1. Introduction
Food allergy (FA) is a hypersensitivity reaction to food mediated by immune system. Clinical symptoms include both local and systemic symptoms that could be life-threating in the most severe cases. In the last decades, prevalence of food allergy has increased, and it is estimated to be about 13% in Europe [
1], becoming a public health issue. In routine clinical settings, diagnosis of FA is based on clinical presentation and in vitro specific IgE to whole-allergen extracts. The oral provocation test, the gold standard for the diagnosis of FA, is performed only by expert allergists due to its safety profile. Indeed, it is limited to cases with an inconclusive clinical pathway. The development of component resolved diagnostics has gained much attention in the last years, specifically in poly-sensitized patients where the sensitization patterns are complex, and recognition of primary sensitization and cross-reactivity remains challenging. In the Mediterranean area, many food allergies are due to lipid-transfer proteins (LTPs) allergens contained in fresh fruits, nuts, wheat and vegetables.
LTPs are non-glycosylated proteins anchored to plasma membrane. Molecular structure of LTPs is highly conserved and comprises of four alpha helices stabilized by disulphide bridges with a characteristic hydrophobic internal cavity. Biological role of LTPs is extracellular transfer and deposition of various lipids to form complex barrier macromolecules on the surface of leaves, roots, and seeds [
2]. LTPs sensitization has not reported in America and Africa, while it is highly prevalent in Mediterranean area. For example, it has been demonstrated that the prevalence of Pru p 3 sensitization is about 9-12% in countries such as Italy or Spain [
3,
4]. Sensitization can be asymptomatic in some cases, but it can also elicit severe and systemic reactions. Overall, variability of clinical presentation and different geographic distribution of sensitization profiles have led to significant uncertainty about the clinical reliability of in vitro LTPs allergen molecular diagnostics [
5]. Although the accuracy of specific IgE to LTPs allergens remains unclear, its use in clinical practice is increasing due to the availability of new in vitro diagnostic systems. Indeed, diagnostic accuracy of component resolved diagnostics has been evaluated in several metanalysis, but few data about LTPs have been reported [6-8]. The aim of this systematic review was to describe the diagnostic accuracy of molecular-based LTPs allergens serological tests (specific IgE to LTPs) for the diagnosis of FA in symptomatic patients using oral food challenge as diagnostic reference standard.
2. Results
2.1. Study Characteristics and Risk of Bias
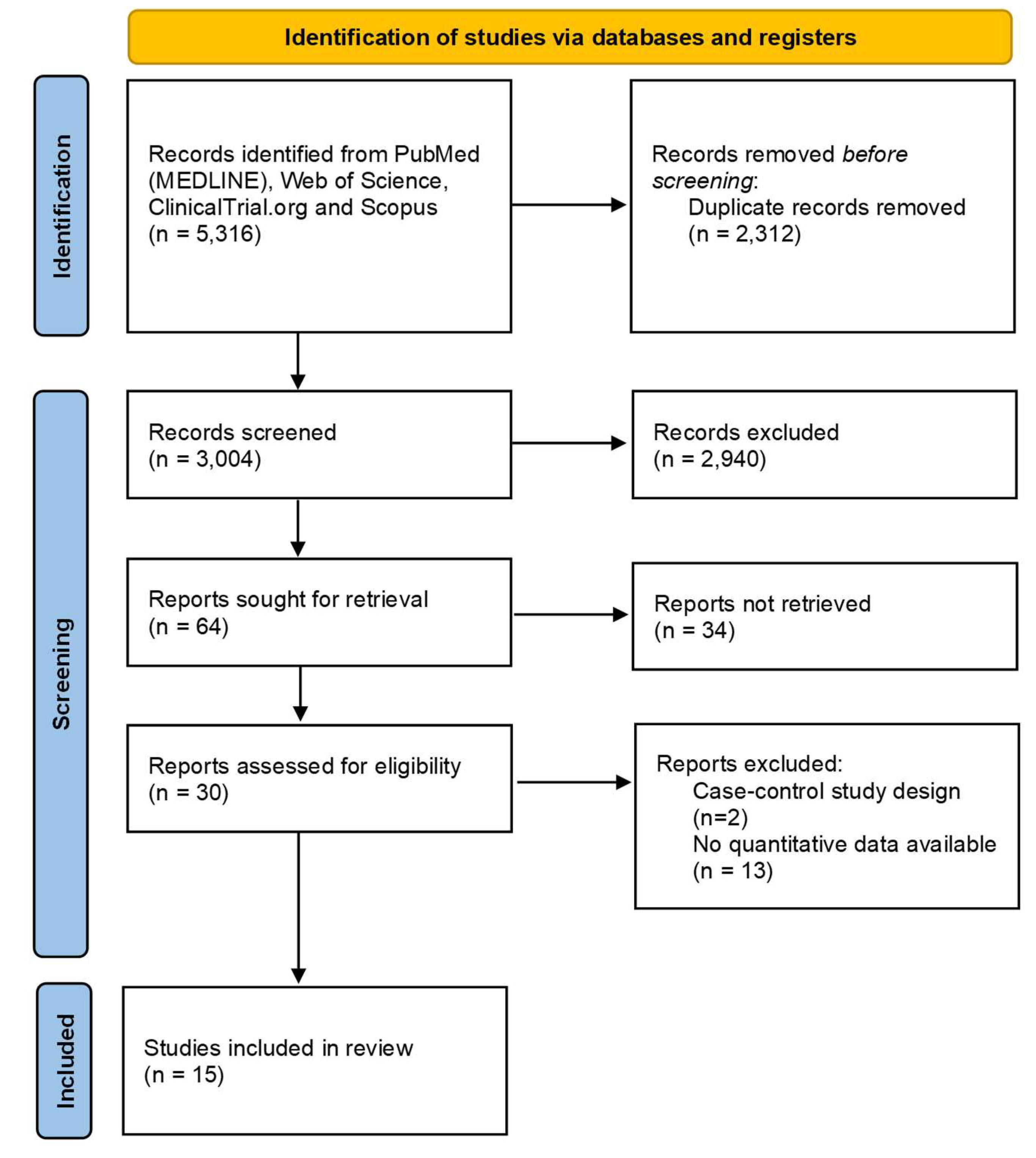
After removing duplicates, 3 004 abstracts were screened for inclusion. 64 full-text articles were assessed for eligibility. We excluded 34 of these, leaving 30 publications to be included. Fifteen articles were further excluded because no quantitative data were available (n=13) or for wrong study design (n=2). Finally, 15 articles were included in the review [16-30]. The selection process is described in
Figure 1.
All the included studies had a cross-sectional design. Overall, this systematic review includes 2 395 subjects, among which 1 312 had allergy to peanut (9 studies), 149 to peach (2 study), and 934 to hazelnut (3 studies). Most patients were pediatric, with only three studies including adult patients (age ranging from18 to 46 years). In most cases patients had only local symptoms, but in 6 studies also patients with systemic symptoms were included. In only 5 studies all patients underwent to OFC, in the others the percentage of participants undergone to OFC was quite variable, ranging from 15% to 98%. In these cases, the reason for not performing OFC was history of anaphylaxis after the ingestion of the culprit food or denying informed consent. Component resolved diagnostics was performed for all participants in most of the studies, except for 4 studies in which the test was available for 16-98% of participants. Single-plex ImmunoCAP was used in most of the studies, except for two, in whom multiplex assay was used. In five studies researchers received funding from manufacturer of laboratory reagents. Main study characteristics are summarized in
Table 1.
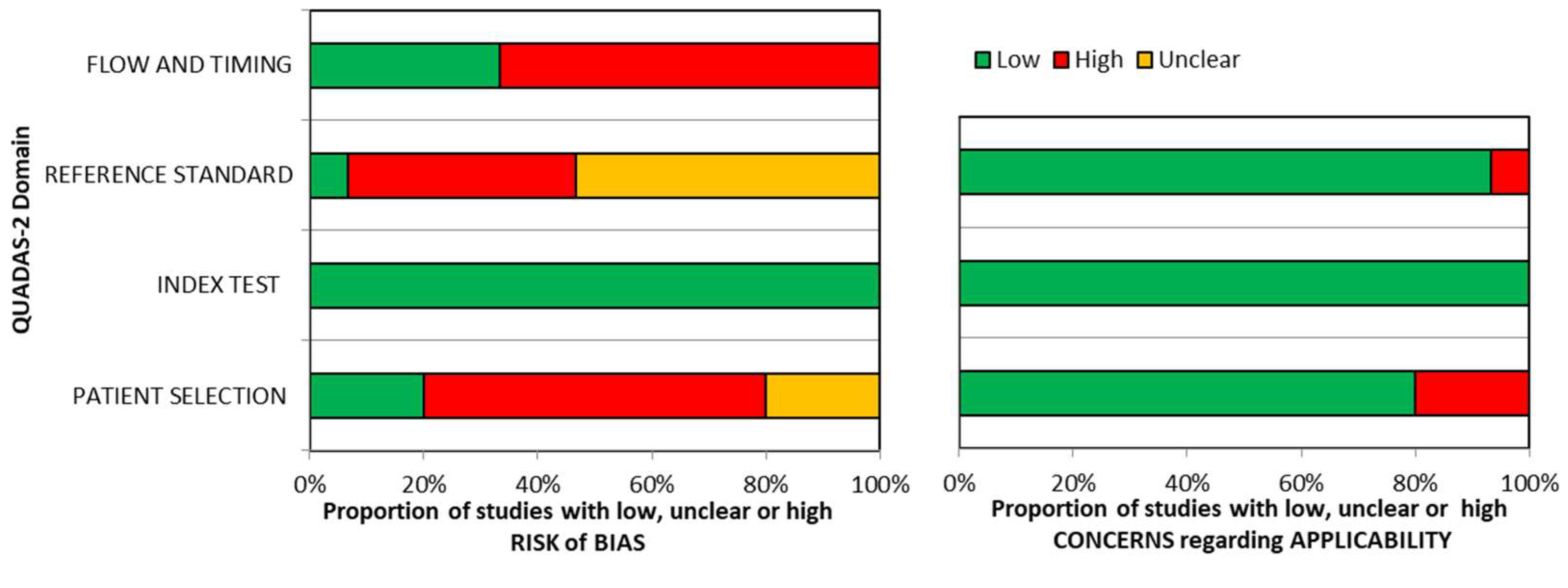
Quality assessment of included studies is summarized in
Figure 2.
No risk of bias was detected for the domain “index test”. Similarly, there were no applicability concerns for this domain. Few commercial sIgE assays are available at present and the threshold is pre-specified in almost all studies, so there is a high grade of concordance between included studies and review question on this domain. “Reference standard” domain was the mostly affected by risk of bias, probably because most of the included studies used an open challenge to diagnose food allergy instead of DBPCFC, considered the gold standard for food allergy diagnosis. In 10 studies only a variable fraction of the enrolled patients (15-98%) underwent to oral food challenge, affecting the risk of bias for the domain “patient selection”. Concerns regarding applicability were high in a small proportion of studies (<10%), probably because review question was not specific about diagnostic criteria for food allergy so results from most studies could be applicable.
2.2. Peanut
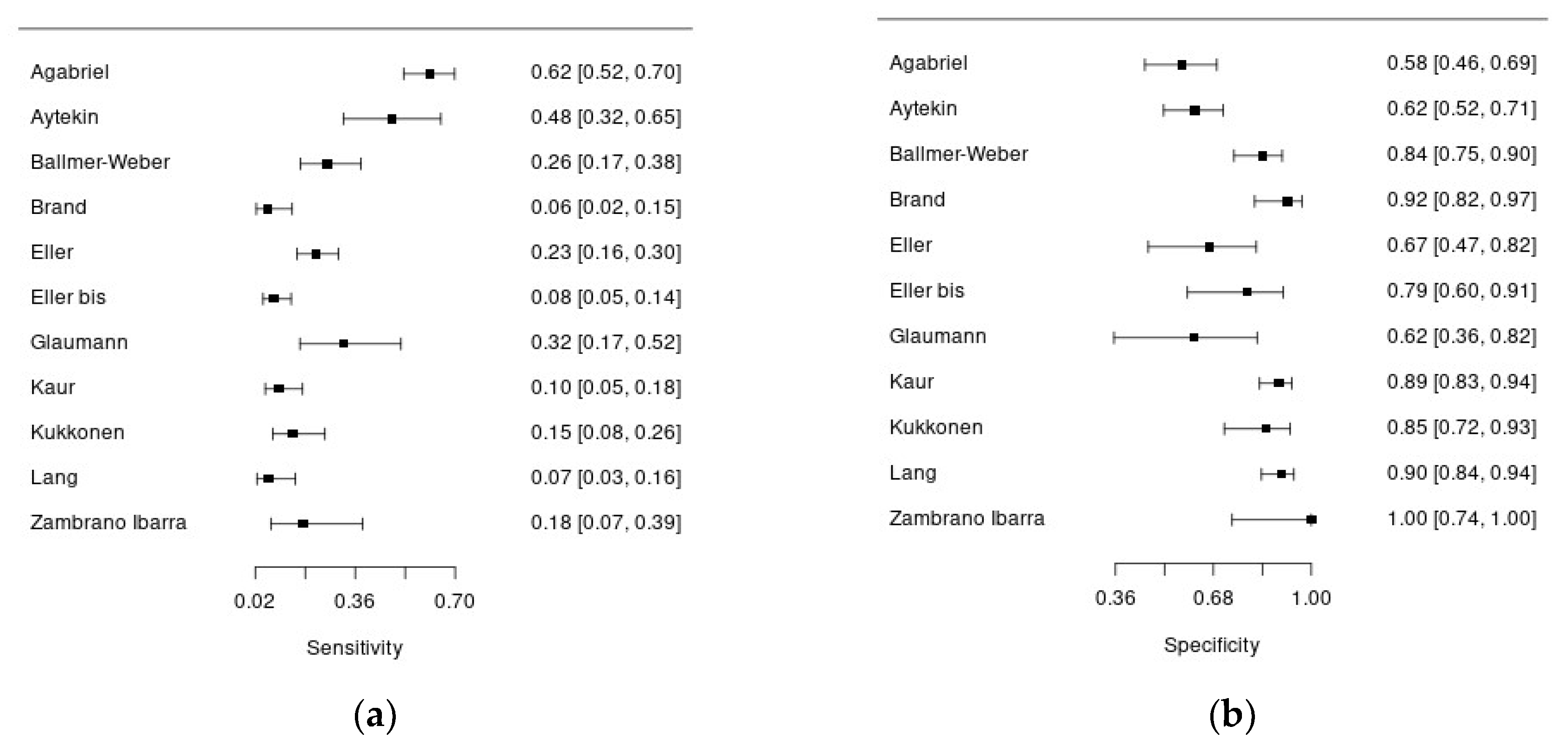
Overall, 10 studies assessed diagnostic accuracy of Ara h 9, using 0.35 kU/l (7 studies) and/or 0.1 kU/l (4 studies) cut-offs. All the studies had a cross-sectional design and were conducted in Europe, except for the study of Kaur et al. that was conducted in Australia and the one of Aytekin et al. in Africa.
Figure 3a and 3b show forest plots summarizing sensibility and specificity of Ara h 9 for the diagnosis of peanut allergy.
Overall, 1 312 patients with suspected peanut allergy were included in this analysis, and in 806 of them the disease was confirmed. Systemic symptoms were reported in 14-54%, but this information was not available for 7 studies. In almost all the studies pediatric patients were enrolled, except for the study of Ballmer-Weber who recruited adult patients (mean age 26 years). The percentage of males ranged from 40% to 68%. There was significant heterogeneity regarding the reference standard to diagnose peanut allergy: 6 studies adopted an open food challenge while 4 studies the double blind-placebo controlled one. Moreover, the percentage of patients undergoing to food challenge was quite variable ranging from 15 to 100%. Regarding the index test, all the studies used ImmunoCAP assay (Thermo Fisher) for serum Ara h 9 measurement, in singleplex or multiplex, except for two studies. The sensitivity of Ara h 9 for peanut allergy diagnosis was quite low, ranging from 6 to 61%, with acceptable specificity, ranging from 57 to 100%.
2.3. Hazelnut
Three studies enrolled patients with suspected hazelnut allergy.
Figure 4a and 4b show forest plots summarizing sensibility and specificity of Cor a 8 for the diagnosis of hazelnut allergy in the included studies.
Overall, 934 patients were included in this analysis, with 171 diagnosis of hazelnut allergy. One study included children, the others adult patients (mean age ≅ 33 years). Percentage of males ranged from 29% to 70%. In all the study DBPCFC was used to diagnose food allergy, but the percentage of patients undergoing to the challenge was variable (17%-87%). The cut-off was always pre-specified. In the study of Lyons et al, the authors assessed diagnostic accuracy of component resolved diagnostics using both 0.1 kU/l and 0.35 kU/l cut-offs, and found that in both cases specificity of Cor a 8 for hazelnut allergy was high, but overall diagnostic accuracy was poor [
25]. The sensitivity of Cor a 8 for the diagnosis of hazelnut allergy was low, ranging from 11 to 43%, with higher specificity, ranging from 59 to 94%.
2.4. Peach
Although the role of Pru p3 in the diagnosis of peach allergy has been evaluated in many studies, only two studies fulfilled inclusion criteria for this systematic review [
18,
29]. In both studies, OFC was conducted administering peeled peach pulp to patients selected for having clear history of hypersensitivity reaction after contact or ingestion of peach, with the aim to exclude the effect of Pru p 3, mainly expressed in the peach peel, and considered as a confounder for the clinical diagnostic pathway. For this reason, diagnostic accuracy of Pru p 3 arising from these studies should be considered with caution because this result could be affected by the OFC protocol.
3. Discussion
This systematic review describes diagnostic accuracy of serum specific IgE against LTPs in food allergy. Overall, most of the evidence regards peanut, hazelnut and peach allergy. The main findings of this analysis are that specific IgE against LTPs has very low sensitivity and acceptable specificity, making this test suitable for confirming food allergy when food challenge, considered the gold standard for food allergy diagnosis, is not practicable or not informative. For this reason, results confirm the role of this test as an add-on one in the clinical pathway of food allergy in adjunction with standard diagnostic procedures including skin prick test, and oral food challenge.
During the last years, clinical laboratory in allergology has experiencing a technological improvement leading to the release of new assays for serum IgE that are specific for molecular allergenic components. This revolution, based on increasing knowledge of the biochemical properties of allergens and on their geographic distribution, is enhancing the comprehension of IgE-mediated allergies towards precision medicine. Until today, hundreds of molecular allergens have been identified, isolated, and characterized, and it is expected that this number will increase rapidly. In allergic reactions, alongside species-specific molecules, ubiquitous molecules could be involved too. These molecules are expressed in many botanical or animal species. Inevitably, IgE sensitization to these components, often referred to as panallergens, can evoke varying degrees of cross-reactivity. LTPs can be considered panallergens because they are expressed in several pollens and plant foods belonging to both the widespread Rosaceae family and many other distantly related species and can elicit systemic reactions in LTPs-sensitized subjects. Often, the primary source of immunization from LTPs is Pru p 3, the peach specific LTP, as arises from epidemiological studies mainly from the Mediterranean area, such as Italy, Spain, and Greece [
30]. Nevertheless, the clinical relevance of LTPs allergens, as well as the primary sensitizer, varies greatly depending on the patient's age and geographic area [
2]. Many studies have described diagnostic accuracy of component resolved diagnostics, but no one has evaluated the specific involvement of sIgE against LTPs allergens. This topic is of particular interest especially in countries of the Mediterranean area, where LTPs-driven allergies are more frequent. In the study of Nilsson et al, diagnostic accuracy of Ara h 9 was evaluated using both 0.10 kU/l and 0.35 kU/l cutoffs. The authors founded a pooled specificity ranging 0.77-0.81 and a pooled sensitivity ranging 0.32-0.19, based on cut-off [
6]. Similarly, Flore Kim et al founded that Ara h 9 had a sensitivity of 0.14 and a specificity of 0.85 at the 0.35 kU/l cut-off [
5]. The present analysis confirms these previous findings, demonstrating that LTPs-component resolved diagnostics is suitable when confirmation of peanut allergy diagnosis is required, but it should be used with caution for excluding the disease. With respect to Cor a 8 and the diagnosis of hazelnut allergy, we founded a pooled sensitivity of 0.18 and specificity of 0.79. In the study of Masthoff et al, sensitivity of Cor a 8 was quite low, 0.06, and with acceptable specificity, 0.96 [
31]. Again, we can confirm that Cor a 8 is a suitable test when diagnosis of hazelnut allergy confirmation is expected, but the test may be misleading for the exclusion of the disease.
The main limit of this systematic review is that risk of bias was high in many studies for the domain “reference standard” due to the use of different challenge protocol (open versus double-blind food challenge) and different proportion of patients undergoing to reference standard. At this regard, it should be noticed that for ethical reason patients with severe systemic symptoms, such as anaphylaxis, or with a clear diagnosis of FA must be excluded from life-threating or unnecessary diagnostic tests, such as oral food challenge.
In conclusion, this review suggests that some LTPs, such as Ara h 9 and Cor a 8, could be useful in the diagnostic pathway of peanut and hazelnut allergies with high specificity, but low sensitivity. Nevertheless, further studies with adequate study design, avoiding the case-control one, and employing DBPCFC as reference standard are needed to define diagnostic accuracy of sIgE for LTPs. Moreover, the role of these tests for diagnosis of food allergy in adults is quite unexplored so requires more investigations.
4. Materials and Methods
4.1. Reporting and Study Protocol Registration
4.2. Eligibility Criteria
The review question was: which is the diagnostic accuracy of molecular-based LTPs allergens serological tests (allergen-based serum IgE) for the diagnosis of food allergies? Due to the nature of the systematic review, namely diagnostic accuracy evaluation, we considered Population, Index test, and Target condition (PIT) to organize our review question instead of the well-established PICO framework, which is specifically intended for intervention studies [
9]. We included studies recruiting patients with symptoms of food allergies, both local (oral allergic syndrome, urticaria, nausea, vomiting, diarrhea, angioedema, itch, rush) and systemic symptoms (anaphylaxis, anaphylactic shock) as population of interest, with no specific restrictions to the context. The index test was specific IgE antibodies against any LTPs, determined using singleplex or multiplex measurement. The target condition was allergy due to LTPs containing food (apple, peach, kiwi, strawberry, pomegranate, walnut, peanut, hazelnut, almond, chestnut, tomato, celery, wheat, lettuce, lentil, corn, grape, olive). Oral provocation test, both open food challenge (OFC) or double-blind, placebo- controlled food challenge (DBPCFC), was the reference standard for food allergy diagnosis. Studies assessing diagnostic accuracy, including cohort, prospective or retrospective cross-sectional studies, were considered. To be considered in the review, studies must include a clear definition of the target population, the target condition, then index test, and the reference standard. Case-control studies were excluded. Narrative reviews, editorials, comments, and any type of paper not presenting quantitative data on the research question were excluded.
4.3. Search Strategy and Information Sources
We searched Pubmed (MEDLINE), Web of Science, Scopus, and ClinicalTrial.org for reports published from database inception to the date of search. The databases were searched on 19 April 2022. Literature search was performed again on the 23 May 2024 to retrieve the most recent reports. The approach used to develop search strategy was adopted from intervention studies considering the non-experimental setting, and specifically defining population of interest, index test, and condition. Literature search was developed using the guidance for describing search strings for systematic reviews in the form of PRISMA-S [
10]. The full search strategy used for MEDLINE was strictly adapted to search the other databases (
Table S2). No restriction for language was applied.
4.4. Selection of Studies
Duplicates were identified and removed by automation tools. Initially, two reviewers (CB, CU) independently reviewed the first 20 records and discussed inconsistency until consensus was reached. Then, two co-authors proceeded screening the remaining records by basing on title and abstract and working independently (CB, CU, LS, CS). Screening was conducted using Rayyan software [
11]. No automation tools were used for study exclusion. Disagreement was resolved by consensus of reviewers. Full texts of the records that passed the first selection were screened by two co-authors working independently (CB, CU, LS, CS) to assure that inclusion criteria were fulfilled. In each step of records screening, the reviewer was blinded to the decision of the other one.
4.5. Data Collection Process
Two reviewers extracted data independently into a standardized Excel format, including any relevant information about study design, sample size, selection criteria, demographics, local and systemic symptoms, culprit food, manufacturer of laboratory reagents, cut-off for test positivity, type of OFC (CB, CU, LS, CS, DSS). The form was piloted and calibration exercises on five records were conducted prior to formal data extraction to ensure consistency between reviewers. Disagreement between collectors was resolved by consensus.
4.6. Quality Assessment
Risk of bias (ROB) and applicability of included studies were evaluated by QUADAS-2 (Quality Assessment of Diagnostic Accuracy Studies) tool [
12]. Briefly, it was conducted in four phases: a) reporting the review question; b) developing review-specific guidance for quality assessment and write a template for ROB assessment for each primary study; c) reviewing a flow-diagram of the primary study or constructing if it was not available; d) evaluate risk of bias and applicability for each item. Regarding review-specific tailoring, the question “Were the index test results interpreted without knowledge of the results of the reference standard?” was omitted from ROB assessment template because index test was a quantitative and objective measure, so no subjective interpretation was requested for positive/negative classification. Regarding the question “Is the reference standard likely to correctly classify the target condition?”, target condition was defined with the highest grade of certainty by double-blind placebo-controlled (DBPC) food challenge [
13], so the answer to the signaling question was “yes” only if DBPC challenge was considered as reference standard, and “no” if open food challenge was used for diagnosis. For the signaling question “Was there an appropriate interval between index tests and reference standard?”, the authors considered that the appropriate interval between index test and reference standard was within one year; if the two tests were performed within the diagnostic work-up the answer was “yes”. As suggested by QUADAS-2 group, ROB was considered “low” if the answers for all signaling questions were “yes”, and “high” if at least one signaling question was answered “no”. Two reviewers (CB, CU, LS, CS) independently assessed the risk of bias for primary studies using the review-specific template constructed in accordance with QUADAS-2 tool. Any disagreement was resolved through discussion.
4.7. Data Synthesis, Analysis and Reporting
Diagnostic accuracy was described by sensitivity and specificity calculated from two-by-two table for each study. Sensitivity and specificity for each study were reported by forest plots. Initially, only studies evaluating diagnostic accuracy of Ara h 9 and Cor a 8 were considered for quantitative analysis because too few studies reported data about other LTPs (Pru p 3) to be included in quantitative analysis. Random effect, hierarchical summary receiver operating characteristic (HS-ROC) model did not converge so narrative synthesis of results was reported. Heterogeneity between studies was investigated by visually inspecting the forest plots of sensitivity and specificity. Studies were grouped according to age of participants, use of DBPC vs oral food challenge, and different cut-offs. Data analysis was performed using MetaDTA v2.1.2 [
14].
Supplementary Materials
The following supporting information can be downloaded at Preprints.org: Table S1: PRISMA checklist; Table S2: search strategy
Author Contributions
Conceptualization, C.B., D.L. and C.G.U.; methodology, C.B, L.S. and C.G.U; formal analysis, C.B, C.S., L.S. and C.G.U; investigation, C.B, D.D.S., C.S. and C.G.U; data curation, C.B., C.S., D.L. and C.G.U; writing—original draft preparation, C.B., D.L., D.D.S., and C.G.U; writing—review and editing, C.B., D.L., and C.G.U. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable
Informed Consent Statement
Not applicable
Data Availability Statement
Dataset available on request from the authors
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Spolidoro, G.C.I.; Amera, Y.T.; Ali, M.M.; Nyassi, S.; Lisik, D.; Ioannidou, A.; Rovner, G.; Khaleva, E.; Venter, C.; van Ree, R.; et al. Frequency of food allergy in Europe: An updated systematic review and meta-analysis. Allergy. Epub 12022 Nov 15567. 2023, 78, 351–368. [Google Scholar] [CrossRef] [PubMed]
- Skypala, I.J.; Asero, R.; Barber, D.; Cecchi, L.; Diaz Perales, A.; Hoffmann-Sommergruber, K.; Pastorello, E.A.; Swoboda, I.; Bartra, J.; Ebo, D.G.; et al. Non-specific lipid-transfer proteins: Allergen structure and function, cross-reactivity, sensitization, and epidemiology. Clin Transl Allergy. 2021, 11, e12010. [Google Scholar] [CrossRef] [PubMed]
- Scala, E.; Alessandri, C.; Bernardi, M.L.; Ferrara, R.; Palazzo, P.; Pomponi, D.; Quaratino, D.; Rasi, C.; Zaffiro, A.; Zennaro, D.; et al. Cross-sectional survey on immunoglobulin E reactivity in 23,077 subjects using an allergenic molecule-based microarray detection system. Clin Exp Allergy 2010, 40, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Barber, D.; de la Torre, F.; Feo, F.; Florido, F.; Guardia, P.; Moreno, C.; Quiralte, J.; Lombardero, M.; Villalba, M.; Salcedo, G.; et al. Understanding patient sensitization profiles in complex pollen areas: a molecular epidemiological study. Allergy. 2008, 63, 1550–1558. [Google Scholar] [CrossRef] [PubMed]
- Flores Kim, J.; McCleary, N.; Nwaru, B.I.; Stoddart, A.; Sheikh, A. Diagnostic accuracy, risk assessment, and cost-effectiveness of component-resolved diagnostics for food allergy: A systematic review. Allergy 2018, 73, 1609–1621. [Google Scholar] [CrossRef]
- Nilsson, C.; Berthold, M.; Mascialino, B.; Orme, M.E.; Sjölander, S.; Hamilton, R.G. Accuracy of component-resolved diagnostics in peanut allergy: Systematic literature review and meta-analysis. Pediatr Allergy Immunol. Epub 12020 Jan 13228. 2020, 31, 303–314. [Google Scholar] [CrossRef]
- Maesa, J.M.; Dobrzynska, A.; Baños-Álvarez, E.; Isabel-Gómez, R.; Blasco-Amaro, J.A. ImmunoCAP ISAC in food allergy diagnosis: a systematic review of diagnostic test accuracy. Clin Exp Allergy. Epub 12021 Apr 13812. 2021, 51, 778–789. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Deeks, J.; Bossuyt, P.; Leeflang, M.; Takwoingi, Y.; Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy. Version 2.0. Available online: https://training.cochrane.org/handbook-diagnostic-test-accuracy (accessed on July 2023).
- Rethlefsen, M.L.; Kirtley, S.; Waffenschmidt, S.; Ayala, A.P.; Moher, D.; Page, M.J.; Koffel, J.B. PRISMA-S: an extension to the PRISMA Statement for Reporting Literature Searches in Systematic Reviews. Syst Rev. 2021, 10, 39. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan-a web and mobile app for systematic reviews. Syst Rev 2016, 5, 210. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M.; Group, Q.-. . QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011, 155, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.F.; Riggioni, C.; Agache, I.; Akdis, C.A.; Akdis, M.; Alvarez-Perea, A.; Alvaro-Lozano, M.; Ballmer-Weber, B.; Barni, S.; Beyer, K.; et al. EAACI guidelines on the diagnosis of IgE-mediated food allergy. Allergy. Epub 12023 Oct 15910. 2023, 78, 3057–3076. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Cooper, N.; Freeman, S.; Sutton, A. Graphical enhancements to summary receiver operating characteristic plots to facilitate the analysis and reporting of meta-analysis of diagnostic test accuracy data. Res Synth Methods. Epub 2020 Aug 1012. 2021, 12, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Agabriel, C.; Ghazouani, O.; Birnbaum, J.; Liabeuf, V.; Porri, F.; Gouitaa, M.; Cleach, I.; Grob, J.; Bongrand, P.; Sarles, J.; et al. Ara h 2 and Ara h 6 sensitization predicts peanut allergy in Mediterranean pediatric patients. Pediatr Allergy Immunol 2014, 25, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Ballmer-Weber, B.; Lidholm, J.; Fernández-Rivas, M.; Seneviratne, S.; Hanschmann, K.; Vogel, L.; Bures, P.; Fritsche, P.; Summers, C.; Knulst, A.; et al. IgE recognition patterns in peanut allergy are age dependent: perspectives of the EuroPrevall study. Allergy 2015, 70, 391–407. [Google Scholar] [CrossRef]
- Boyano-Martínez, T.; Pedrosa, M.; Belver, T.; Quirce, S.; García-Ara, C. Peach allergy in Spanish children: tolerance to the pulp and molecular sensitization profile. Pediatr Allergy Immunol 2013, 24, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Brand, H.; Schreurs, M.; Emons, J.; Gerth van Wijk, R.; de Groot, H.; Arends, N. Peanut components measured by ISAC: comparison with ImmunoCap and clinical relevance in peanut allergic children. Clin Mol Allergy 2021, 19, 14. [Google Scholar] [CrossRef]
- Buyuktiryaki, B.; Cavkaytar, O.; Sahiner, U.; Yilmaz, E.; Yavuz, S.; Soyer, O.; Sekerel, B.; Tuncer, A.; Sackesen, C.; Buyuktiryaki, B.; et al. Cor a 14, Hazelnut-Specific IgE, and SPT as a Reliable Tool in Hazelnut Allergy Diagnosis in Eastern Mediterranean Children. J Allergy Clin Immunol Pract 2016, 4, 265–272. [Google Scholar] [CrossRef]
- Datema, M.; Zuidmeer-Jongejan, L.; Asero, R.; Barreales, L.; Belohlavkova, S.; de Blay, F.; Bures, P.; Clausen, M.; Dubakiene, R.; Gislason, D.; et al. Hazelnut allergy across Europe dissected molecularly: A EuroPrevall outpatient clinic survey. J Allergy Clin Immunol 2015, 136, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Glaumann, S., N. A., Johansson, S.G.O., Rudengren, M., Borres, M.P., Nilsson, C. Basophil allergen threshold sensitivity, CD-sens, IgE-sensitization and DBPCFC in peanut-sensitized children. Allergy 2012, 67. [Google Scholar] [CrossRef]
- Kaur, N.; Mehr, S.; Katelaris, C.; Wainstein, B.; Altavilla, B.; Saad, R.; Valerio, C.; Codarini, M.; Burton, P.; Perram, F.; et al. Added Diagnostic Value of Peanut Component Testing: A Cross-Sectional Study in Australian Children. J Allergy Clin Immunol Pract 2021, 9, 245–253.e244. [Google Scholar] [CrossRef] [PubMed]
- Kukkonen, A.; Pelkonen, A.; Makinen-Kiljunen, S.; Voutilainen, H.; Makela, M.; Kukkonen, A.K.; Pelkonen, A.S.; Makinen-Kiljunen, S.; Voutilainen, H.; Makela, M.J. Ara h 2 and Ara 6 are the best predictors of severe peanut allergy: a double-blind placebo-controlled study. Allergy 2015, 70, 1239–1245. [Google Scholar] [CrossRef] [PubMed]
- Lyons, S.; Welsing, P.; Hakobyan, M.; Kansen, H.; Knol, E.; Otten, H.; van Ree, R.; Knulst, A.; Le, T.; Lyons, S.A.; et al. Measurement of IgE to hazelnut allergen components cannot replace hazelnut challenge in Dutch adults. Allergy 2022, 77, 1559–1569. [Google Scholar] [CrossRef] [PubMed]
- Zambrano Ibarra, G.; Fuentes Aparicio, V.; Infante Herrero, S.; Blanca, M.; Zapatero Remon, L. Peanut Allergy in Spanish Children: Comparative Profile of Peanut Allergy versus Tolerance. Int Arch Allergy Immunol 2019, 178, 370–376. [Google Scholar] [CrossRef]
- Eller, E.; Baumann, K.; Skov, P.S.; Bindslev-Jensen, C. Measurement of Ara h 6 can improve diagnosis in patients suspected of peanut allergy. Clin Exp Allergy. Epub 12023 Apr 14318. 2023, 53, 683–685. [Google Scholar] [CrossRef]
- Aytekin, E.S.; Soyer, O.; Sahiner, U.M.; Wieser, S.; Lupinek, C.; Sekerel, B.E. Diagnostic accuracy of the ALEX(2) test in peanut-sensitized children. Clin Exp Allergy. Epub 12023 May 14329. 2023, 53, 1041–1044. [Google Scholar] [CrossRef]
- Vílchez-Sánchez, F.; Rodríguez-Pérez, R.; Gómez-Traseira, C.; Dominguez-Ortega, J.; Hernández-Rivas, L.; García, I.L.; Quirce, S.; Pedrosa, M. Sensitisation to peach allergen Pru p 7 is associated with severe clinical symptoms in a Spanish population. Pediatr Allergy Immunol. 2023, 34, e14030. [Google Scholar] [CrossRef]
- Lang, A.; Balmert, L.C.; Weiss, M.; Pongracic, J.A.; Singh, A.M. Real world use of peanut component testing among children in the Chicago metropolitan area. Allergy Asthma Proc. 2022, 43, 226–233. [Google Scholar] [CrossRef]
- Asero, R.; Mistrello, G.; Roncarolo, D.; Amato, S.; Caldironi, G.; Barocci, F.; van Ree, R. Immunological cross-reactivity between lipid transfer proteins from botanically unrelated plant-derived foods: a clinical study. Allergy. 2002, 57, 900–906. [Google Scholar] [CrossRef] [PubMed]
- Masthoff, L.J.; Mattsson, L.; Zuidmeer-Jongejan, L.; Lidholm, J.; Andersson, K.; Akkerdaas, J.H.; Versteeg, S.A.; Garino, C.; Meijer, Y.; Kentie, P.; et al. Sensitization to Cor a 9 and Cor a 14 is highly specific for a hazelnut allergy with objective symptoms in Dutch children and adults. J Allergy Clin Immunol 2013, 132, 393–399. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).