTA. In Monte Carlo Transport of Electrons and Photons; Jenkins, T.M., Nelson, W.R., Rindi, A., Nahum, A.E., Rogers, D.W.O., Eds.; Plenum Press: New York, NY, USA, 1988; pp. 115–137. [Google Scholar]
- Nutbrown, R.F.; Shipley, D.R. Calculation of the Response of a NE2561 Ion Chamber in a Water Phantom for High Energy Photons; NPL Report No. CIRM 40; NPL: Teddington, UK, 2000. [Google Scholar]
- Abramowitz, M.; Stegun, I.A. (Eds.) Handbook of Mathematical Functions with Formulae, Graphs, and Mathematical Tables; National Bureau of Standards: Washington, DC, USA, 1964; Applied Mathematics Series 55. [Google Scholar]
- Siuly, S.; Zhang, Y. Medical Big Data: Neurological Diseases Diagnosis Through Medical Data Analysis. Data Science and Engineering 2016, 1, 54–64. [Google Scholar] [CrossRef]
- Sinha, G.R. CAD Based Medical Image Processing: Emphasis to Breast Cancer Detection. i-Manager’s Journal on Software Engineering 2017, 12, 15. [Google Scholar]
- Abe, K.; Takeo, H.; Nagai, Y.; Kuroki, Y.; Nawano, S. Creation of New Artificial Calcification Shadows for Breast Cancer and Verification of Effectiveness of CAD Development Technique That Uses No Actual Cases. In Proceedings of the 14th International Workshop on Breast Imaging (IWBI 2018), 10718, 1071817; International Society for Opticsand Photonics, 2018. [Google Scholar]
- Sathish, D.; Kamath, S.; Rajagopal, K.V.; Prasad, K. Medical Imaging Techniques and Computer Aided Diagnostic Approaches for the Detection of Breast Cancer with an Emphasis on Thermography—A Review. International Journal of Medical Engineering and Informatics 2016, 8, 275–299. [Google Scholar] [CrossRef]
- Moon, W.K.; Chen, I.-L.; Chang, J.M.; Shin, S.U.; Lo, C.-M.; Chang, R.-F. The Adaptive Computer-Aided Diagnosis System Based on Tumor Sizes for the Classification of Breast Tumors Detected at Screening Ultrasound. Ultrasonics 2017, 76, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Tang, W.; Xu, D.M.; Jirapatnakul, A.C.; Reeves, A.P.; Henschke, C.I.; Yankelevitz, D. Low-dose CT Screening for Lung Cancer: Computer-Aided Detection of Missed Lung Cancers. Radiology 2016, 281, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Chon, A.; Balachandar, N.; Lu, P. Deep Convolutional Neural Networks for Lung Cancer Detection. Standford University 2017. [Google Scholar]
- Giannini, V.; Rosati, S.; Regge, D.; Balestra, G. Specificity Improvement of a CAD System for Multiparametric MR Prostate Cancer Using Texture Features and Artificial Neural Networks. Health and Technology 2017, 7, 71–80. [Google Scholar] [CrossRef]
- Campa, R.; Del Monte, M.; Barchetti, G.; Pecoraro, M.; Salvo, V.; Ceravolo, I.; Indino, E.L.; Ciardi, A.; Catalano, C.; Panebianco, V. Improvement of Prostate Cancer Detection Combining a Computer-Aided Diagnostic System with TRUS-MRI Targeted Biopsy. Abdominal Radiology 2019, 44, 264–271. [Google Scholar] [CrossRef]
- Lemaitre, G.; Martí, R.; Rastgoo, M.; Mériaudeau, F. Computer-Aided Detection for Prostate Cancer Detection Based on Multi-Parametric Magnetic Resonance Imaging. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 3138–3141; IEEE, 2017. [Google Scholar]
- Rasti, R.; Rabbani, H.; Mehridehnavi, A.; Hajizadeh, F. Macular OCT Classification Using a Multi-Scale Convolutional Neural Network Ensemble. IEEE Transactions on Medical Imaging 2017, 37, 1024–1034. [Google Scholar] [CrossRef]
- Brinton, T.J.; Ali, Z.; Di Mario, C.; Hill, J.; Whitbourn, R.; Gotberg, M.; Illindala, U.; Maehara, A.; Van Mieghem, N.; Meredith, I.; et al. Performance of the Lithoplasty System in Treating Calcified Coronary Lesions Prior to Stenting: Results from the Disrupt CAD OCT Sub-Study. Journal of the American College of Cardiology 2017, 69, 1121. [Google Scholar] [CrossRef]
- Usman, M.; Fraz, M.M.; Barman, S.A. Computer Vision Techniques Applied for Diagnostic Analysis of Retinal OCT Images: A Review. Archives of Computational Methods in Engineering 2017, 24, 449–465. [Google Scholar] [CrossRef]
- Charfi, S.; El Ansari, M.; Balasingham, I. Computer-Aided Diagnosis System for Ulcer Detection in Wireless Capsule Endoscopy Images. IET Image Processing 2019, 13, 1023–1030. [Google Scholar] [CrossRef]
- Jani, K.K.; Srivastava, S.; Srivastava, R. Computer Aided Diagnosis System for Ulcer Detection in Capsule Endoscopy Using Optimized Feature Set. Journal of Intelligent & Fuzzy Systems 2019, Preprint, 1–8. [Google Scholar]
- García-Zapirain, B.; Elmogy, M.; El-Baz, A.; Elmaghraby, A.S. Classification of Pressure Ulcer Tissues with 3D Convolutional Neural Network. Medical & Biological Engineering & Computing 2018, 56, 2245–2258. [Google Scholar]
- Su, M.-C.; Cheng, C.-Y.; Wang, P.-C. A Neural-Network-Based Approach to White Blood Cell Classification. The Scientific World Journal 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Young, I.T. The Classification of White Blood Cells. IEEE Transactions on Biomedical Engineering 1972, 4, 291–298. [Google Scholar] [CrossRef]
- Ongun, G.; Halici, U.; Leblebicioglu, K.; Atalay, V.; Beksaç, M.; Beksaç, S. Feature Extraction and Classification of Blood Cells for an Automated Differential Blood Count System. In Proceedings of the IJCNN’01. International Joint Conference on Neural Networks; IEEE, 2001; Volume 4, pp. 2461–2466. [Google Scholar]
- Solomon, C.; Breckon, T. Fundamentals of Digital Image Processing: A Practical Approach with Examples in Matlab. John Wiley & Sons, 2011. [Google Scholar]
- Flusser, J.; Suk, T. Pattern Recognition by Affine Moment Invariants. Pattern Recognition 1993, 26, 167–174. [Google Scholar] [CrossRef]
- Lezoray, O.; Elmoataz, A.; Cardot, H.; Gougeon, G.; Lecluse, M.; Revenu, M. Segmentation of Cytological Image Using Color and Mathematical Morphology. 1998. [Google Scholar]
- Sinha, N.; Ramakrishnan, A.G. Automation of Differential Blood Count. In Proceedings of the TENCON 2003. Conference on Convergent Technologies for Asia-Pacific Region; IEEE, 2003; Volume 2, pp. 547–551. [Google Scholar]
- Hu, C.; Jiang, L.-J.; Bo, J. Wavelet Transform and Morphology Image Segmentation Algorism for Blood Cell. In Proceedings of the 2009 4th IEEE Conference on Industrial Electronics and Applications; IEEE, 2009; pp. 542–545. [Google Scholar]
- Shao, L.; Zhu, F.; Li, X. Transfer Learning for Visual Categorization: A Survey. IEEE Transactions on Neural Networks and Learning Systems 2014, 26, 1019–1034. [Google Scholar] [CrossRef]
- Huh, M.; Agrawal, P.; Efros, A.A. What Makes ImageNet Good for Transfer Learning? arXiv preprint 2016, arXiv:1608.08614 2016. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. International Journal of Computer Vision 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Gopalakrishnan, K.; Khaitan, S.K.; Choudhary, A.; Agrawal, A. Deep Convolutional Neural Networks with Transfer Learning for Computer Vision-Based Data-Driven Pavement Distress Detection. Construction and Building Materials 2017, 157, 322–330. [Google Scholar] [CrossRef]
- Ide, H.; Kurita, T. Improvement of Learning for CNN with ReLU Activation by Sparse Regularization. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), 2684–2691; IEEE, 2017. [Google Scholar]
- Mittal, S. A Survey of FPGA-Based Accelerators for Convolutional Neural Networks. Neural Computing and Applications 2018, 1–31. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. arXiv preprint 2015, arXiv:1502.03167 2015. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-Based Learning Applied to Document Recognition. Proceedings of the IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Chen, F.; Chen, N.; Mao, H.; Hu, H. Assessing Four Neural Networks on Handwritten Digit Recognition Dataset (MNIST). arXiv preprint 2018, arXiv:1811.08278 2018. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv preprint 2014, arXiv:1409.1556 2014. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 770–778; 2016. [Google Scholar]
- Liu, K.; Zhang, M.; Pan, Z. Facial Expression Recognition with CNN Ensemble. In Proceedings of the 2016 International Conference on Cyberworlds (CW), 163–166; IEEE, 2016. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 1–9; 2015. [Google Scholar]
- Suzuki, S. Topological Structural Analysis of Digitized Binary Images by Border Following. Computer Vision, Graphics, and Image Processing 1985, 30, 32–46. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. In Proceedings of the IEEE International Conference on Computer Vision, 2961–2969; 2017. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich Feature Hierarchies for Accurate Object Detection and Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2014; pp. 580–587. [Google Scholar]
- RCNN, Faster; He, K.; Girshick, R.; Sun, J. Towards Real-Time Object Detection with Region Proposal Networks. Kaiming He, Ross Girshick, and Jian Sun 2015.
- Girshick, R. Fast R-CNN. In Proceedings of the IEEE International Conference on Computer Vision, 1440–1448; 2015. [Google Scholar]
- Mohan, V.S.; Vinayakumar, R.; Sowmya, V.; Soman, K.P. Deep Rectified System for High-Speed Tracking in Images. Journal of Intelligent & Fuzzy Systems 2019, 36, 1957–1965. [Google Scholar]
- Wang, Y.; Liu, J.; Mišić, J.; Mišić, V.B.; Lv, S.; Chang, X. Assessing Optimizer Impact on DNN Model Sensitivity to Adversarial Examples. IEEE Access 2019, 7, 152766–152776. [Google Scholar] [CrossRef]
- Shopsin, B.; Friedmann, R.; Gershon, S. Lithium and Leukocytosis. Clinical Pharmacology & Therapeutics 1971, 12, 923–928. [Google Scholar]
- Boxer, L.A.; Greenberg, M.S.; Boxer, G.J.; Stossel, T.P. Autoimmune Neutropenia. New England Journal of Medicine 1975, 293, 748–753. [Google Scholar] [CrossRef]
- Devriendt, K.; Kim, A.S.; Mathijs, G.; Frints, S.G.M.; Schwartz, M.; Van den Oord, J.J.; Verhoef, G.E.G.; Boogaerts, M.A.; Fryns, J.P.; You, D.; et al. Constitutively Activating Mutation in WASP Causes X-Linked Severe Congenital Neutropenia. Nature Genetics 2001, 27, 313–317. [Google Scholar] [CrossRef]
- Dale, D.C.; Bolyard, A.A.; Aprikyan, A. Cyclic Neutropenia. Seminars in Hematology 2002, 39, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Baehner, R.L.; Nathan, D.G. Quantitative Nitroblue Tetrazolium Test in Chronic Granulomatous Disease. New England Journal of Medicine 1968, 278, 971–976. [Google Scholar] [CrossRef] [PubMed]
- Kuijpers, T.W.; Van Lier, R.A.; Hamann, D.; de Boer, M.; Thung, L.Y.; Weening, R.S.; Verhoeven, A.J.; Roos, D. Leukocyte Adhesion Deficiency Type 1 (LAD-1)/Variant. A Novel Immunodeficiency Syndrome Characterized by Dysfunctional Beta2 Integrins. The Journal of Clinical Investigation 1997, 100, 1725–1733. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A.; Hanson, C.A.; Inwards, D.J. How to Interpret and Pursue an Abnormal Complete Blood Cell Count in Adults. Mayo Clinic Proceedings 2005, 80, 923–936. [Google Scholar] [CrossRef]
- Twig, G.; Afek, A.; Shamiss, A.; Derazne, E.; Tzur, D.; Gordon, B.; Tirosh, A. White Blood Cells Count and Incidence of Type 2 Diabetes in Young Men. Diabetes Care 2013, 36, 276–282. [Google Scholar] [CrossRef]
- Bøyum, A. Separation of White Blood Cells. Nature 1964, 204, 793–794. [Google Scholar] [CrossRef]
- Jha, K.K.; Das, B.K.; Dutta, H.S. Detection of Abnormal Blood Cells on the Basis of Nucleus Shape and Counting of WBC. In Proceedings of the 2014 International Conference on Green Computing Communication and Electrical Engineering (ICGCCEE); IEEE, 2014; pp. 1–5. [Google Scholar]
- Marchesi, V.T.; Gowans, J.L. The Migration of Lymphocytes Through the Endothelium of Venules in Lymph Nodes: An Electron Microscope Study. Proceedings of the Royal Society of London. Series B. Biological Sciences 1964, 159, 283–290. [Google Scholar]
- Guo, Y.; Şengür, A.; Ye, J. A Novel Image Thresholding Algorithm Based on Neutrosophic Similarity Score. Measurement 2014, 58, 175–186. [Google Scholar] [CrossRef]
- Shahin, A.I.; Guo, Y.; Amin, K.M.; Sharawi, A.A. A Novel White Blood Cells Segmentation Algorithm Based on Adaptive Neutrosophic Similarity Score. Health Information Science and Systems 2018, 6, 1. [Google Scholar] [CrossRef]
- Sadeghian, F.; Seman, Z.; Ramli, A.R.; Kahar, B.H.A.; Saripan, M.-I. A Framework for White Blood Cell Segmentation in Microscopic Blood Images Using Digital Image Processing. Biological Procedures Online 2009, 11, 196. [Google Scholar] [CrossRef]
- Liu, F.; Zhao, B.; Kijewski, P.K.; Wang, L.; Schwartz, L.H. Liver Segmentation for CT Images Using GVF Snake. Medical Physics 2005, 32, 3699–3706. [Google Scholar] [CrossRef] [PubMed]
- Zack, G.W.; Rogers, W.E.; Latt, S.A. Automatic Measurement of Sister Chromatid Exchange Frequency. Journal of Histochemistry & Cytochemistry 1977, 25, 741–753. [Google Scholar]
- Hegde, R.B.; Prasad, K.; Hebbar, H.; Singh, B.M.K. Development of a Robust Algorithm for Detection of Nuclei and Classification of White Blood Cells in Peripheral Blood Smear Images. Journal of Medical Systems 2018, 42, 110. [Google Scholar] [CrossRef] [PubMed]
- Shahin, A.I.; Guo, Y.; Amin, K.M.; Sharawi, A.A. White Blood Cells Identification System Based on Convolutional Deep Neural Learning Networks. Computer Methods and Programs in Biomedicine 2019, 168, 69–80. [Google Scholar] [CrossRef]
- Kutlu, H.; Avci, E.; Özyurt, F. White Blood Cells Detection and Classification Based on Regional Convolutional Neural Networks. Medical Hypotheses 2020, 135, 109472. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.-Y.; Berg, A.C. SSD: Single Shot Multibox Detector. In Proceedings of the European Conference on Computer Vision, 21–37; Springer, 2016. [Google Scholar]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level Accuracy with 50x Fewer Parameters and <0.5 MB Model Size. arXiv preprint 2016, arXiv:1602.07360 2016. [Google Scholar]
- Tiwari, P.; Qian, J.; Li, Q.; Wang, B.; Gupta, D.; Khanna, A.; Rodrigues, J.J.P.C.; de Albuquerque, V.H.C. Detection of Subtype Blood Cells Using Deep Learning. Cognitive Systems Research 2018, 52, 1036–1044. [Google Scholar] [CrossRef]
- Baydilli, Y.Y.; Atila, Ü. Classification of White Blood Cells Using Capsule Networks. Computerized Medical Imaging and Graphics 2020, 101699. [Google Scholar] [CrossRef]
- Sabour, S.; Frosst, N.; Hinton, G.E. Dynamic Routing Between Capsules. In Advances in Neural Information Processing Systems; 2017; pp. 3856–3866. [Google Scholar]
- Tran, T.; Kwon, O.-H.; Kwon, K.-R.; Lee, S.-H.; Kang, K.-W. Blood Cell Images Segmentation Using Deep Learning Semantic Segmentation. In Proceedings of the 2018 IEEE International Conference on Electronics and Communication Engineering (ICECE), 13–16; IEEE, 2018. [Google Scholar]
- Tareef, A.; Song, Y.; Feng, D.; Chen, M.; Cai, W. Automated Multi-Stage Segmentation of White Blood Cells via Optimizing Color Processing. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), 565–568; IEEE, 2017. [Google Scholar]
- Schalkoff, R.J. Digital Image Processing and Computer Vision; Wiley: New York, NY, USA, 1989. [Google Scholar]
- Wachowiak, M.P.; Elmaghraby, A.S.; Smolikova, R.; Zurada, J.M. Classification and Estimation of Ultrasound Speckle Noise with Neural Networks. In Proceedings of the IEEE International Symposium on Bio-Informatics and Biomedical Engineering, 245–252; IEEE, 2000. [Google Scholar]
- Zhu, H.; Sencan, I.; Wong, J.; Dimitrov, S.; Tseng, D.; Nagashima, K.; Ozcan, A. Cost-effective and Rapid Blood Analysis on a Cell-phone. Lab on a Chip 2013, 13, 1282–1288. [Google Scholar] [CrossRef]
- Blaus, B. Medical Gallery of Blausen Medical 2014. Wiki J Med 2014, 1, 10. [Google Scholar]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Transactions on Knowledge and Data Engineering 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
Figure 1.
Illustration of WBC types: (a) Neutrophils, (b) Lymphocytes, (c) Eosinophils, and (d) Monocytes.
Figure 1.
Illustration of WBC types: (a) Neutrophils, (b) Lymphocytes, (c) Eosinophils, and (d) Monocytes.
Figure 2.
3D rendering of the four WBC types [
85].
Figure 2.
3D rendering of the four WBC types [
85].
Figure 3.
Stages of the color-based ROI extraction: (a) original image, (b) blurred image, (c) HSV space image, (d) binarized image, and (e) extracted contour.
Figure 3.
Stages of the color-based ROI extraction: (a) original image, (b) blurred image, (c) HSV space image, (d) binarized image, and (e) extracted contour.
Figure 4.
The Mask R-CNN architecture, an enhancement of Faster R-CNN with an additional branch for binary mask generation, employs the ROI Align algorithm to accurately map the generated masks to the original image, enhancing precise ROI extraction.
Figure 4.
The Mask R-CNN architecture, an enhancement of Faster R-CNN with an additional branch for binary mask generation, employs the ROI Align algorithm to accurately map the generated masks to the original image, enhancing precise ROI extraction.
Figure 5.
Architecture of the first customized CNN model, illustrating the four operational blocks and layer configurations.
Figure 5.
Architecture of the first customized CNN model, illustrating the four operational blocks and layer configurations.
Figure 6.
Detailed view of the second customized CNN model’s main building block and comprehensive model architecture.
Figure 6.
Detailed view of the second customized CNN model’s main building block and comprehensive model architecture.
Figure 7.
IoU distribution for the color-based extraction method on the test dataset.
Figure 7.
IoU distribution for the color-based extraction method on the test dataset.
Figure 8.
Comparative visualization of bounding boxes: algorithm-generated (black) vs. ground truth (blue).
Figure 8.
Comparative visualization of bounding boxes: algorithm-generated (black) vs. ground truth (blue).
Figure 9.
IoU distribution for Mask R-CNN ROI extraction, comparing (a)VGG16 and (b)ResNet50.
Figure 9.
IoU distribution for Mask R-CNN ROI extraction, comparing (a)VGG16 and (b)ResNet50.
Figure 10.
Illustration of ROI extraction using Mask R-CNN: algorithm-generated (orange) vs. ground truth (blue).
Figure 10.
Illustration of ROI extraction using Mask R-CNN: algorithm-generated (orange) vs. ground truth (blue).
Figure 11.
Sample images at varying levels of Gaussian noise.
Figure 11.
Sample images at varying levels of Gaussian noise.
Figure 12.
Model accuracy at different levels of Gaussian noise.
Figure 12.
Model accuracy at different levels of Gaussian noise.
Table 1.
Number of images in each WBC type for both training and testing datasets.
Table 1.
Number of images in each WBC type for both training and testing datasets.
| WBC Type |
Training |
Testing |
| Eosinophils |
2,497 |
623 |
| Lymphocytes |
2,483 |
620 |
| Monocytes |
2,478 |
620 |
| Neutrophils |
2,499 |
624 |
Table 2.
Summary statistics of IoU for the color-based ROI extraction.
Table 2.
Summary statistics of IoU for the color-based ROI extraction.
| Metric |
Value |
| Mean IoU |
0.59 |
| Minimum IoU |
0.25 |
| Maximum IoU |
0.91 |
| Standard Deviation |
0.12 |
| Median IoU |
0.6 |
Table 3.
IoU mean, minimum, maximum, standard deviation, and median for Mask R-CNN using VGG16 and ResNet50.
Table 3.
IoU mean, minimum, maximum, standard deviation, and median for Mask R-CNN using VGG16 and ResNet50.
| Metric |
VGG16 |
ResNet50 |
| Mean IoU |
0.71 |
0.69 |
| Minimum IoU |
0.06 |
0.002 |
| Maximum IoU |
0.92 |
0.93 |
| Standard Deviation |
0.11 |
0.11 |
| Median IoU |
0.73 |
0.7 |
Table 4.
Confusion matrix for the first customized model (percentages based on row totals).
Table 4.
Confusion matrix for the first customized model (percentages based on row totals).
| Predicted label |
True Label |
| EOSINOPHIL |
LYMPHOCYTE |
MONOCYTE |
NEUTROPHIL |
| EOSINOPHIL |
62.22% |
4.17% |
17.5% |
16.11% |
| LYMPHOCYTE |
5.51% |
91.22% |
1.38% |
1.89% |
| MONOCYTE |
2.76% |
6.67% |
89.66% |
0.92% |
| NEUTROPHIL |
17.44% |
4.13% |
12.78% |
65.65% |
Table 5.
Confusion matrix for the second customized model (percentages based on column totals).
Table 5.
Confusion matrix for the second customized model (percentages based on column totals).
| Predicted label |
True Label |
| EOSINOPHIL |
LYMPHOCYTE |
MONOCYTE |
NEUTROPHIL |
| EOSINOPHIL |
88.39% |
5.78% |
8.06% |
4.97% |
| LYMPHOCYTE |
4.84% |
91.81% |
4.68% |
5.29% |
| MONOCYTE |
5.16% |
0.64% |
78.71% |
2.88% |
| NEUTROPHIL |
1.61% |
1.77% |
8.55% |
86.86% |
Table 6.
The confusion matrix for the first customized architecture with extracted ROIs (percentages based on row totals).
Table 6.
The confusion matrix for the first customized architecture with extracted ROIs (percentages based on row totals).
| Predicted label |
True Label |
| EOSINOPHIL |
LYMPHOCYTE |
MONOCYTE |
NEUTROPHIL |
| EOSINOPHIL |
91.22% |
0.00% |
0.00% |
8.78% |
| LYMPHOCYTE |
0.00% |
99.67% |
0.33% |
0.00% |
| MONOCYTE |
0.00% |
0.75% |
96.48% |
2.76% |
| NEUTROPHIL |
6.80% |
0.00% |
20.25% |
72.95% |
Table 7.
The confusion matrix for the second customized architecture with extracted ROIs (percentages based on row totals).
Table 7.
The confusion matrix for the second customized architecture with extracted ROIs (percentages based on row totals).
| Predicted label |
True Label |
| EOSINOPHIL |
LYMPHOCYTE |
MONOCYTE |
NEUTROPHIL |
| EOSINOPHIL |
89.35% |
5.58% |
2.97% |
2.09% |
| LYMPHOCYTE |
6.37% |
89.28% |
3.85% |
0.50% |
| MONOCYTE |
0.89% |
4.65% |
86.40% |
8.05% |
| NEUTROPHIL |
0.54% |
2.71% |
1.63% |
95.12% |
Table 8.
Performance comparison of ROI extraction methods.
Table 8.
Performance comparison of ROI extraction methods.
| Metric |
Mask R-CNN (VGG16) |
Color-Based |
| Mean IoU |
0.71 |
0.59 |
| Minimum IoU |
0.06 |
0.25 |
| Maximum IoU |
0.92 |
0.91 |
| Standard Deviation |
0.11 |
0.12 |
| Median IoU |
0.73 |
0.6 |
Table 9.
Model performance with whole images vs. extracted ROI.
Table 9.
Model performance with whole images vs. extracted ROI.
| Model |
Whole Images |
Extracted ROI |
| VGG16 |
48.4% |
64% |
| ResNet50 |
74.6% |
84% |
| Inception |
43% |
55% |
| First Custom Model |
80% |
88% |
| Second Custom Model |
89.5% |
92% |
Table 10.
Comparison of model performance in the literature.
Table 10.
Comparison of model performance in the literature.
| Study |
Sample Size |
Accuracy |
| ongun2001feature[30] |
108 |
91% |
| sinha2003automation[34] |
84 |
94% |
| tiwari2018detection[77] |
13,000 |
88% |
| baydilli2020classification[78] |
263 |
92% |
| Proposed Approach |
12,444 |
92.5% |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).