Submitted:
26 September 2024
Posted:
29 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
2.1. Biological Background
2.2. The Agent-Based Model
2.2.1. World Structure
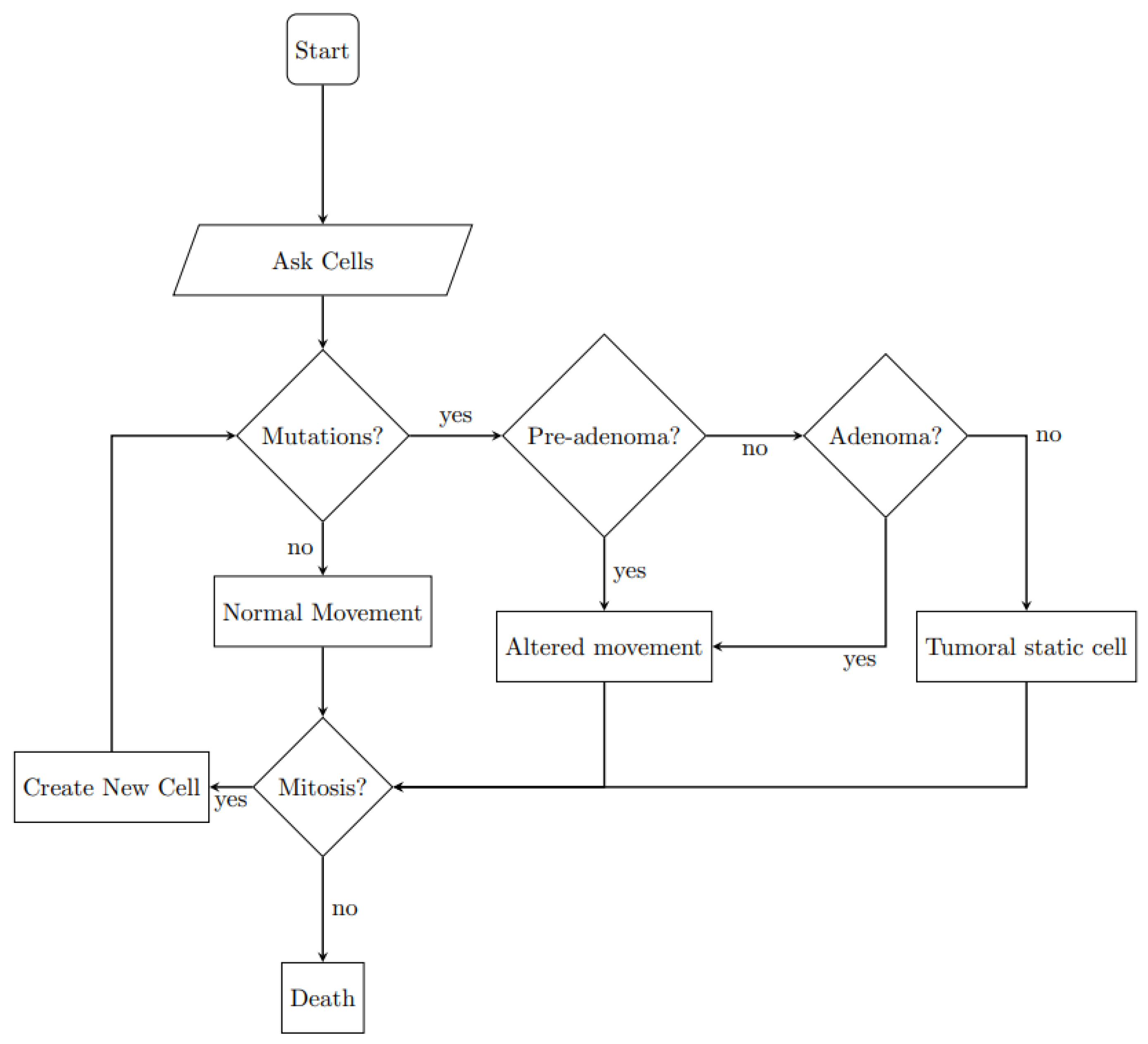
2.2.2. Agents Behavior
2.2.3. Genetic Features and Mutation Effects
2.2.4. Micro-Environmental Features
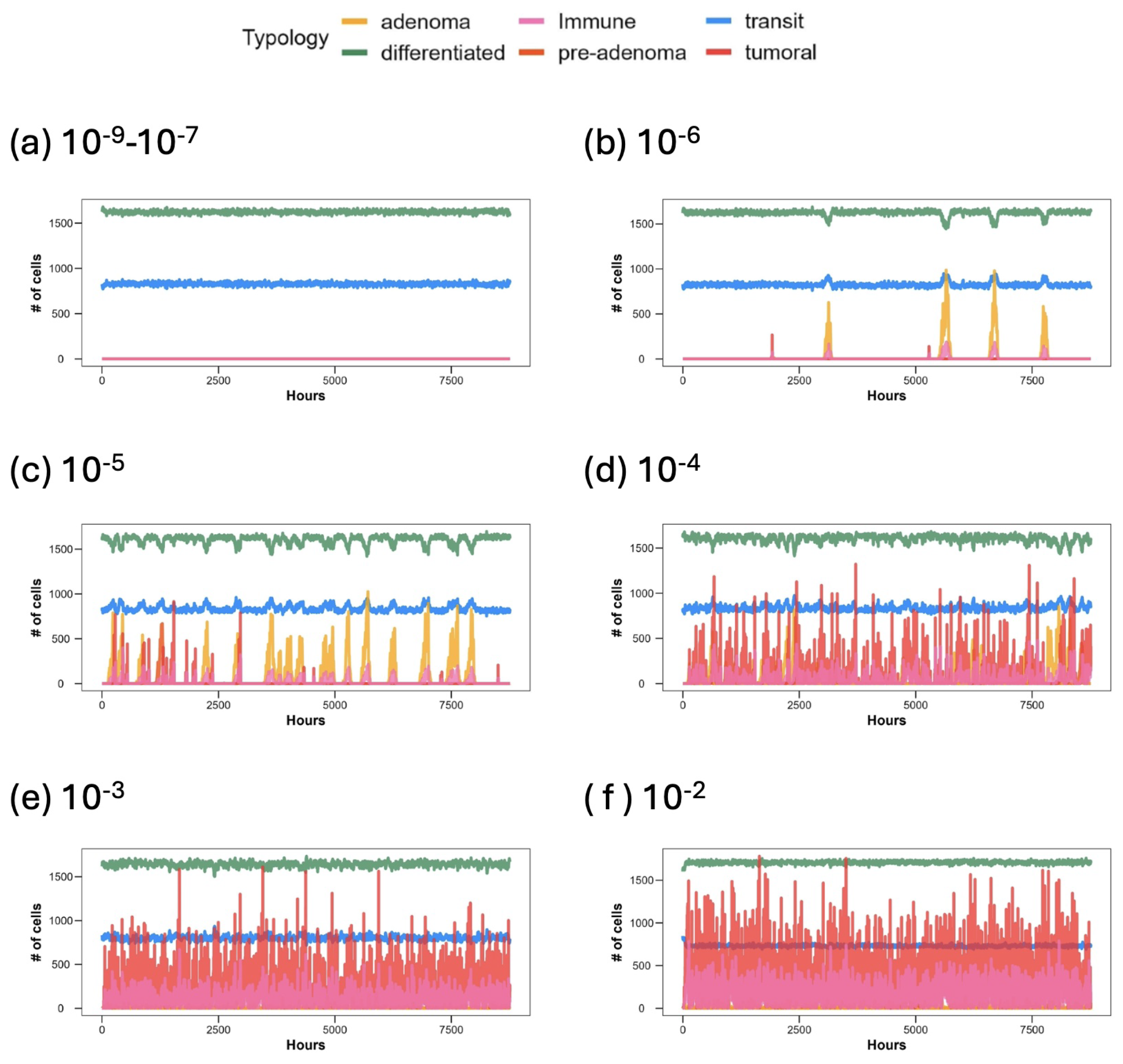
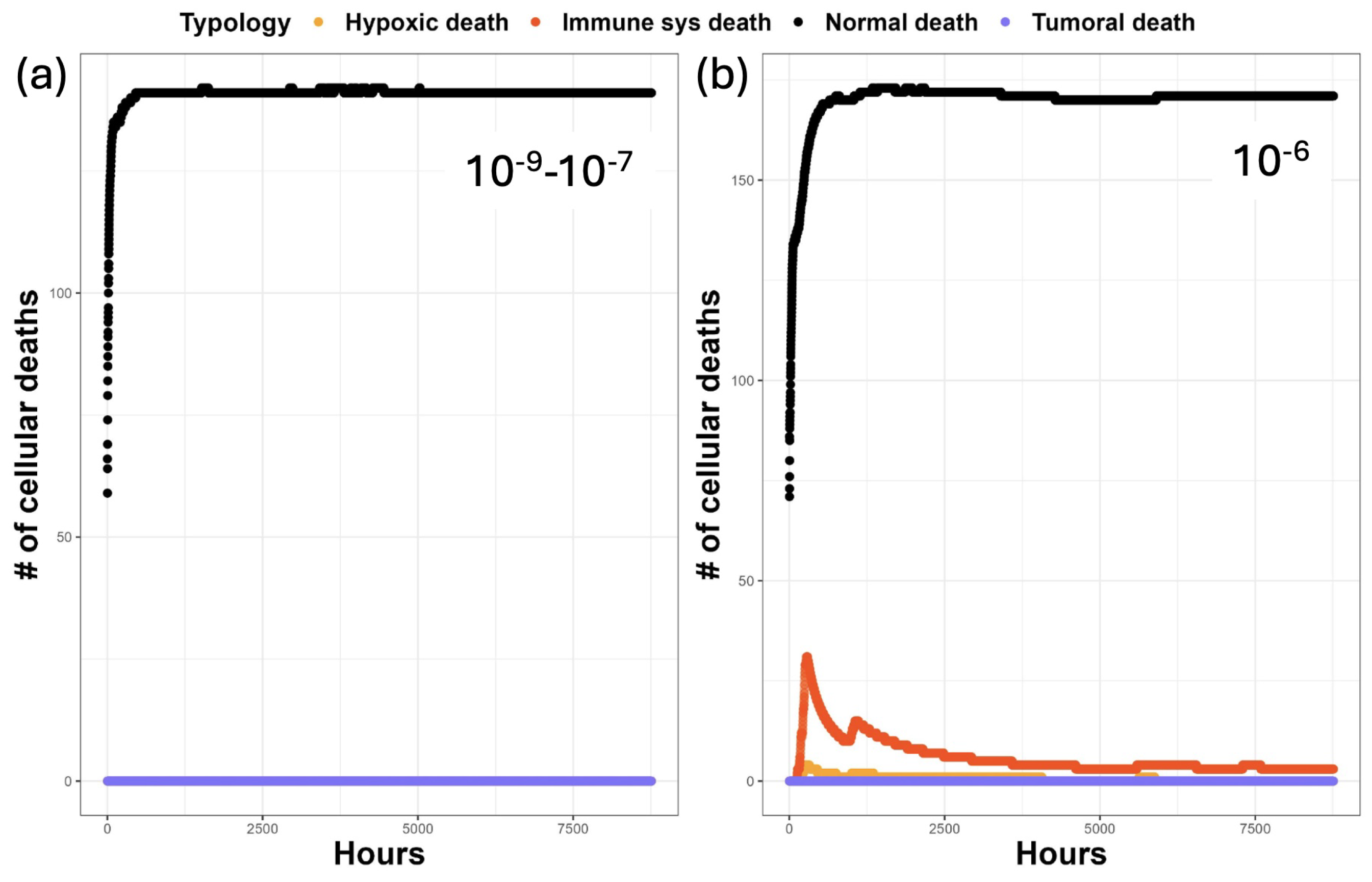
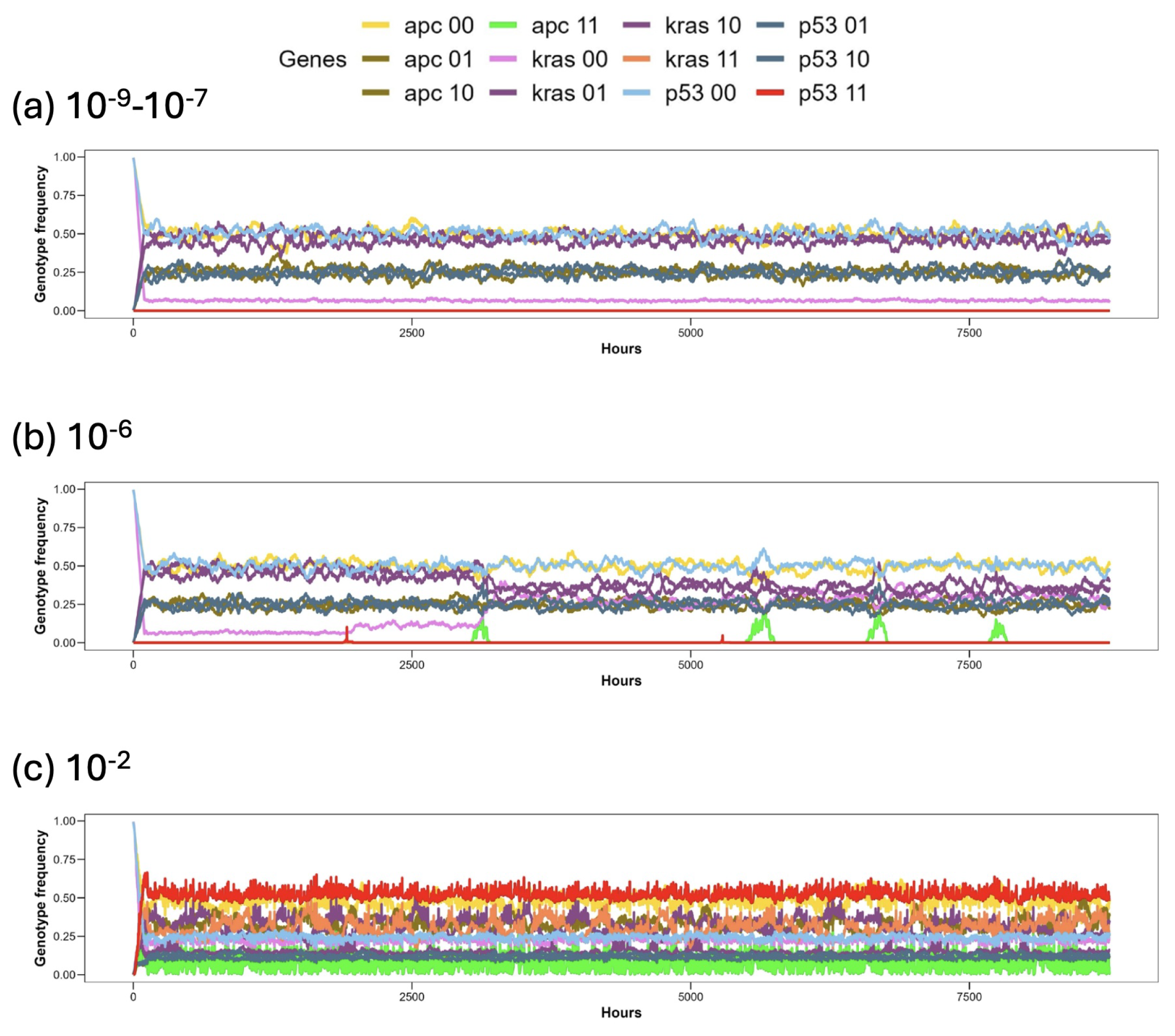
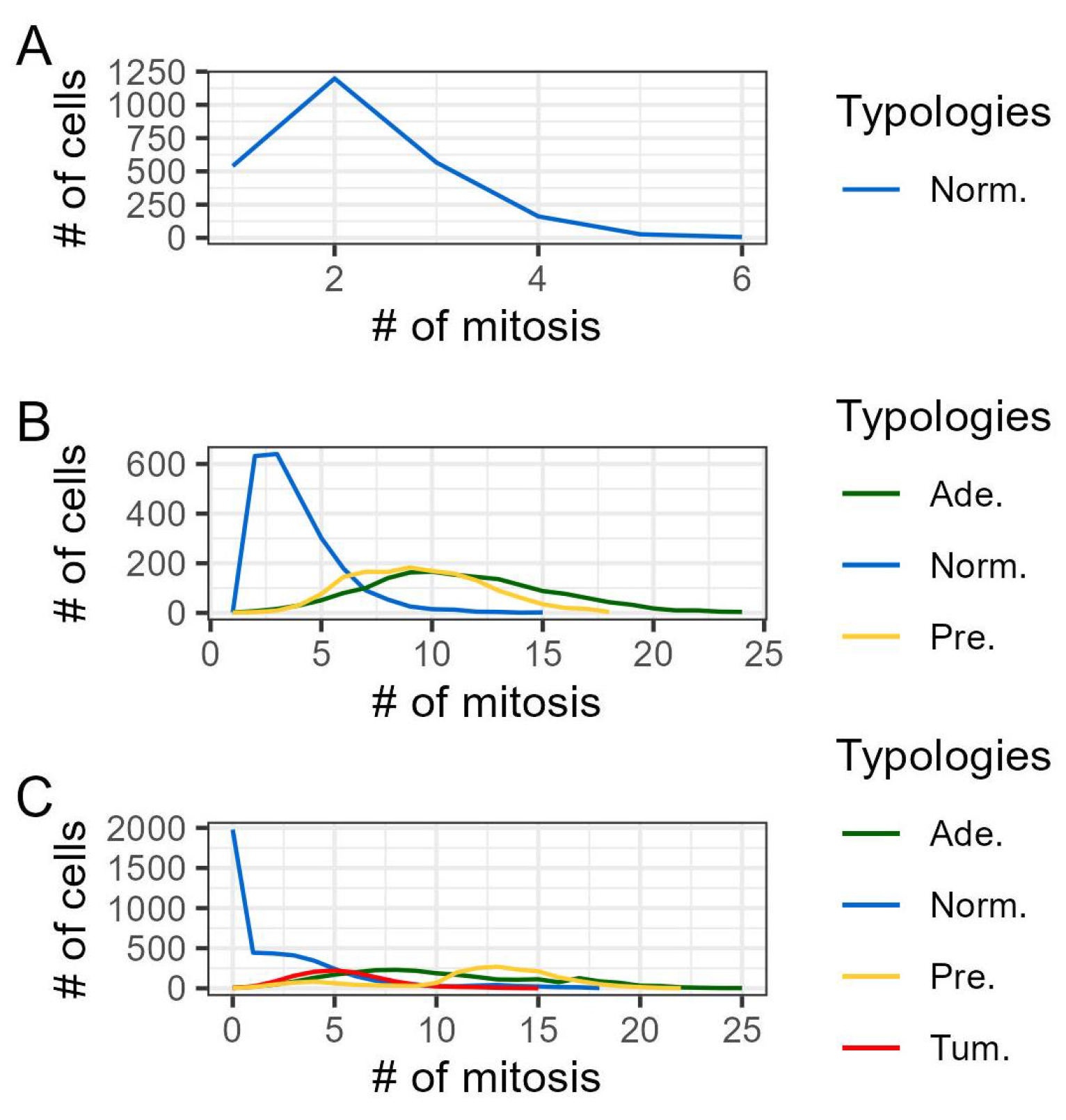
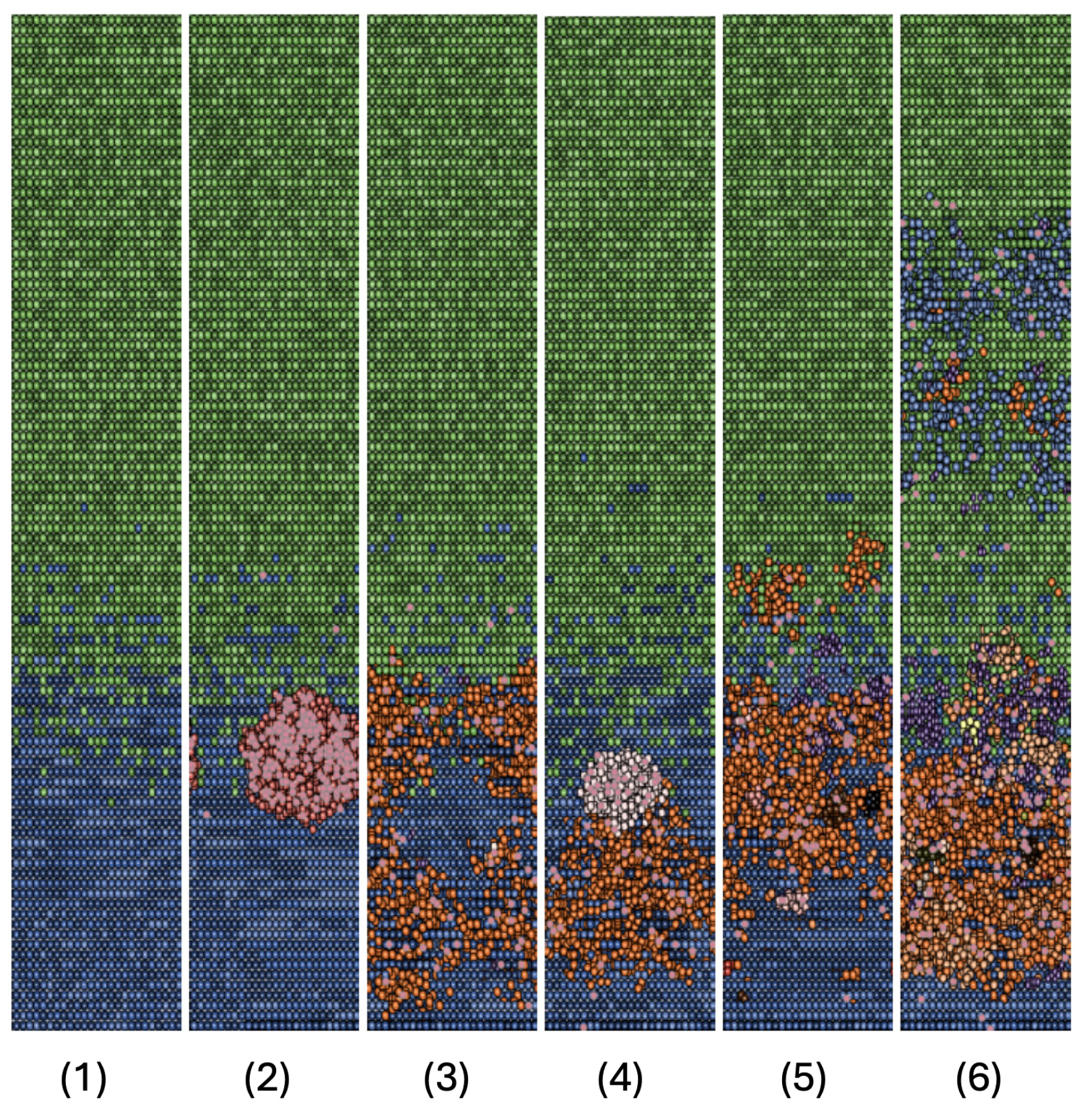
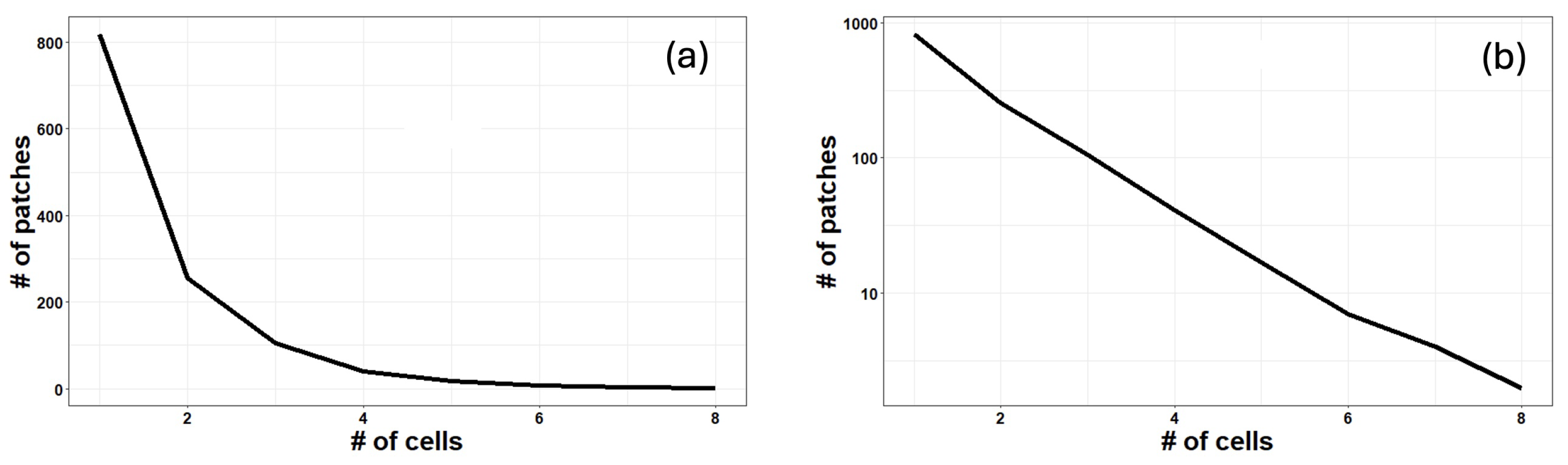
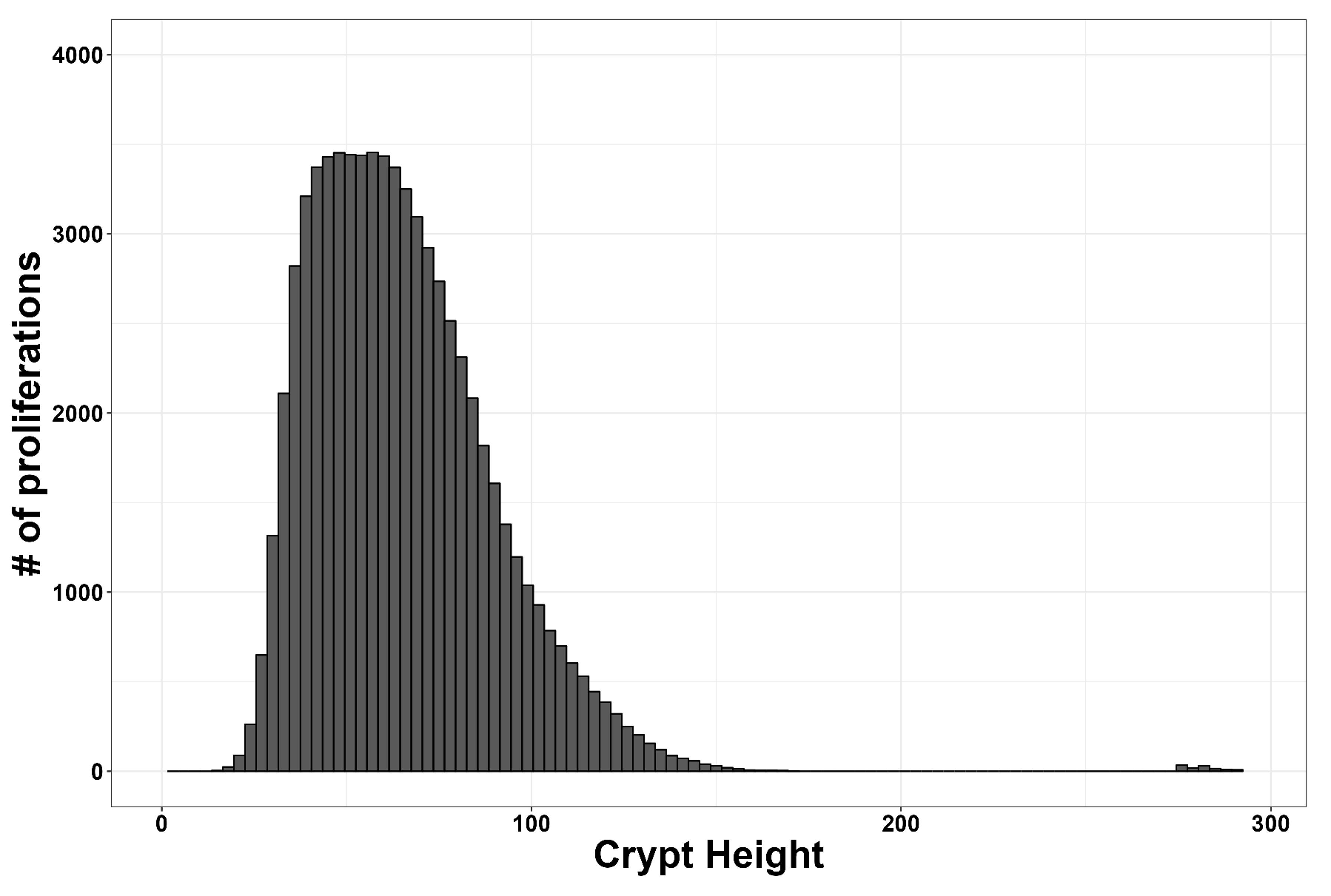
3. Results
- number of immune system’s cells: 1 / hour
- number of cells that a killer cells eliminates before die: 5 cells
- hypoxic threshold: 5 cells / patch
- immune-system threshold: 5 cells / patch
- probability of starting genotype: P = 0.5
3.1. Discussion
4. Conclusions
Author Contributions
Data Availability Statement
Conflicts of Interest
Abbreviations
| CRC | Colorectal Cancer |
| SMT | Somatic Mutation Theory |
| TOFT | Tissue Organization Field Theory |
| TME | Tumor Micro Environment |
References
- Plutynski, A. How is cancer complex? Eur. J. Philos. Sci. 2021, 11, 1–30. [Google Scholar] [CrossRef]
- Weinberg, R. A. The Biology of Cancer. Unpublished 2013. [Google Scholar]
- Fearon, E. R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Kinzler, K. W. Cancer genes and the pathways they control. Nat. Med. 2004, 10, 789–799. [Google Scholar] [CrossRef]
- Grady, W. M.; Carethers, J. M. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 2008, 135, 1079–1099. [Google Scholar]
- Nguyen, H. T.; Duong, H.-Q. The molecular characteristics of colorectal cancer: Implications for diagnosis and therapy. Oncol. Lett. 2018, 16, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Bürtin, F.; Mullins, C. S.; Linnebacher, M. Mouse models of colorectal cancer: Past, present and future perspectives. World J. Gastroenterol. 2020, 26, 1394. [Google Scholar] [CrossRef] [PubMed]
- Williams, A. C.; Hague, A.; Manning, A. M.; Van der Stappen, J. W.; Paraskeva, C. In vitro models of human colorectal cancer. Cancer Surveys 1993, 16, 15–29. [Google Scholar]
- Chen, H.; Cheng, Y.; Wang, X.; Wang, J.; Shi, X.; Li, X.; Tan, W.; Tan, Z. 3D printed in vitro tumor tissue model of colorectal cancer. Theranostics 2020, 10, 12127. [Google Scholar] [CrossRef]
- Johnston, M. D.; Edwards, C. M.; Bodmer, W. F.; Maini, P. K.; Chapman, S. J. Mathematical modeling of cell population dynamics in the colonic crypt and in colorectal cancer. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 4008–4013. [Google Scholar] [CrossRef]
- Michor, F.; Iwasa, Y.; Lengauer, C.; Nowak, M. A. Dynamics of colorectal cancer. In Seminars in Cancer Biology; Elsevier, 2005; Vol. 15, pp 484–493.
- Meineke, F.A.; Potten, C.S.; Loeffler, M. Cell migration and organization in the intestinal crypt using a lattice-free model. In Cell Proliferation, Wiley Online Library, 2001, 34, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Haupt, S.; Gleim, N.; Ahadova, A.; Bl"aker, H.; von Knebel Doeberitz, M.; Kloor, M.; Heuveline, V. A computational model for investigating the evolution of colonic crypts during Lynch syndrome carcinogenesis. In Computational and Systems Oncology, Wiley Online Library, 2021, 1, e1020. [Google Scholar] [CrossRef]
- Van Leeuwen, I.M.M.; Mirams, G.R.; Walter, A.; Fletcher, A.; Murray, P.; Osborne, J.; Varma, S.; Young, S.J.; Cooper, J.; Doyle, B.; <i>, *!!! REPLACE !!!*; et al. An integrative computational model for intestinal tissue renewal. In Cell Proliferation, Wiley Online Library, 2009, 42, 617–636. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Quirke, P. Experimental models of colorectal cancer. Dis. Colon Rectum 1998, 41, 490–505. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, R.E.; Buczacki, S.J.A.; Arends, M.J.; Adams, D.J. Mouse models of colorectal cancer as preclinical models. In Bioessays, Wiley Online Library, 2015, 37, 909–920. [Google Scholar] [CrossRef]
- Leystra, A.A.; Clapper, M.L. Gut microbiota influences experimental outcomes in mouse models of colorectal cancer. Genes 2019, 10, 900. [Google Scholar] [CrossRef]
- Sensi, F.; D’Angelo, E.; D’Aronco, S.; Molinaro, R.; Agostini, M. Preclinical three-dimensional colorectal cancer model: The next generation of in vitro drug efficacy evaluation. Journal of Cellular Physiology 2019, 234, 181–191. [Google Scholar] [CrossRef]
- Metzcar, J.; Wang, Y.; Heiland, R.; Macklin, P. A review of cell-based computational modeling in cancer biology. JCO Clin. Cancer Inform. 2019, 2, 1–13. [Google Scholar] [CrossRef]
- Ingham-Dempster, T.; Walker, D. C.; Corfe, B. M. An agent-based model of anoikis in the colon crypt displays novel emergent behaviour consistent with biological observations. R. Soc. Open Sci. 2017, 4, 160858. [Google Scholar] [CrossRef]
- Pluchino, A.; Rapisarda, A.; Garofalo, C. Efficient promotion strategies in hierarchical organizations. Physica A: Statistical Mechanics and its Applications 2011, 390, 3496–3511. [Google Scholar] [CrossRef]
- Camillen, F.; Caprií, S.; Garofalo, C.; Ignaccolo, M.; Inturri, G.; Pluchino, A.; Rapisarda, A.; Tudisco, S. Multi agent simulation of pedestrian behavior in closed spatial environments. Journal of Artificial Societies and Social Simulations 2014, 17, 16. [Google Scholar]
- Le Pira, M.; Inturri, G.; Ignaccolo, M.; Pluchino, A.; Rapisarda, A. Finding shared decisions in stakeholder networks: An agent-based approach. In Physica A: Statistical Mechanics and its Applications, Elsevier, 2017, 466, 277–287. [Google Scholar] [CrossRef]
- Fichera, A.; Pluchino, A.; Volpe, R. A multi-layer agent-based model for the analysis of energy distribution networks in urban areas. In Physica A: Statistical Mechanics and its Applications, Elsevier, 2018, 508, 710–725. [Google Scholar] [CrossRef]
- Fazio, M.; Pluchino, A.; Inturri, G.; Le Pira, M.; Giuffrida, N.; Ignaccolo, M. Exploring the impact of mobility restrictions on the COVID-19 spreading through an agent-based approach. In Journal of Transport & Health, Elsevier, 2022, 25, 101373. [Google Scholar]
- Conti, E.; Di Mauro, L.S.; Pluchino, A.; Mulder, C. Testing for top-down cascading effects in a biomass-driven ecological network of soil invertebrates. In Ecology and Evolution, Wiley Online Library, 2020, 10, 7062–7072. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; DeAngelis, D.L. An overview of agent-based models in plant biology and ecology. In Annals of Botany, Oxford University Press US, 2020, 126, 539–557. [Google Scholar] [CrossRef] [PubMed]
- Glen, C.M.; Kemp, M.L.; Voit, E.O. Agent-based modeling of morphogenetic systems: Advantages and challenges. In PLoS Computational Biology, Public Library of Science, 2019, 15, e1006577. [Google Scholar] [CrossRef]
- An, G.; Mi, Q.; Dutta-Moscato, J.; Vodovotz, Y. Agent-based models in translational systems biology. WIREs Syst. Biol. Med. 2009, 1, 159–171. [Google Scholar] [CrossRef]
- Ingham-Dempster, T. A.; Rosser, R.; Corfe, B. M.; Walker, D. C. From cell to multi-crypt: Agent-based models of the human colon suggests novel processes of Field cancerisation. J. Comput. Sci. 2020, 41, 101066. [Google Scholar] [CrossRef]
- Mirams, G.R.; Fletcher, A.G.; Maini, P.K.; Byrne, H.M. A theoretical investigation of the effect of proliferation and adhesion on monoclonal conversion in the colonic crypt. In Journal of Theoretical Biology, Elsevier, 2012, 312, 143–156. [Google Scholar] [CrossRef]
- Niida, A.; Mimori, K.; Shibata, T.; Miyano, S. Modeling colorectal cancer evolution. J. Hum. Genet. 2021, 66, 869–878. [Google Scholar] [CrossRef]
- Doyle, J.J. Cell types as species: Exploring a metaphor. In Frontiers in Plant Science, Frontiers, 2022, 13, 868565. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H. What is a cell type and how to define it? Cell 2022, 185, 2739–2755. [Google Scholar] [CrossRef] [PubMed]
- Tëmkin, I.; Eldredge, N. Networks and hierarchies: Approaching complexity in evolutionary theory. In Macroevolution; Springer, 2015; pp 183–226.
- Bernstein, C.; Facista, A.; Nguyen, H.; Zaitlin, B.; Hassounah, N.; Loustaunau, C.; Payne, C.M.; Banerjee, B.; Goldschmid, S.; Tsikitis, V.L.; <i>, *!!! REPLACE !!!*; et al. Cancer and age-related colonic crypt deficiencies in cytochrome c oxidase I. In World Journal of Gastrointestinal Oncology, Baishideng Publishing Group Inc, 2010, 2, 429. [Google Scholar] [CrossRef] [PubMed]
- Humphries, A.; Wright, N. A. Colonic crypt organization and tumorigenesis. Nat. Rev. Cancer 2008, 8, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Van Der Flier, L.G.; Clevers, H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. In Annual Review of Physiology, Annual Reviews, 2009, 71, 241–260. [Google Scholar] [CrossRef]
- Wilensky, U. NetLogo; http://ccl.northwestern.edu/netlogo/, 1999.
- Potten, C.S.; Chwalinski, S.; Swindell, R.; Palmer, M. The spatial organization of the hierarchical proliferative cells of the crypts of the small intestine into clusters of ‘synchronized’ cells. In Cell Proliferation, Wiley Online Library, 1982, 15, 351–370. [Google Scholar] [CrossRef]
- Potten, C.S.; Kellett, M.; Rew, D.A.; Roberts, S.A. Proliferation in human gastrointestinal epithelium using bromodeoxyuridine in vivo: Data for different sites, proximity to a tumour, and polyposis coli. In Gut, BMJ Publishing Group, 1992, 33, 524. [Google Scholar] [CrossRef]
- Bravo, R.; Axelrod, D.E. A calibrated agent-based computer model of stochastic cell dynamics in normal human colon crypts useful for in silico experiments. In Theoretical Biology and Medical Modelling, Springer, 2013, 10, 1–24. [Google Scholar] [CrossRef]
- Zhao, R.; Michor, F. Patterns of proliferative activity in the colonic crypt determine crypt stability and rates of somatic evolution. In PLoS Computational Biology, Public Library of Science, 2013, 9, e1003082. [Google Scholar] [CrossRef]
- Yatabe, Y.; Tavaré, S.; Shibata, D. Investigating stem cells in human colon by using methylation patterns. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 10839–10844. [Google Scholar] [CrossRef] [PubMed]
- Nowak, M. A. Evolutionary dynamics: Exploring the equations of life. Harvard Univ. Press 2006. [Google Scholar]
- Baker, A.-M.; Cereser, B.; Melton, S.; Fletcher, A. G.; Rodriguez-Justo, M.; Tadrous, P. J.; Humphries, A.; Elia, G.; McDonald, S. A. C.; Wright, N. A.; et al. Quantification of crypt and stem cell evolution in the normal and neoplastic human colon. Cell Rep. 2014, 8, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Loeffler, M.; Stein, R.; Wichmann, H.-E.; Potten, C.S.; Kaur, P.; Chwalinski, S. Intestinal cell proliferation. I. A comprehensive model of steady-state proliferation in the crypt. Cell Proliferation 1986, 19, 627–645. [Google Scholar] [CrossRef] [PubMed]
- Di Gregorio, A.; Bowling, S.; Rodriguez, T. A. Cell competition and its role in the regulation of cell fitness from development to cancer. Dev. Cell 2016, 38, 621–634. [Google Scholar] [CrossRef]
- Nowak, M.A.; Komarova, N.L.; Sengupta, A.; Jallepalli, P.V.; Shih, I.-M.; Vogelstein, B.; Lengauer, C. The role of chromosomal instability in tumor initiation. Proceedings of the National Academy of Sciences, National Acad Sciences 2002, 99, 16226–16231. [Google Scholar] [CrossRef]
- Michor, F.; Iwasa, Y.; Nowak, M.A. Dynamics of cancer progression. In Nature Reviews Cancer, Nature Publishing Group UK, 2004, 4, 197–205. [Google Scholar] [CrossRef]
- Liu, X.; Jakubowski, M.; Hunt, J.L. KRAS gene mutation in colorectal cancer is correlated with increased proliferation and spontaneous apoptosis. In American Journal of Clinical Pathology, Oxford University Press, 2011, 135, 245–252. [Google Scholar] [CrossRef]
- Mantovani, F.; Collavin, L.; Del Sal, G. Mutant p53 as a guardian of the cancer cell. In Cell Death & Differentiation, Nature Publishing Group UK London, 2019, 26, 199–212. [Google Scholar]
- Rivlin, N.; Brosh, R.; Oren, M.; Rotter, V. Mutations in the p53 tumor suppressor gene: Important milestones at the various steps of tumorigenesis. In Genes & Cancer, SAGE Publications, 2011, 2, 466–474. [Google Scholar]
- Anderson, N.M.; Simon, M.C. The tumor microenvironment. In Current Biology, Elsevier, 2020, 30, R921–R925. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, S.; Guo, C. Crosstalk between macrophages and natural killer cells in the tumor microenvironment. In International Immunopharmacology, Elsevier, 2021, 101, 108374. [Google Scholar] [CrossRef]
- Al Tameemi, W.; Dale, T.P.; Al-Jumaily, R.M.Kh.; Forsyth, N.R. Hypoxia-modified cancer cell metabolism. In Frontiers in Cell and Developmental Biology, Frontiers Media SA, 2019, 7, 4. [Google Scholar] [CrossRef]
- Guo, G.; Wang, Y.; Zhou, Y.; Quan, Q.; Zhang, Y.; Wang, H.; Zhang, B.; Xia, L. Immune cell concentrations among the primary tumor microenvironment in colorectal cancer patients predicted by clinicopathologic characteristics and blood indexes. In Journal for ImmunoTherapy of Cancer, BioMed Central, 2019, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sieber, O.; Heinimann, K.; Tomlinson, I. Genomic stability and tumorigenesis. In Seminars in Cancer Biology, Elsevier, 2005, 15, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Woodward, J.; Woodward, J.F. Making Things Happen: A Theory of Causal Explanation; Oxford University Press, 2005.
- Woodward, J. Causation in biology: Stability, specificity, and the choice of levels of explanation. In Biology & Philosophy, Springer, 2010, 25, 287–318. [Google Scholar]
- Rondeau, E.; Larmonier, N.; Pradeu, T.; Bikfalvi, A. Characterizing causality in cancer. In Elife, eLife Sciences Publications, Ltd, 2019, 8, e53755. [Google Scholar] [CrossRef] [PubMed]
- Bechtel, W. Explicating top-down causation using networks and dynamics. In Philosophy of Science, Cambridge University Press, 2017, 84, 253–274. [Google Scholar] [CrossRef]
- Ellis, G.F.R. Top-down causation and emergence: Some comments on mechanisms. In Interface Focus, The Royal Society, 2012, 2, 126–140. [Google Scholar] [CrossRef]
- Zheng, Z.; Yu, T.; Zhao, X.; Gao, X.; Zhao, Y.; Liu, G. Intratumor heterogeneity: A new perspective on colorectal cancer research. In Cancer Medicine, Wiley Online Library, 2020, 9, 7637–7645. [Google Scholar] [CrossRef]
- Bertolaso, M. Philosophy of Cancer; Springer, 2016.
- Soto, A.M.; Sonnenschein, C. The tissue organization field theory of cancer: A testable replacement for the somatic mutation theory. In Bioessays, Wiley Online Library, 2011, 33, 332–340. [Google Scholar] [CrossRef] [PubMed]
| 1 |
Anoikis is a particular form of programmed cell death, triggered when a tissue cell detaches from the extracellular matrix. |
| 2 | There is still an important ongoing debate about necessary and sufficient conditions to assess whether it is possible to talk about cellular species or cellular types [34]. However, in this work the terminology will be used since the simulated cells are conceived as general asexual organisms, missing all the complex molecular, genetic and morphological characteristics that make them so difficult to be classified in real organisms. |
| 3 | Mitosis is the process of replication of a single cell, which produces two genetically identical cells. We opted to model the distribution of mitosis time as approximately Normal, influenced by its implementation in other computational models, see [20,43]. This choice was also prompted by the limited availability of specific data on the population distribution of this feature in the literature, despite the existence of numerous studies exploring the life cycle and division timing of individual cells, see for example [31,37,47]. |
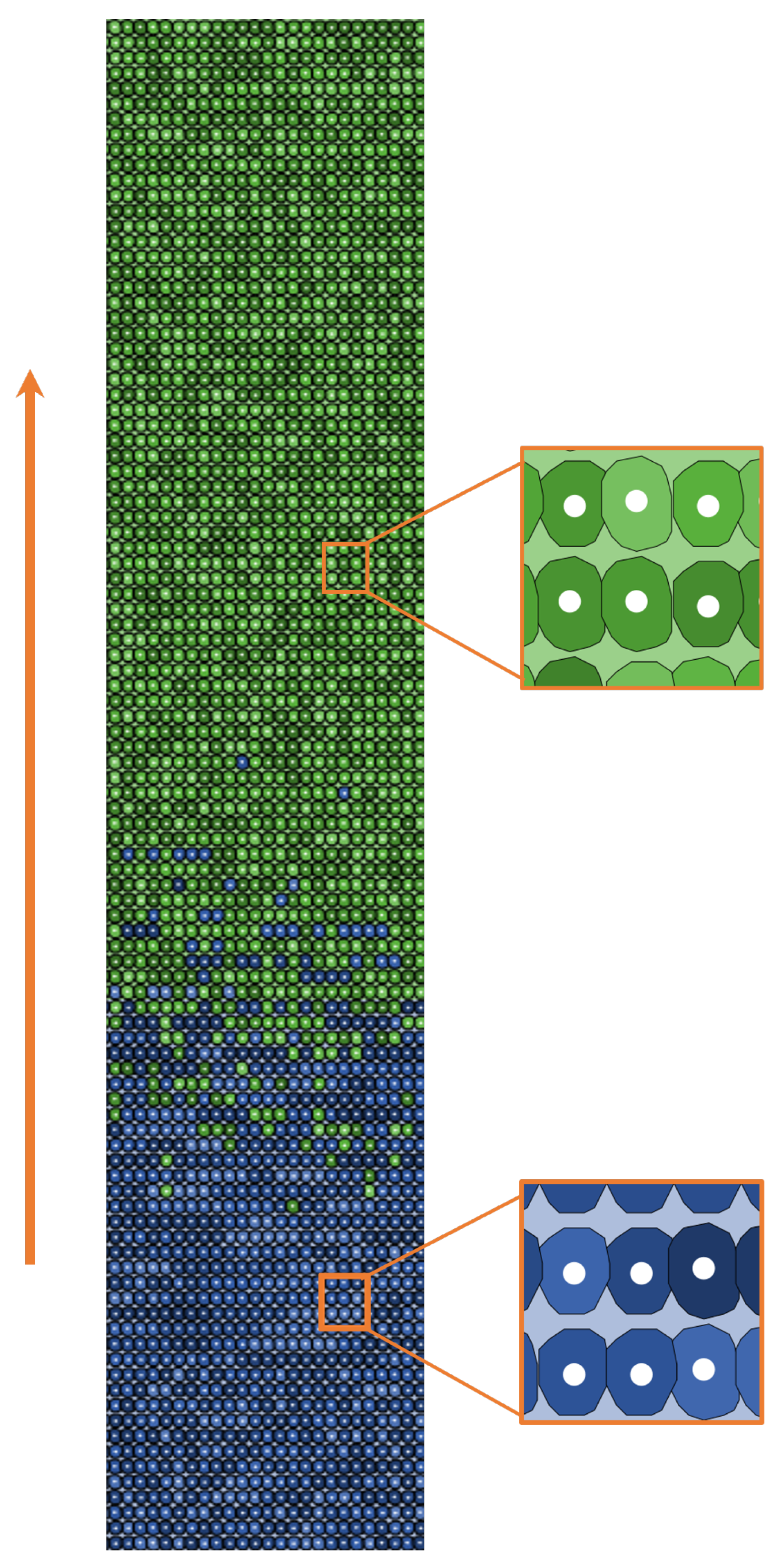
| Main Cell Variables | Values |
|---|---|
| Lifetime | physiological = min 96 h (hours), max 120 h; Tumoral = min 96 h, max 150 h. |
| Mitosis time3 | Normal distribution, mean = Mitosis mean time, variance 1. |
| Mitosis mean time | physiological (transit-amplifying, differentiated cells) = 24 h; with KRAS heterozygote = 12 h; with typology tumoral = 10 h. |
| -cat | Descendent gradient: 1 at the bottom, 0 at the top of the crypt. |
| Neoplastic typologies | pre-adenoma: true if APC = [1 1] |
| adenoma: true if APC = [1 1] and K-ras = [1 0] or [0 1] | |
| tumoral: true if APC = [1 1], K-ras = [1 0] and TP53 = [1 1] | |
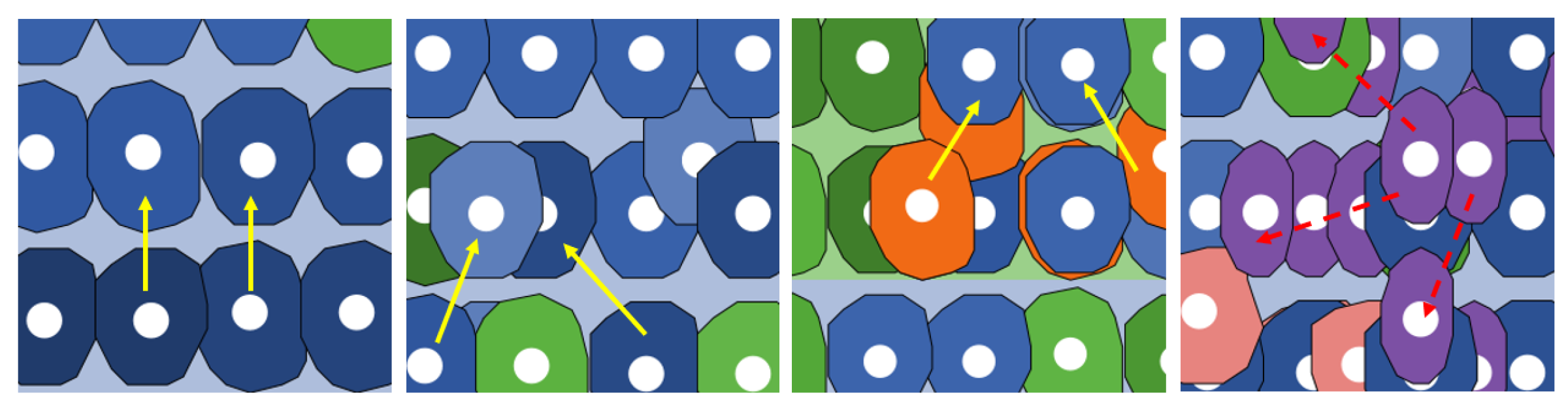
| Movement | Physiological cells = ↑ |
| pre-adenoma = | |
| adenoma = | |
| tumoral = no movement | |
| Proliferation directions | Physiological cells = ⟷ |
| pre-adenoma = | |
| adenoma = | |
| tumoral = |
| Genes | State |
|---|---|
| APC | wild type = [0 0], heterozygote = [1 0] [0 1], |
| mutated = [1 1] (trigger pre-adenoma typology) | |
| KRAS | wild type = [0 0], heterozygote = [1 0] [0 1] |
| (trigger adenoma typology) | |
| TP53 | wild type = [0 0], heterozygote = [1 0] [0 1], |
| mutated = [1 1] (trigger tumoral typology) | |
| Regulation genes (N = 50) | N = [ [0 0], [0 1], ..., [1 0] ] |
| if a given threshold x is passed | |
| and TP53 = [1 1], trigger cells death |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





