Submitted:
25 September 2024
Posted:
26 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Experimental
2.1. General
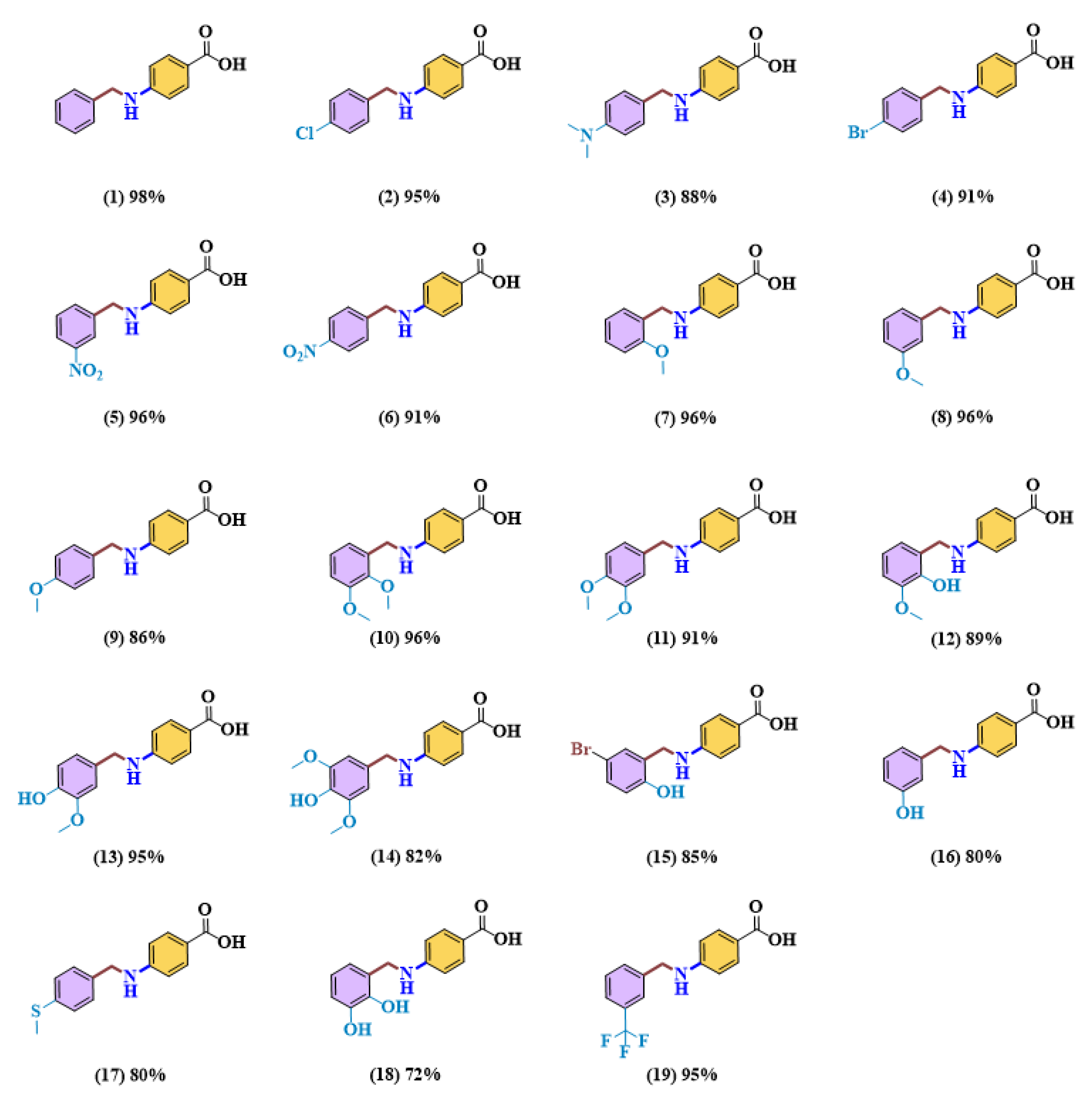
2.2. General Procedure for Reductive Amination Reaction between 4-Aminobenzoic Acid and Benzaldehyde Derivatives to Produce 4-(benzylamino) Benzoic Acid Derivatives
2.2.1. Synthesis of 4-(benzylamino)benzoic Acid (1)
2.2.2. Synthesis of 4-((4-chlorobenzyl)amino)benzoic Acid (2)
2.2.3. Synthesis of 4-((4-(dimethylamino)benzyl)amino)benzoic Acid (3)
2.2.4. Synthesis of 4-((4-bromobenzyl)amino)benzoic Acid (4)
2.2.5. Synthesis of 4-((3-nitrobenzyl)amino)benzoic Acid (5)
2.2.6. Synthesis of 4-((4-nitrobenzyl)amino)benzoic Acid (6)
2.2.7. Synthesis of 4-((2-methoxybenzyl)amino)benzoic Acid (7)
2.2.8. Synthesis of 4-((3-methoxybenzyl)amino)benzoic Acid (8)
2.2.9. Synthesis of 4-((4-methoxybenzyl)amino)benzoic Acid (9)
2.2.10. Synthesis of 4-((2,3-dimethoxybenzyl)amino)benzoic Acid (10)
2.2.11. Synthesis of 4-((3,4-dimethoxybenzyl)amino)benzoic Acid (11)
2.2.12. Synthesis of 4-((2-hydroxy-3-methoxybenzyl)amino)benzoic Acid (12)
2.2.13. Synthesis of 4-((4-hydroxy-3-methoxybenzyl)amino)benzoic Acid (13)
2.2.14. Synthesis of 4-((4-hydroxy-3,5-dimethoxybenzyl)amino)benzoic Acid (14)
2.2.15. Synthesis of 4-((5-bromo-2-hydroxybenzyl)amino)benzoic Acid (15)
2.2.16. Synthesis of 4-((3-hydroxybenzyl)amino)benzoic Acid (16)
2.2.17. Synthesis of 4-((4-(methylthio)benzyl)amino) benzoic Acid (17)
2.2.18. Synthesis of 4-((2,3-dihydroxybenzyl)amino) benzoic Acid (18)
2.2.19. Synthesis of 4-((3-(trifluoromethyl)benzyl)amino) benzoic Acid (19)
2.3. Cell subculture and Growth Conditions
2.4. Cell Viability Assays Using Resazurin Dye Method
2.5. Antibacterial Activity
2.6. Single-Crystal X-ray Diffraction Measurements (SC-XRD)
3. Result and Discussion:
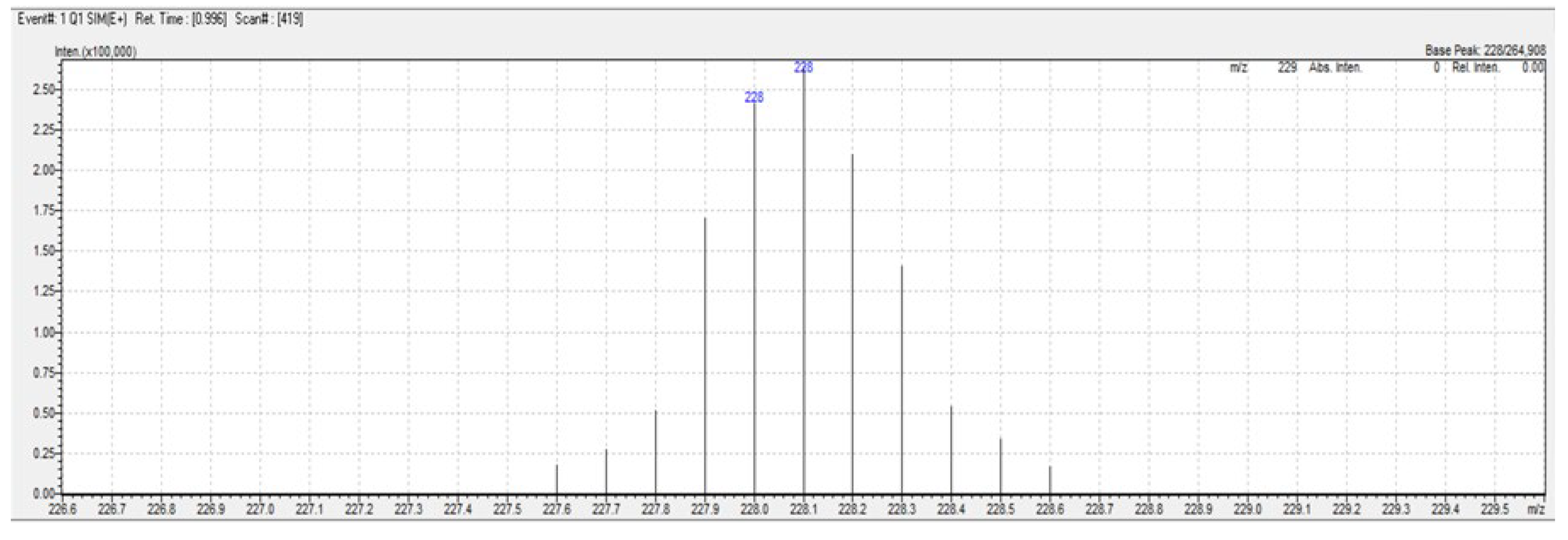
3.1. Elemental Analysis & Mass Spectra
3.2. NMR Spectroscopy
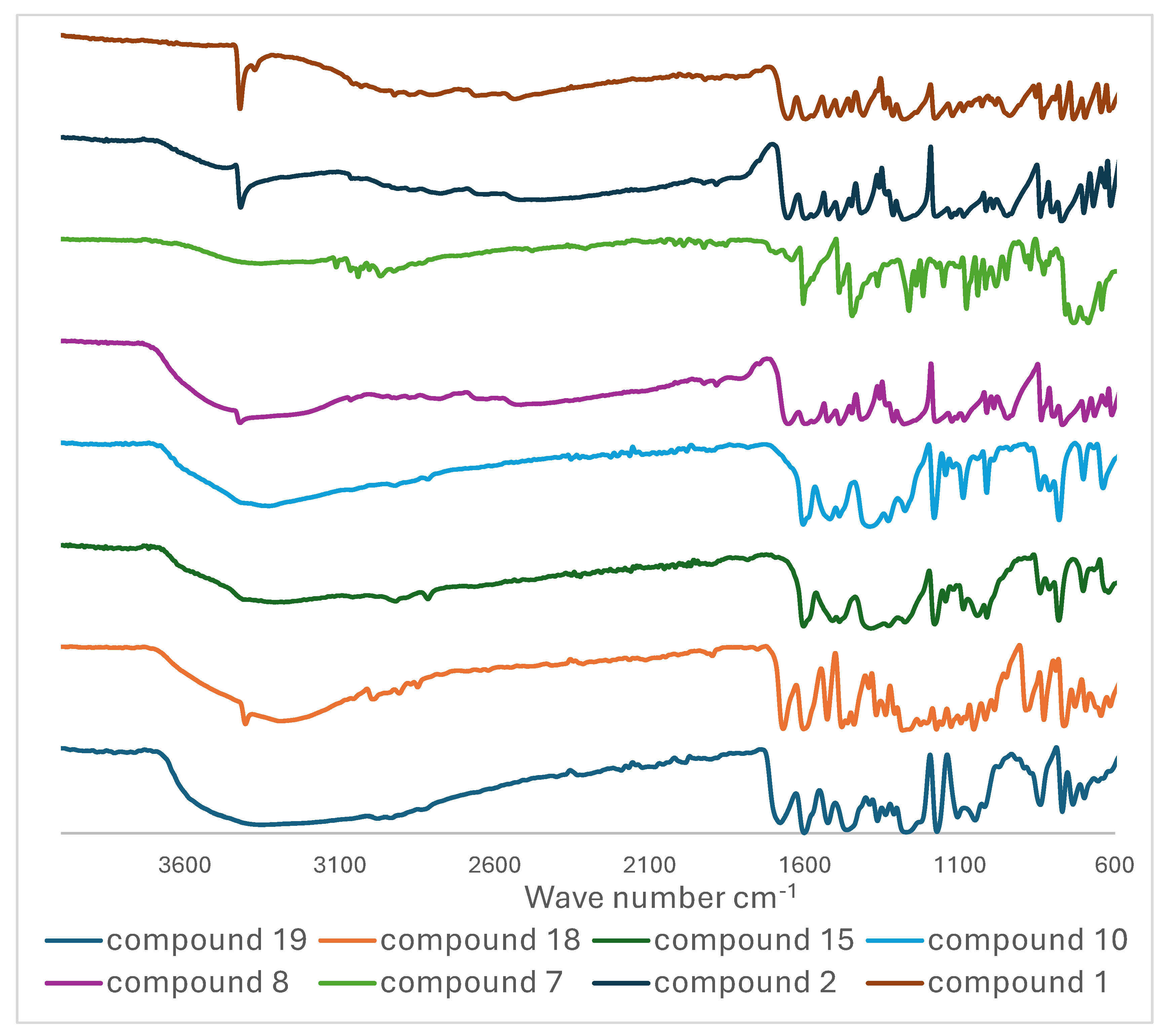
3.3. Infra-Red Spectroscopy
3.4. Molecular Structures
3.5. Anti-Cancer Properties
3.6. Antibacterial Properties
4. Conclusion
Supplementary Materials
Author Contributions
Acknowledgments
References
- K. -J. Lee, G.-H. Lee, H. Kim, M.-S. Oh, I.J. Hwang, J.-y. Lee, A. Choi, C.-i. Kim, H.-M. Park, Determination of heterocyclic amines and acrylamide in agricultural products with liquid chromatography-tandem mass spectrometry, Toxicological Research 2015, 31, 255–264. [CrossRef]
- Y. Liu, X. Li, D. Huang, Y. Liu, H. Wang, D. Di, Comparison of adsorption selectivity for (–)-epigallocatechin gallate and caffeine by porous materials modified with different amino groups, Colloids and Surfaces A: Physicochemical and Engineering Aspects 2017, 520, 166–172. [CrossRef]
- K. Matsuda, F. Hasebe, Y. Shiwa, Y. Kanesaki, T. Tomita, H. Yoshikawa, K. Shin-Ya, T. Kuzuyama, M. Nishiyama, Genome mining of amino group carrier protein-mediated machinery: discovery and biosynthetic characterization of a natural product with unique hydrazone unit, ACS Chemical Biology 2017, 12, 124–131. [CrossRef]
- M. Papageorgiou, D. Lambropoulou, C. Morrison, E. Kłodzińska, J. Namieśnik, J. Płotka-Wasylka, Literature update of analytical methods for biogenic amines determination in food and beverages, TrAC Trends in Analytical Chemistry 2018, 98, 128–142. [CrossRef]
- M. Popko, I. Michalak, R. Wilk, M. Gramza, K. Chojnacka, H. Górecki, Effect of the new plant growth biostimulants based on amino acids on yield and grain quality of winter wheat, Molecules 2018, 23, 470. [CrossRef]
- S. E. Rossiter, M.H. Fletcher, W.M. Wuest, Natural products as platforms to overcome antibiotic resistance, Chemical reviews 2017, 117, 12415–12474. [CrossRef]
- A. I. Ibrahim, H. Abul-Futouh, L.M. Bourghli, M. Abu-Sini, S. Sunoqrot, B. Ikhmais, V. Jha, Q. Sarayrah, D.H. Abulebdah, W.H. Ismail, Design and synthesis of thionated levofloxacin: Insights into a new generation of quinolones with potential therapeutic and analytical applications, Current Issues in Molecular Biology 2022, 44, 4626–4638. [CrossRef]
- X. Shen, X. Chen, J. Chen, Y. Sun, Z. Cheng, Z. Lu, Ligand-promoted cobalt-catalyzed radical hydroamination of alkenes, Nature communications 2020, 11, 783. [CrossRef]
- L. R. Staben, S.G. Koenig, S.M. Lehar, R. Vandlen, D. Zhang, J. Chuh, S.-F. Yu, C. Ng, J. Guo, Y. Liu, Targeted drug delivery through the traceless release of tertiary and heteroaryl amines from antibody–drug conjugates, Nature chemistry 2016, 8, 1112–1119. [CrossRef]
- Y. Tang, D. Lee, J. Wang, G. Li, J. Yu, W. Lin, J. Yoon, Development of fluorescent probes based on protection–deprotection of the key functional groups for biological imaging, Chemical Society Reviews 2015, 44, 5003–5015. [CrossRef]
- Q. Yin, Y. Shi, J. Wang, X. Zhang, Direct catalytic asymmetric synthesis of α-chiral primary amines, Chemical Society Reviews 2020, 49, 6141–6153. [CrossRef]
- S. Kobayashi, H. Ishitani, Catalytic enantioselective addition to imines, Chemical Reviews 1999, 99, 1069–1094. [CrossRef]
- A. Kaithal, B. Chatterjee, C. Gunanathan, Ruthenium-catalyzed selective hydroboration of nitriles and imines, The Journal of Organic Chemistry 2016, 81, 11153–11161. [CrossRef]
- Bhunia, S.R. Sahoo, A. Das, J. Ahmed, P. Sreejyothi, S.K. Mandal, Transition metal-free catalytic reduction of primary amides using an abnormal NHC based potassium complex: integrating nucleophilicity with Lewis acidic activation, Chemical Science 2020, 11, 1848–1854. [Google Scholar] [CrossRef]
- M. Orlandi, D. Brenna, R. Harms, S. Jost, M. Benaglia, Recent developments in the reduction of aromatic and aliphatic nitro compounds to amines, Organic Process Research & Development 2016, 22, 430–445. [CrossRef]
- J. Masuda, S. Kondo, Y. Matsumoto, M. Yamanaka, Gabriel Synthesis of Hexakis (aminomethyl) benzene and Its Derivatization, ChemistrySelect 2018, 3, 6112–6115. [CrossRef]
- C. M. Pearson, J.W. Fyfe, T.N. Snaddon, A regio-and stereodivergent synthesis of homoallylic amines by a one-pot cooperative-catalysis-based allylic alkylation/Hofmann rearrangement strategy, Angewandte Chemie International Edition 2019, 58, 10521–10527. [CrossRef]
- A. K. Ghosh, M. Brindisi, A. Sarkar, The Curtius rearrangement: applications in modern drug discovery and medicinal chemistry, ChemMedChem 2018, 13, 2351–2373. [CrossRef]
- K. Murugesan, M. Beller, R.V. Jagadeesh, Reusable nickel nanoparticles-catalyzed reductive amination for selective synthesis of primary amines, Angewandte Chemie 2019, 131, 5118–5122. [CrossRef]
- K. Murugesan, T. Senthamarai, V.G. Chandrashekhar, K. Natte, P.C. Kamer, M. Beller, R.V. Jagadeesh, Catalytic reductive aminations using molecular hydrogen for synthesis of different kinds of amines, Chemical Society Reviews 2020, 49, 6273–6328. [CrossRef]
- K. Rosenthal, S. Lütz, Recent developments and challenges of biocatalytic processes in the pharmaceutical industry, Current Opinion in Green and Sustainable Chemistry 2018, 11, 58–64. [CrossRef]
- Z. Wu, S. Du, G. Gao, W. Yang, X. Yang, H. Huang, M. Chang, Secondary amines as coupling partners in direct catalytic asymmetric reductive amination, Chemical Science 2019, 10, 4509–4514. [CrossRef]
- H. Zhou, Y. Liu, S. Yang, L. Zhou, M. Chang, One-Pot N-Deprotection and Catalytic Intramolecular Asymmetric Reductive Amination for the Synthesis of Tetrahydroisoquinolines, Angewandte Chemie 2017, 129, 2769–2773. [CrossRef]
- S. D. Roughley, A.M. Jordan, The medicinal chemist’s toolbox: an analysis of reactions used in the pursuit of drug candidates, Journal of medicinal chemistry 2011, 54, 3451–3479. [CrossRef]
- 25. Yun X-J, Ling C, Deng W, Liu Z-J, Yao Z-J. Half-sandwich Ru (II) complexes with N, O-chelate ligands: diverse catalytic activity for amine synthesis in water. Organometallics. 3: 2020;39(21), 2020; -8. [CrossRef]
- A. Y. Sukhorukov, Catalytic reductive amination of aldehydes and ketones with nitro compounds: new light on an old reaction, Frontiers in Chemistry 2020, 8, 215. [CrossRef]
- M. Krátký, K. Konečná, J. Janoušek, M. Brablíková, O. Janďourek, F. Trejtnar, J. Stolaříková, J. Vinšová, 4-Aminobenzoic acid derivatives: converting folate precursor to antimicrobial and cytotoxic agents, Biomolecules 2019, 10, 9. [CrossRef]
- B. Marbois, L.X. Xie, S. Choi, K. Hirano, K. Hyman, C.F. Clarke, para-Aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae, Journal of Biological Chemistry 2010, 285, 27827–27838. [CrossRef]
- G. J. Basset, E.P. Quinlivan, S. Ravanel, F. Rébeillé, B.P. Nichols, K. Shinozaki, M. Seki, L.C. Adams-Phillips, J.J. Giovannoni, J.F. Gregory III, Folate synthesis in plants: the p-aminobenzoate branch is initiated by a bifunctional PabA-PabB protein that is targeted to plastids, Proceedings of the National Academy of Sciences 2004, 101, 1496–1501. [CrossRef]
- S. Akberova, New biological properties of p-aminobenzoic acid, Biology Bulletin of the Russian Academy of Sciences 2002, 29, 390–393. [CrossRef]
- F. P. Gasparro, M. Mitchnick, J.F. Nash, A review of sunscreen safety and efficacy, Photochemistry and photobiology 1998, 68, 243–256.
- H. Flindt-Hansen, P. Thune, T. Eeg Larsen, The inhibiting effect of PABA on photocarcinogenesis, Archives of dermatological research 1990, 282, 38–41.
- H. M. Patel, V. Bhardwaj, P. Sharma, M.N. Noolvi, S. Lohan, S. Bansal, A. Sharma, Quinoxaline-PABA bipartite hybrid derivatization approach: Design and search for antimicrobial agents, Journal of Molecular Structure 2019, 1184, 562–568. [CrossRef]
- S. Nikfar, M. S. Nikfar, M. Jaberidoost, Dyes and colorants, Dyes and Colorants. In Encyclopedia of Toxicology: Third Edition (pp. 252–261). 2014. [Google Scholar] [CrossRef]
- S. Gutiérrez-Tarriño, S. Rojas-Buzo, C.W. Lopes, G. Agostini, J.J. Calvino, A. Corma, P. Oña-Burgos, Cobalt nanoclusters coated with N-doped carbon for chemoselective nitroarene hydrogenation and tandem reactions in water, Green Chemistry 2021, 23, 4490–4501. [CrossRef]
- K. Long, T.A. Edwards, A.J. Wilson, Microwave assisted solid phase synthesis of highly functionalized N-alkylated oligobenzamide α-helix mimetics, Bioorganic & medicinal chemistry 2013, 21, 4034–4040. [CrossRef]
- T. Strassmaier, S.R. Kirk, T. Banerji, J.W. Karpen, Block of cyclic nucleotide-gated channels by tetracaine derivatives: role of apolar interactions at two distinct locations, Bioorganic & medicinal chemistry letters 2008, 18, 645–649. [CrossRef]
- E. Dikusar, Synthesis of (E)-2-methoxy-6-(R-imino) methylphenols and 2-methoxy-6-(R-amino) methylphenols, Russian Journal of General Chemistry 2012, 82, 693–696. [CrossRef]
- H. M. König, R. Abbel, D. Schollmeyer, A.F. Kilbinger, Solid-phase synthesis of oligo (p-benzamide) foldamers, Organic Letters 2006, 8, 1819–1822. [CrossRef]
- J. -M. Yang, R. Jiang, L. Wu, X.-P. Xu, S.-Y. Wang, S.-J. Ji, In (OTf) 3 catalyzed N-benzylation of amines utilizing benzyl alcohols in water, Tetrahedron 2013, 69, 7988–7994. [CrossRef]
- Abusara, S. Freeman, H. S. Aojula, Pentapeptides for the treatment of small cell lung cancer: Optimisation by Nind-alkyl modification of the tryptophan side chain, European journal of medicinal chemistry 2017, 137, 221–232. [Google Scholar] [CrossRef]
- H. Abumansour, O.H. H. Abumansour, O.H. Abusara, W. Khalil, H. Abul-Futouh, A.I. Ibrahim, M.K. Harb, D.H. Abulebdah, W.H. a: Ismail, Biological evaluation of levofloxacin and its thionated derivatives, 2024; -11. [Google Scholar] [CrossRef]
- K Hijazi, Z. A Taha, A. M Ajlouni, W. M Al-Momani, I. M Idris, E. A Hamra, Synthesis and biological activities of lanthanide (III) nitrate complexes with N-(2-hydroxynaphthalen-1-yl) methylene) nicotinohydrazide Schiff Base, Medicinal Chemistry 2017, 13, 77–84. [Google Scholar] [CrossRef]
- A. K. Hijazi, Z.A. Taha, N.J. Abuhamad, W.M. Al-Momani, Synthesis, and characterization of some Cu (I) complexes having propionitrile and pyridine moieties: An investigation on their antibacterial properties, Journal of Saudi Chemical Society 2021, 25, 101387. [CrossRef]
- Dolomanov, L.J. Bourhis, R. J. Gildea, J.A. Howard, H. Puschmann, OLEX2: a complete structure solution, refinement and analysis program, Journal of applied crystallography 2009, 42, 339–341. [Google Scholar] [CrossRef]
- G. M. Sheldrick, Crystal structure refinement with SHELXL, Acta Crystallographica Section C: Structural Chemistry 2015, 71, 3–8. [CrossRef]
- A. H. Delcour, Outer membrane permeability and antibiotic resistance, Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics 2009, 1794, 808–816.
- Z.A. Taha, A.K. Z.A. Taha, A.K. Hijazi, W.M. S: Al Momani, Lanthanide complexes of the tridentate Schiff base ligand salicylaldehyde-2-picolinoylhydrazone, 1220. [Google Scholar] [CrossRef]
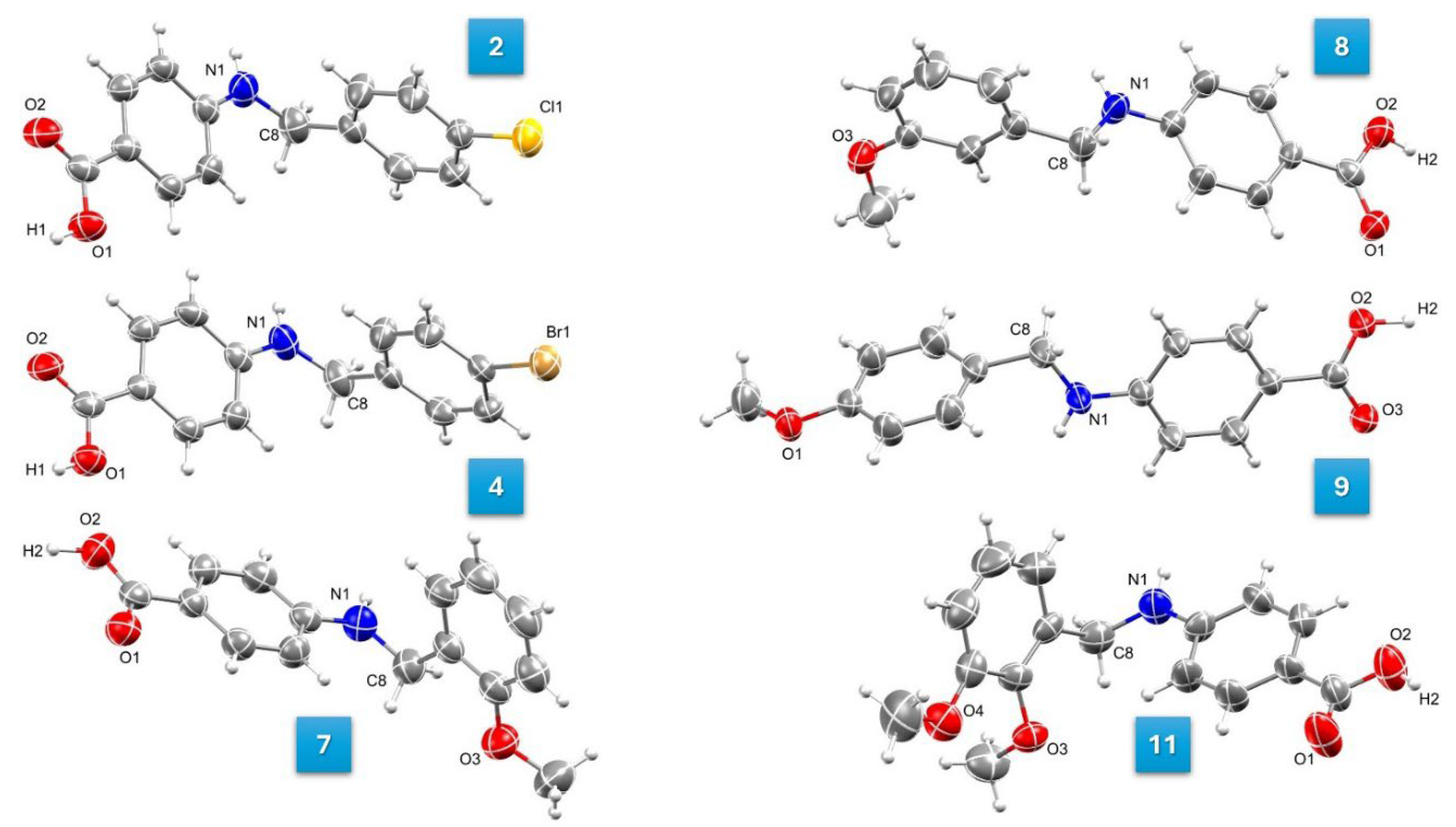
| Compound | 2 | 4 | 7 | 8 | 9 | 11 |
|---|---|---|---|---|---|---|
| CCDC deposition number | 2379919 | 2379920 | 2379921 | 2379922 | 2379923 | 2379924 |
| Empirical formula | C14H12ClNO2 | C14H12BrNO2 | C15H15NO3 | C15H15NO3 | C15H15NO3 | C16H17NO4 |
| Formula weight | 261.70 | 306.16 | 257.28 | 257.28 | 257.28 | 287.30 |
| Crystal system | triclinic | triclinic | monoclinic | monoclinic | triclinic | monoclinic |
| Space group | P-1 | P-1 | P21/c | P21/c | P-1 | P21/c |
| Cell metric a/Å | 5.9490(4) | 5.9204(2) | 12.0311(8) | 8.2327(16) | 8.082(2) | 10.763(4) |
| b/Å | 8.6353(5) | 8.7920(3) | 10.0807(6) | 28.167(5) | 10.565(3) | 10.915(4) |
| c/Å | 12.4159(7) | 12.3226(5) | 12.2667(9) | 11.239(2) | 16.078(4) | 25.978(9) |
| α/° | 97.416(3) | 95.916(2) | 90 | 90 | 93.781(10) | 90 |
| β/° | 96.441(3) | 97.411(2) | 118.669(3) | 93.263(7) | 103.741(9) | 101.871(7) |
| γ/° | 103.954(3) | 100.743(2) | 90 | 90 | 96.597(10) | 90 |
| Cell volume/Å3 | 606.98(6) | 619.57(4) | 1305.34(15) | 2602.0(9) | 1318.5(6) | 2986.5(18) |
| Molecules per cell Z | 2 | 2 | 4 | 8 | 4 | 4 |
| ρcalcg/cm3 | 1.432 | 1.641 | 1.309 | 1.314 | 1.296 | 0.639 |
| μ/mm-1 | 0.307 | 3.310 | 0.092 | 0.092 | 0.091 | 0.046 |
| Electron per cell F(000) | 272.0 | 308.0 | 544.0 | 1088.0 | 544.0 | 608.0 |
| Crystal size/mm3 | 0.853 × 0.341 × 0.28 | 0.758 × 0.694 × 0.414 | ? × ? × ? | 0.993 × 0.364 × 0.362 | 0.73 × 0.531 × 0.524 | 0.767 × 0.572 × 0.258 |
| Radiation | MoKα (λ = 0.71073) | MoKα (λ = 0.71073) | MoKα (λ = 0.71073) | MoKα (λ = 0.71073) | MoKα (λ = 0.71073) | MoKα (λ = 0.71073) |
| 2Θ range for data collection/° | 4.926 to 41.582 | 4.756 to 46.896 | 5.588 to 43.176 | 4.642 to 38.322 | 5.244 to 46.684 | 4.062 to 34.272 |
| Index ranges | -5 ≤ h ≤ 5, -8 ≤ k ≤ 8, -12 ≤ l ≤ 12 | -6 ≤ h ≤ 6, -9 ≤ k ≤ 9, -13 ≤ l ≤ 13 | -12 ≤ h ≤ 12, -10 ≤ k ≤ 10, -12 ≤ l ≤ 12 | -7 ≤ h ≤ 7, -25 ≤ k ≤ 25, -10 ≤ l ≤ 10 | -8 ≤ h ≤ 9, -11 ≤ k ≤ 11, -17 ≤ l ≤ 17 | -8 ≤ h ≤ 8, -9 ≤ k ≤ 9, -21 ≤ l ≤ 20 |
| Reflections collected | 7350 | 12314 | 9379 | 29492 | 28182 | 9063 |
| Independent reflections | 1254 [Rint = 0.0267, Rsigma = 0.0185] | 1809 [Rint = 0.0292, Rsigma = 0.0180] | 1510 [Rint = 0.0413, Rsigma = 0.0266] | 2127 [Rint = 0.0450, Rsigma = 0.0174] | 3766 [Rint = 0.0427, Rsigma = 0.0247] | 1742 [Rint = 0.0554, Rsigma = 0.0387] |
| Data/restraints/parameters | 1254/0/168 | 1809/0/168 | 1510/0/178 | 2127/0/351 | 3766/0/354 | 1742/0/393 |
| Goodness-of-fit on F2 | 1.084 | 1.123 | 1.086 | 1.118 | 1.046 | 1.071 |
| Final R indexes [I>=2σ (I)] | R1 = 0.0281, wR2 = 0.0669 | R1 = 0.0237, wR2 = 0.0530 | R1 = 0.0346, wR2 = 0.0849 | R1 = 0.0354, wR2 = 0.0882 | R1 = 0.0384, wR2 = 0.0896 | R1 = 0.0341, wR2 = 0.0779 |
| Final R indexes [all data] | R1 = 0.0348, wR2 = 0.0711 | R1 = 0.0295, wR2 = 0.0565 | R1 = 0.0552, wR2 = 0.0996 | R1 = 0.0459, wR2 = 0.0991 | R1 = 0.0577, wR2 = 0.1024 | R1 = 0.0564, wR2 = 0.0896 |
| Largest diff. peak/hole / e Å-3 | 0.13/-0.13 | 0.27/-0.35 | 0.13/-0.14 | 0.13/-0.13 | 0.15/-0.14 | 0.11/-0.13 |
| Compound | IC50 (± SEM) µM; n=3 | ||
|---|---|---|---|
| HGF (Fibroblasts; normal cell line) |
A549 Non-Small Cell Lung Cancer cell line |
H69 Small Cell Lung Cancer cell line |
|
| 1 | >100 | >100 | >100 |
| 2 | |||
| 3 | |||
| 4 | |||
| 5 | |||
| 6 | |||
| 7 | |||
| 8 | |||
| 9 | |||
| 10 | |||
| 11 | |||
| 12 | |||
| 13 | |||
| 14 | |||
| 15 | |||
| 16 | |||
| 17 | |||
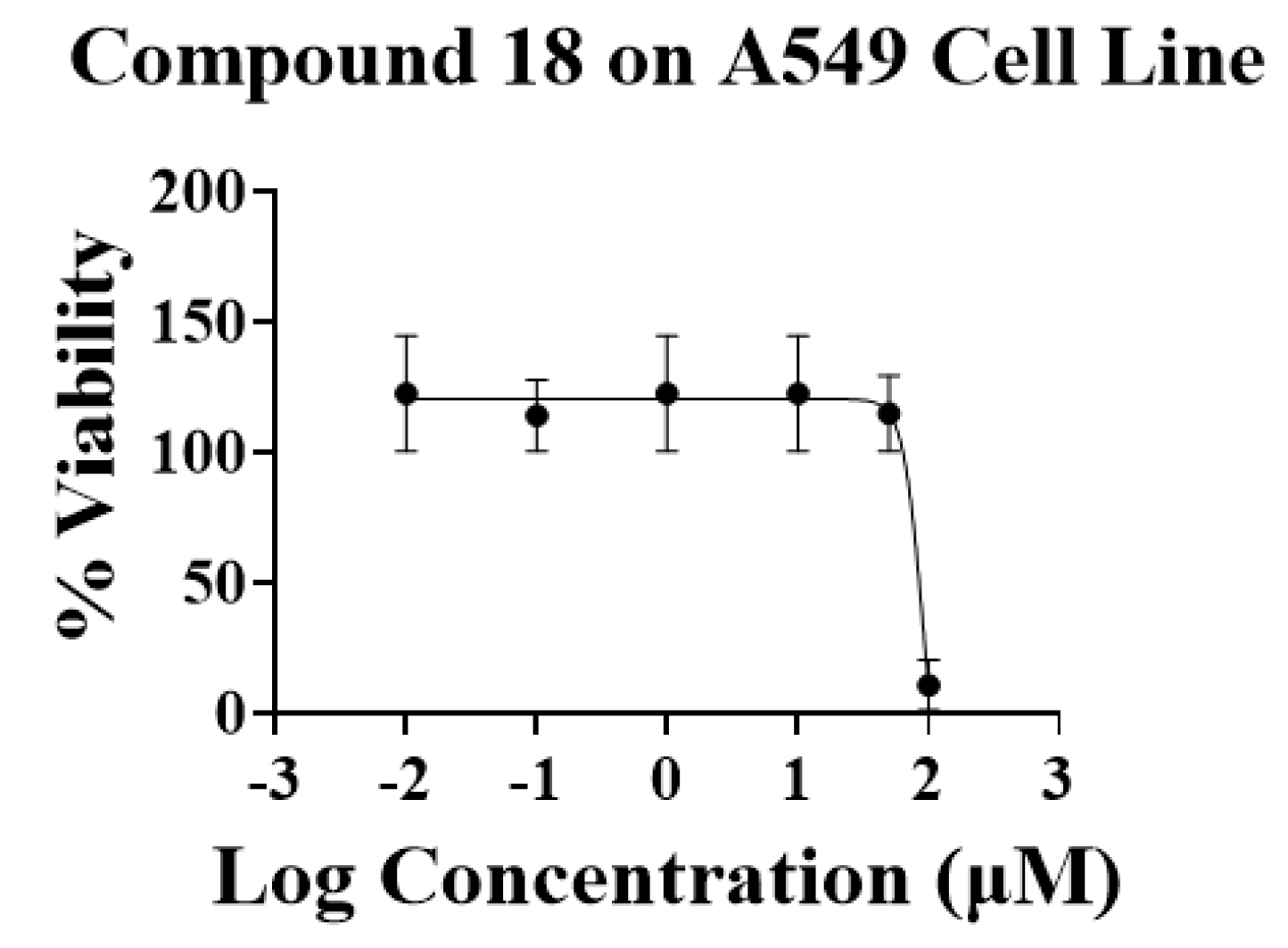
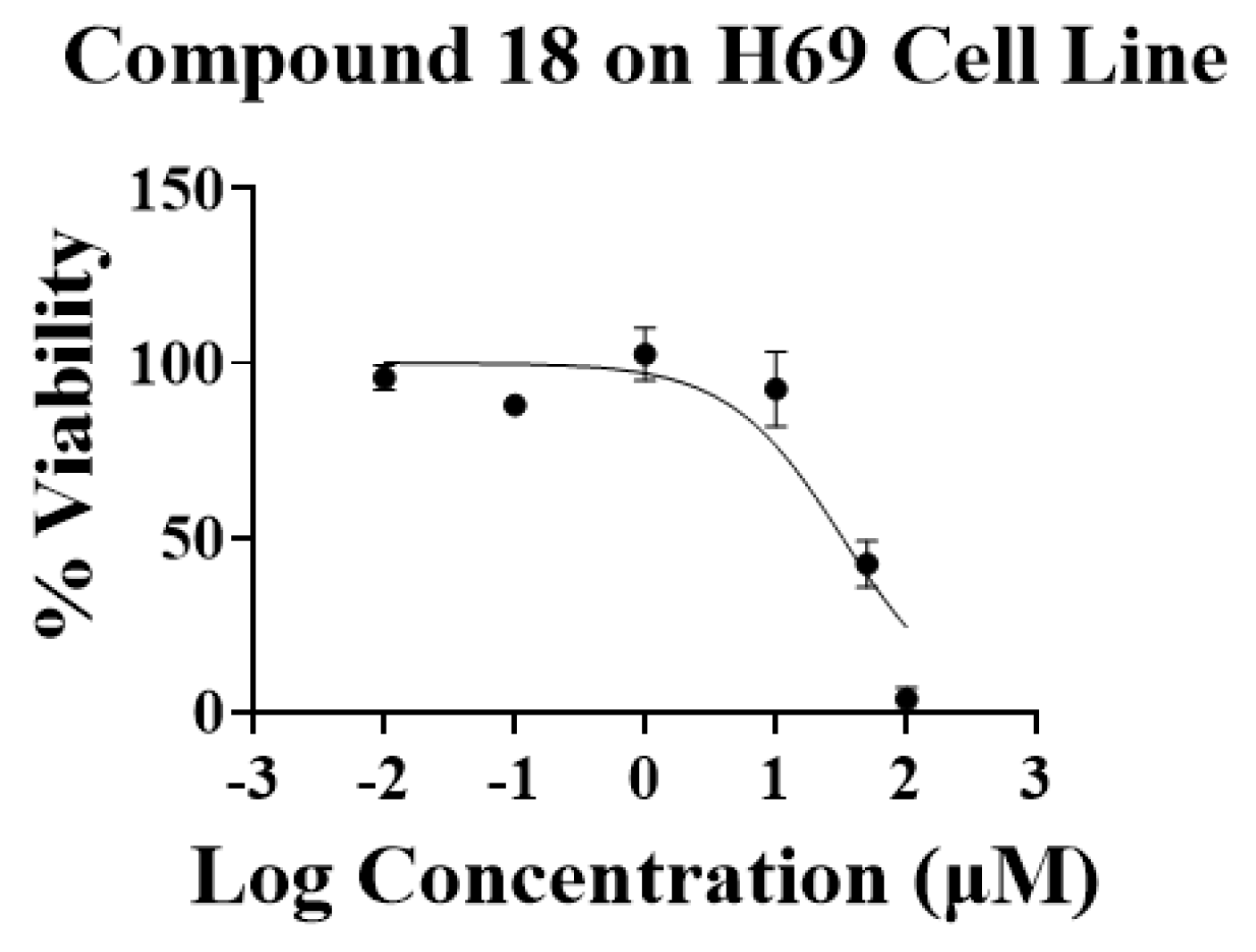
| 18 | 90.69 ± 10.23 | 32.22 ± 7.08 | |
| 19 | >100 | >100 | |
| Compound | Minimum Inhibitory Concentration (MIC, µg/ mL) | |||||
|---|---|---|---|---|---|---|
| Gram-Positive | Gram-Negative | |||||
| En | Sa | MRSA | Ec | Kp | Se | |
| 1 | N | N | N | N | N | N |
| 2 | 128 | N | N | N | 64 | 64 |
| 3 | N | N | N | N | 64 | 64 |
| 4 | N | N | N | N | N | N |
| 5 | N | N | N | N | 64 | 64 |
| 6 | N | N | N | N | N | N |
| 7 | 128 | N | N | N | 64 | 64 |
| 8 | 256 | N | N | N | 64 | 64 |
| 9 | N | N | N | N | N | N |
| 10 | 256 | N | N | N | 64 | 64 |
| 11 | N | N | N | N | 64 | 64 |
| 12 | N | N | N | N | N | N |
| 13 | N | N | N | N | 64 | 64 |
| 14 | N | N | N | N | 64 | 64 |
| 15 | 64 | N | N | N | 64 | 128 |
| 16 | N | N | N | N | 64 | 64 |
| 17 | N | N | N | N | 256 | N |
| 18 | N | N | N | N | 256 | N |
| 19 | N | N | N | N | 256 | N |
| DMSO (-ve control) | N | N | N | N | N | N |
| Amoxicillin (+ve control) | 16 | 16 | 16 | 32 | 16 | 16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





