1. Introduction
Sunflower (
Helianthus annuus L.) is one of the major oil crops in the world. It is widely cultivated in various countries and ranks third among oilseeds and fourth among vegetable oils in terms of global production. Sunflower belongs to both oil and protein species due to its high content of oil (about 44%) and protein (about 16%). It is a rich source of healthy nutrients, minerals, antioxidants, and vitamins [
1,
2,
3,
4]. Therefore, sunflower is often utilized as a common ingredient in many processed food products such as butter, granola, cereals, breads, bakery products, trail mix, pasta, etc. In addition, sunflower ingredients may be present in processed foods due to contamination, which is a concern. However, sunflower belongs to food allergens. Ingestion of sunflower food by sensitized individuals can trigger a variety of symptoms ranging from mild to severe. Moreover, sunflower might elicit life-threatening anaphylactic reactions [
5,
6,
7,
8]. Accurate information on the presence of sunflower in food can prevent health problems for consumers who are sensitive to sunflower.
It should be noted that sunflowers are mainly distributed for its edible oil, the content and quality of which is the most desirable. Sunflower oil is used in salads and cooking foods or can be hardened to make margarine. In addition, sunflower oil has been found as an undeclared additive in occasionally contaminated and adulterated oils [
9,
10]. Thus tracking sunflower in highly processed foods and oils is particularly important for safe food production.
Edible oils represent an important part of the food system and are widely used in the human diet. They can provide essential fatty acids and vitamins. Oils are commonly used in industrial food production and home cooking worldwide. Edible oils are produced from oilseed crops such as soybean, rapeseed, sunflower, palm, corn, etc. In addition, there is a wide variety of blended oils [
11]. Oils from different plants vary in quality, toxicity, health benefits, and price. The modern food industry faces a big challenge regarding the authenticity and adulteration of oils. Oil Contamination or fraudulent alteration is the oil industry's biggest problem. Cross-contamination of oils can occur accidentally during the manufacture of oils. In addition, low-cost oils are intentionally mixed with expensive oils to increase profits in the oil business. [
9,
10,
12]. An important health aspect related to the different allergenicity of oilseeds should be noted. Allergenic oil crops can cause allergic disease in susceptible individuals. Allergenic proteins are mostly degraded during oil production, although available data indicate that even trace amounts of allergenic ingredients may trigger allergic reactions in sensitized persons [
13,
14].
In recent years, attention to the authenticity of edible oils has increased due to the emergence of recycled used cooking oils (UCO) in edible oils. UCO or WCO (waste cooking oils) is collected from the food industry and includes restaurant fryer oil, residential cooking oil, and blended oils. They contain toxic and dangerous compounds like aflatoxin, benzopyrene, etc. [
15,
16,
17,
18]. Thus, the presence of UCO in edible oil poses a significant threat to consumer health. To defend food safety and consumer-free choice, international regulations require monitoring and labeling of food ingredients. Thus, checking the authenticity of oil is a growing demand to protect laws and human health.
Today one of the most widespread and commercially viable raw materials for biodiesel production is UCO. Biodiesel is considered one of the most effective, renewable, environmentally friendly, and carbon-neutral alternative biofuels. Biodiesel production has been steadily increasing, especially in the 21st century and biodiesel is gaining more share of the fuel market worldwide [
19]. Biodiesel is a high-quality fuel too; therefore, biodiesel can be used in any industry segment where the mineral, i.e. petroleum-based diesel fuel is used, including the largest component, the internal combustion engines, in the transport sector [
20,
21].
Biodiesel can significantly reduce the air pollution and prevent environmental disasters. Another important advantage of biodiesel is the fact that biodiesel is biodegradable in both soil and water. Almost 89% of the carbon compounds contained in biodiesel can be biodegraded within one month [
22]. Biodiesel is a very effective solution for maintaining a healthier atmosphere in urban areas, for it can reduce 85% of emissions of polycyclic aromatic, which are identified as, extremely harmful to human health, carcinogenic compounds [
23].
It has been reported that 95% of global biodiesel is produced from various edible oils, including UCO (10%). In recent years, more attention has been paid to using UCO due to the scarcity of edible oils, which are important for the food industry [
24]. The UCO comprises different plant oils used in the food industry. The edible oils have different characteristics, such as types and ratios of the fatty acids, density, flash point, and kinematics viscosity which ultimately affect the quality of biodiesel made from these oils as well as the process of transesterification which is the most common method for biodiesel production in the industry. Therefore, the production of biodiesel from plant oils needs well planning and careful analysis of the raw materials i.e. UCO and it is crucial to determine the composition of UCO before starting the biodiesel production process [
24,
25,
26].
Sunflower ranks second (13 %) among single edible oils in terms of global biodiesel production [
24]. Due to the widespread sunflower cooking oil, it is one of the main components of UCO, therefore, quick and reliable analysis of UCO on the detection of sunflower oil is very important for planning the production of biodiesel, namely, adjusting the catalysts for the transesterification process which can determine the smooth, high quality and economically viable process for biodiesel production, thus being very important for the industry. Based on the described above reliable and efficient detection of sunflower in any oil is crucial for safe and high-quality food production as well as for biodiesel production.
To ensure the authenticity of the oil, it is necessary to verify the identity of the oily ingredients. To date, various approaches have been developed to determine the authenticity of edible oils. Analytical methods of detection often include nuclear magnetic resonance spectroscopy, high-performance liquid chromatography, gas chromatography, and mass spectrometry [
9,
27,
28,
29,
30]. These methods are based on edible oils' physical and chemical properties and analyze the content of various components such as fatty acids, tocopherols, amino acids, and sterols. However, data analysis in these methods is complicated because the chemical composition of edible oil depends on cultivars, seasons, and growing areas. Moreover, the detection limits of these methods are not sufficient to ensure the authenticity of the edible oils [
31,
32,
33,
34].
Molecular methods include DNA and protein-based techniques. DNA-based polymerase chain reaction (PCR) technology has been demonstrated as a promising tool for verifying the authenticity and traceability of processed foods. It has an advantage over protein-based methods because DNAs are more stable molecules than proteins in food processing. PCR allows accurate identification of species through their specific DNA sequences and does not depend on cultivars or environmental conditions. In recent years, PCR methods have been successfully used for the identification of oilseeds and the traceability of edible oils [
12,
33,
34,
35,
36,
37,
38,
39,
40,
41,
42].
Despite existing methods, there are still significant challenges in effective oil authenticity detection due to the diversity of oilseeds and edible oils, DNA degradation, and small amounts of DNA in the oil. Available data indicate that oil identification methods depend on the texture, chemical and molecular properties, and characteristics of the oilseed [
34]. Thus, the novel application of a DNA-based approach to the detection of each species of oilseeds is of particular interest.
A few works previously described PCR-based detection of sunflower in processed foods [
43,
44,
45] and edible oils [
10,
34]. However, a method for identifying sunflowers in UCO has not yet been reported. Due to the demands of the food and biodiesel industry, there is a need for more sensitive and efficient detection of sunflower in oils.
In this study, a comprehensive investigation of sunflower oils (crude, cold pressed, extra virgin, refined, and UCO) was carried out using different methods of DNA extraction and PCR amplification. A rapid and inexpensive CTAB protocol was developed for the efficient extraction of genomic DNA from oils. Uniplex PCR systems targeting the sunflower-specific helianthinin gene allow the identification of new specific DNA markers. An efficient nested PCR method was developed and optimized for accurate and rapid detection of sunflower in both edible and used cooking oil.
4. Discussion
Accurate detection of sunflower in oils is an urgent necessity to ensure the sustainable development of both the food and biodiesel industries. In this study, PCR-based technology was selected as a widely used approach to efficiently and reliably detect ingredients in processed foods. The analytical procedure for PCR detection consists of several steps: sample preparation, genomic DNA extraction, and PCR amplification. Effective performance of each step is an important prerequisite for successful PCR detection. Sufficient amounts of amplifiable genomic DNA and efficient PCR systems are important aspects of PCR sensitivity. In the present study, a comprehensive investigation and optimization of the critical factors of PCR detection was carried out to achieve highly sensitive identification of sunflowers in strongly processed oils.
Used (waste) cooking oil together with edible oils was chosen as the research objects due to the special importance of UCO, in particular, the adulteration of edible oils with UCO is a huge problem for the food industry while the use of UCO in biodiesel production is very beneficial. This study describes for the first time the identification of oilseed species in the UCO. The UCO is obtained from refined or other oils after use in cooking, so a more efficient detection technology than for refined oil analysis was required. Due to the strong treatment of refined oil and UCO, only trace amounts of highly degraded DNA were expected to be present in the oil samples, especially in UCO.
The process of producing oils from seeds involves mechanical, chemical, and/or thermal treatment, which varies according to the plant species, the end-use of the oil and manufacturing companies. Different types of oils are obtained from the seeds by adding one or more treatment modes. It was observed the loss of extracted DNA in parallel with the progress of the oil refining process [
37]. The crude, cold-pressed, virgin, and extra-virgin oils are generally produced without high-temperature treatment, although there are different means of their production. Even crude oil can be obtained by mechanical or chemical processing.
In the present work, four types of sunflower oil (crude, cold pressed, extra virgin, and refined) as well as UCO were investigated to determine the critical factors of oil characteristics and processing that affect PCR detection. It should be noted that the oils were produced by different companies and had different characteristics. In particular, crude, cold pressed and used cooking oils were produced locally by different small manufactures and were provided very quickly after production, they had characteristic odor and taste. In addition, visible precipitation was observed after storage. The extra virgin and refined oils were products of large industrial enterprises, and they were imported from other countries. Thus, they had to undergo more rigorous processing in order to obtain a final product with acceptable organoleptic properties and maintain quality for a long time. After storage, there was no smell or visible pallet in their bottles.
Proper sampling and DNA extraction are considered obstacles for PCR analysis of oil [
12,
35,
52]. Selecting the appropriate volume of oil is important for correct sampling. In this study, different volumes (between 500 µl and 300 ml) of four types of sunflower oil (crude, cold pressed, extra virgin, and refined) were investigated to select the correct volume for successful PCR amplification. Electrophoretic analysis of uniplex PCR products of pre-concentrated oils showed that despite the amplificability of all DNAs of the oils, the detection efficiency of sunflower by uniplex PCR was different due to the different amounts of DNA in the oil extracts. In particular, the crude oil sample contained more genomic DNA than the cold-pressed oil, and the cold-pressed oil sample had more DNA than the extra virgin or refined oil samples. Moreover, similar PCR efficiency was observed in pre-concentrated 12 mL crude oil and 48 mL cold pressed oil samples, suggesting that more processing was used on cold pressed oil than on crude oil due to different production by different companies. In addition, PCR detection of sunflower failed even when the volume of refined oil was increased to 300 ml.
Significant differences between the PCR results of cold-pressed and refined oils were previously reported in other works and were explained by severe processing during oil refining [
34,
35,
37]. In the present study, an unexpected significant difference was observed between the PCR results of cold pressed and extra virgin oils. In particular, DNAs from extra-virgin and refined oils showed similar PCR efficiency, but it was much lower than that of cold-pressed oil DNAs. This indicated that despite non-thermal processing of extra virgin oil it has undergone additional cleaning and processing in industrial production, as it was intended for transportation and long-term storage. While cold-pressed oils were relatively poorly processed in small local enterprises. Thus, the PCR efficiency of oil DNA depends on both the type of oil and the form of processing used in production.
A sufficient amount of amplifiable genomic DNA is a crucial prerequisite for successful PCR detection. Extracting DNA from plants is complicated because plant tissue is rich in primary and secondary metabolites. Due to the unique chemical composition of plant species, the extraction procedure needs to be optimized for each species [
53]. In addition, harsh processing in oil production affects the integrity of DNA and leads to high degradation of DNA fragments [
10,
12,
34,
35,
37]. Due to the low amount of DNA in the oils, pre-concentration [
35] or DNA enrichment [
50,
52,
54] procedures were often applied to extract DNA from large volume (10 ml – 500 ml) oil samples. Therefore, available extraction methods are often very laborious, long, expensive, and may fail to generate amplifiable DNA.
Two pre-concentration protocols were successfully used in the present work. It should be emphasized that the pre-concentration protocol 2 is a very simple, rapid and inexpensive procedure developed in this study. It has the advantage of using centrifugation in a 2 mL mini-centrifuge tube for 20 minutes at room temperature and is convenient for concentrating relatively small to medium volume (1 mL – 48 mL) samples. While pre-concentration 1 [
35] requires a large refrigerated centrifuge with larger tubes (≥ 25 mL) and is more suitable for the concentration of large volume samples.
Despite the availability of commercial kits, various modifications of CTAB-based methods are still widely used to extract DNA from plant foods, including oils [
34,
35,
42,
50,
52,
54,
55]. This is due to the availability of additional optimization of CTAB methods and the relatively low cost compared to expensive ready-made DNA isolation kits. Sunflower contains high concentrations of polyphenols, polysaccharides, and tannins that prevent amplifiable DNA isolation and are considered as PCR inhibitors. Earlier works described optimized procedures using the QIAamp DNA Stool Mini Kit (Qiagen) [
10] and modified CTAB [
34] for DNA extraction from 15 ml and 30 ml sunflower oil without sample pretreatment.
In this study, two (standard and our modified) CTAB protocols were tested for DNA extraction from sunflower seeds and crude oil. To increase the amount of DNA isolated from the oil, a modified CTAB protocol was developed that lacked the final purification steps of the standard CTAB method. The results showed that amplifiable DNA was obtained from the seeds only by the standard CTAB method and from the oil only by the modified protocol.
This suggests that vigorous purification by the standard CTAB method is important for seed DNA because its extract is rich in PCR inhibitors. While a short modified protocol supports the isolation of more DNA from oil samples, in which the main challenge is the small amount of DNA. It is important to note that our modified CTAB protocol ensures fast, cheap, and easy extraction of oil DNA and facilitates oil PCR detection. As far as we know
Figure 2A represents a unique picture of a visible band of oil DNA that has not yet been previously obtained.
Examination of commercial kits of DNA extraction revealed Nucleospin, Qiagen food and Oil kit suitable for sunflower oil as well as Qiagen plant kit useful for sunflower seeds. These kits were applied to sunflower oil for the first time. Our results are consistent with previous findings of the effectiveness of the traditional CTAB method in removing polyphenols, but its inability to extract DNA from oil. In addition, earlier reports on the effectiveness of the NucleoSpin and Qiagen Food kits in extracting amplifying DNA from oils were confirmed [
12,
35,
56].
This study is the first to present spectrophotometric data for sunflower oils. The results obtained coincide with previous reports about the dependence of DNA yields and purity on the extraction method used for oil DNA isolation [
12,
35,
52,
54]. The low ratios A260/A230 between 0.18 and 0.62 coincide with previous data [
12] and indicate remaining organic compounds such as polysaccharides, phenols, etc. in oil DNA extracts. Comparison and interpretation of electrophoretic and spectrophotometric results obtained for DNAs from 700 µl of oils suggested that all DNAs extracts were amplifiable and generated expected amplicons by both the double and nested PCRs despite low DNA concentration (about 1.3 ng/ µl) and low purity (ratios A260/A280 about 1.1 and A260/A230 about 0.18). Our outcomes confirm previous findings that the contaminants in the oil DNA extracts may not inhibit the PCR [
12,
35].
The PCR approach and the number of copies of the target DNA largely determine the sensitivity of the PCR assay. In this study, a nested PCR approach was chosen as a highly sensitive method for DNA detection. Nested and double PCR allows for increased PCR sensitivity by direct re-amplification of the product from the primary PCR with a second PCR [
57]. This technique is considered a promising tool for the analysis of processed foods. Its application to trace virgin olive oil [
58], as well as transgenic soybean, corn, and canola oils [
42] allowed to reduce the oil sample volume to 200 μl and 2 ml, respectively.
The PCR approach for species identification is based on the detection of a specific DNA sequence characteristic of that species. Debode et al. [
37] reported that the use of species-specific sequences of chloroplastic DNA and ribosomal RNA genes as high copy number targets gave better results than low cellular copy number targets in PCR detection of oils. Notably, multicopy targets provide a better signal, but they are not suitable for quantitative purposes because the copy number can vary in the cells [
37]. Previous studies used PCRs targeting sunflower-specific sequences of multicopy rDNA and plastid DNA to detect sunflower in foods [
44,
45] and in oils [
10], respectively.
In this study, two eukaryote-specific PCR systems producing 140 bp [
49] and 167 bp fragments [
50] of the multicopy18S RNA gene were used to check the amplifiability of oil DNAs. It should be noted that the comparative analysis of 18S-140 and 18S-167 PCR products in concentrated oils contributed to the development of pre-concentration II and modified CTAB methods, as well as to the identification of critical factors for PCR detection of oil. These PCR systems were able to generate amplified products by uniplex PCR, even in DNAs from 700 μl of oils. Our results confirmed previous reports on the successful use of primers 18S-140f/18S-140r [Zhang, 2018] to amplify DNAs from processed foods and primers 18S-167f/18S-167r to amplify DNAs from maize and soybean oils [
50].
Commonly, species-specific protein genes are utilized as low copy number targets in PCR for reliable identification and quantification of species DNA. In this study, the Helianthinin gene was chosen as a target gene to develop PCR systems for sunflower detection because of the available data on the sunflower species-specificity of Helianthinins proteins [
51]. Moreover, a PCR assay targeting the 60 bp sequence of the
helianthinin gene was developed by Hernandez et al. [
43] and then used to identify sunflower in edible oils by He et al. [
34]. In the present work, three uniplex PCR systems were developed for the detection of sunflower using newly designed primer pairs. Moreover, two pairs of them, producing amplicons of 188 bp and 162 bp, were successfully used in nested and double PCR systems.
The use of both double and nested PCRs dramatically changed the sensitivity of sunflower detection in oils. Uniplex PCRs allowed the detection of sunflower only in concentrated 12 ml crude and 48 ml cold-pressed oil samples, but these methods failed to analyze extra virgin and refined oils. However, double and nested PCRs enabled sunflower tracking in 700 µl of all studied oils extracted by NucleoSpin, oil kit, Qiagen Food, or modified CTAB methods. It should be emphasized that nested PCR exhibited a higher sensitivity than double PCR. The absence of amplified products in DNAs of maize and soybean seeds and oils as well as in water-negative controls showed high specificity and purity of PCR assays. Thus, the application of the nested PCR method resulted in efficient and reliable detection of sunflower in edible and used cooking oils. The ability to detect sunflower in 700 μl of refined and used cooking oils improved the sensitivity of previous methods that detected sunflower in 15 ml and 30 ml of edible oil [
10,
34].
Further experiments are needed to validate the developed technology for detecting sunflowers in blended oils. It would also be possible to develop a similar technology to detect other oilseeds in oils.
Noteworthy, the use of modified CTAB in combination with nested PCR provides cheap, fast, and efficient tracking of sunflowers in oils. Compared to existing methods of oil authentication, the main advantages of the technology developed in this study are higher sensitivity, less amount of oil samples, and a simple DNA extraction method. Furthermore, this is the first report of sunflower detection in UCO, which may be useful for both food and biodiesel production.
The present work meets critical challenges of sustainable food and biofuel systems, such as food security, food waste utilization, and eco-friendly fuel production. These challenges are closely related to sunflower oils, which are widely used in food preparation and result in waste cooking oils. The use of WCO in biodiesel production is an excellent example of the use of food waste and contributes to the sustainability of the food and biofuel industries.
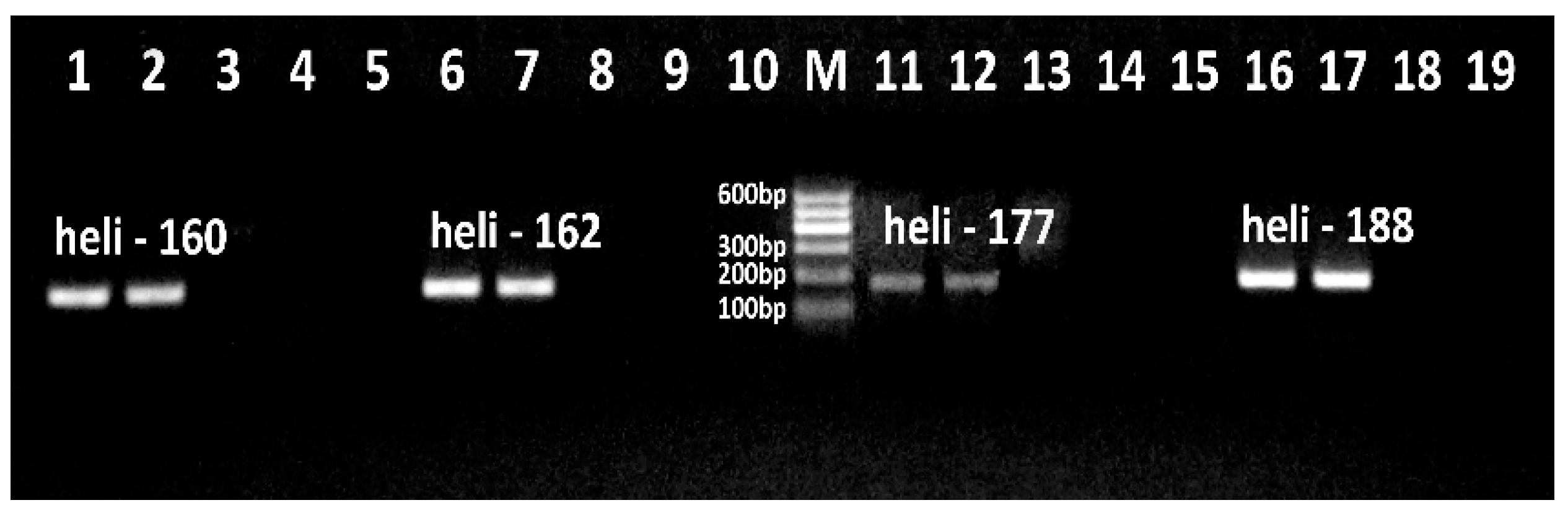
Figure 1.
PCR detection of sunflower Helianthinin gene using primer pair heli160f/heli160r (lanes 1-5), heli162f/heli162r (lanes 6-10), heli177f/ heli177r (lanes 11-15) and heli188f/heli188r (lanes 16-19). Samples: lanes 1-2, 6-7, 11-12, 16-17. Sunflower; lanes 3, 8, 13, 18. Maize; lanes 4, 9, 14,19. Soybean; lanes 5, 10, 15. water (negative control).
Figure 1.
PCR detection of sunflower Helianthinin gene using primer pair heli160f/heli160r (lanes 1-5), heli162f/heli162r (lanes 6-10), heli177f/ heli177r (lanes 11-15) and heli188f/heli188r (lanes 16-19). Samples: lanes 1-2, 6-7, 11-12, 16-17. Sunflower; lanes 3, 8, 13, 18. Maize; lanes 4, 9, 14,19. Soybean; lanes 5, 10, 15. water (negative control).
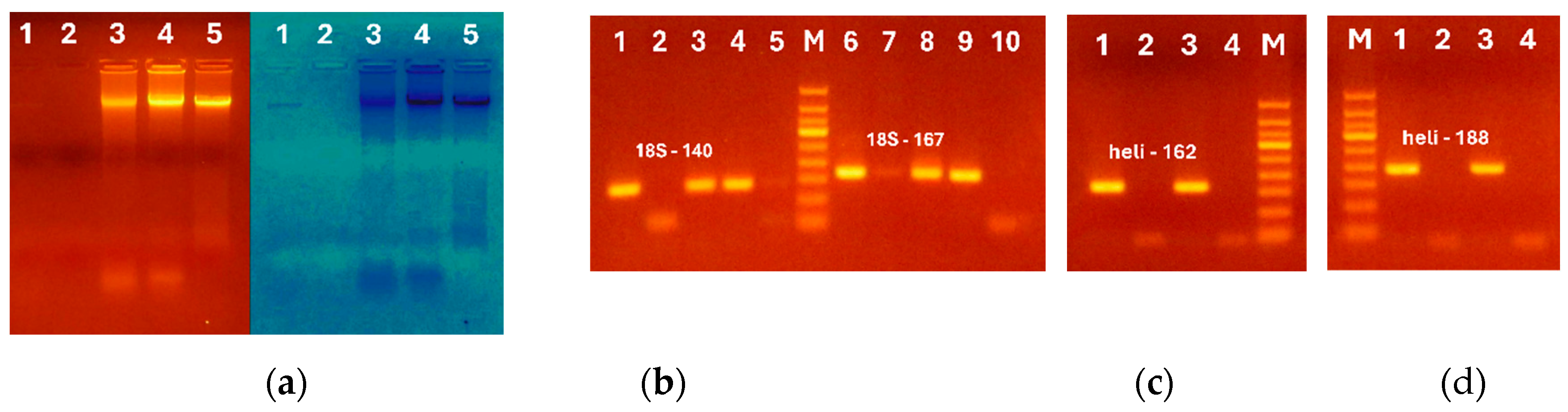
Figure 2.
Comparison of CTAB protocols for DNA extraction from sunflower seeds and oils: (a) Genomic DNAs from crude oil (lanes 1-2) and seeds (lanes 3-5) extracted by modified CTAB ( lanes 1, 5) and standard CTAB (lanes 2-4); (a-d) PCR amplification using primers 18S-140f/18S-140r (b, lanes 1-5), 18S-167f 18S-167r (b, lanes 6-10); heli162f/ heli162r (c) and heli188f/heli188r (d); Samples: DNA from seeds (b, lanes 1-2, 6-7, c, d. lane 1) and crude oil (b, lanes 3-5, 8-9; c, d. 2-3) extracted by standard CTAB (b, lanes 1, 5, 6; c, d, 1-2) and modified CTAB (b, lanes 2-4, 7-9; c, d. lanes 3-4). Water-negative control (b, lane 10, c, d, lane 4). M. Molecular weight marker (Qiagen GelPilot 100 bp ladder). .
Figure 2.
Comparison of CTAB protocols for DNA extraction from sunflower seeds and oils: (a) Genomic DNAs from crude oil (lanes 1-2) and seeds (lanes 3-5) extracted by modified CTAB ( lanes 1, 5) and standard CTAB (lanes 2-4); (a-d) PCR amplification using primers 18S-140f/18S-140r (b, lanes 1-5), 18S-167f 18S-167r (b, lanes 6-10); heli162f/ heli162r (c) and heli188f/heli188r (d); Samples: DNA from seeds (b, lanes 1-2, 6-7, c, d. lane 1) and crude oil (b, lanes 3-5, 8-9; c, d. 2-3) extracted by standard CTAB (b, lanes 1, 5, 6; c, d, 1-2) and modified CTAB (b, lanes 2-4, 7-9; c, d. lanes 3-4). Water-negative control (b, lane 10, c, d, lane 4). M. Molecular weight marker (Qiagen GelPilot 100 bp ladder). .
Figure 3.
Comparison of Oil kit and NucleoSpin for DNA extraction from sunflower oil. PCR amplification using primer pairs 18S-140f/18S-140r (a, lanes 1-6); heli160f/ heli160r (b, lanes 1-14); heli162f/ heli162r (b, lanes 5-8) and heli188f/heli188r (b, lanes 9-12). Samples: DNAs from crude oil (a, lanes 1-4, b. lanes 1-2, 5-6, 9-10) and seeds (a, lane 5; b, lanes 3, 7, 11) extracted by Oil kit (a, lanes 1-2, b, 1, 5, 9), by NucleoSpin (a, lanes 3-4; b. 2, 6, 10) and by Qiagen plant (a, lane 5; b, lanes 3, 7, 11). Water – negative control (a, lane 6, b. lanes 4, 8, 12). M. Molecular weight markers: Qiagen GelPilot 50 bp ladder) (a) and Qiagen GelPilot 100 bp ladder (b).
Figure 3.
Comparison of Oil kit and NucleoSpin for DNA extraction from sunflower oil. PCR amplification using primer pairs 18S-140f/18S-140r (a, lanes 1-6); heli160f/ heli160r (b, lanes 1-14); heli162f/ heli162r (b, lanes 5-8) and heli188f/heli188r (b, lanes 9-12). Samples: DNAs from crude oil (a, lanes 1-4, b. lanes 1-2, 5-6, 9-10) and seeds (a, lane 5; b, lanes 3, 7, 11) extracted by Oil kit (a, lanes 1-2, b, 1, 5, 9), by NucleoSpin (a, lanes 3-4; b. 2, 6, 10) and by Qiagen plant (a, lane 5; b, lanes 3, 7, 11). Water – negative control (a, lane 6, b. lanes 4, 8, 12). M. Molecular weight markers: Qiagen GelPilot 50 bp ladder) (a) and Qiagen GelPilot 100 bp ladder (b).
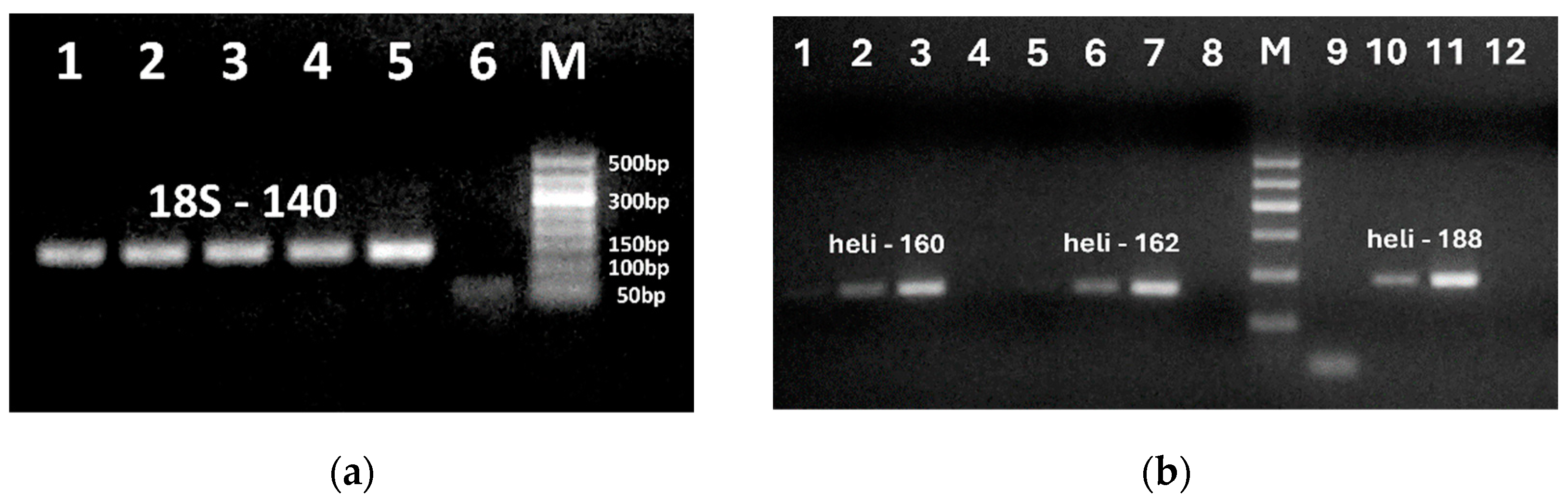
Figure 4.
PCR testing of various pre-concentrated sunflower oils using primers: heli188f/heli188r (a, lanes 1-6; c, lanes 1-6); heli162f/ heli162r (b, lanes 1-4); 18S-140f/18S-140r (c, lanes 7-12). Samples: crude (a, lane 1, b, lane 1, c, lanes 5, 11); cold press (a, lane 2-3, b. lane 2, c, lanes 1-2. 7-8); extra virgin (c, lanes 3-4, 9-10), refined (a, lane 4-5, B. 3) extracted by modified CTAB (a, lane 1-5, B, 1-3); NucleoSpin (c, lanes 1-5, 7-11). Water – negative control (a, lane 6, b. lane 4, c, lanes 6, 12). M. Molecular weight markers: Qiagen GelPilot 50 bp ladder).
Figure 4.
PCR testing of various pre-concentrated sunflower oils using primers: heli188f/heli188r (a, lanes 1-6; c, lanes 1-6); heli162f/ heli162r (b, lanes 1-4); 18S-140f/18S-140r (c, lanes 7-12). Samples: crude (a, lane 1, b, lane 1, c, lanes 5, 11); cold press (a, lane 2-3, b. lane 2, c, lanes 1-2. 7-8); extra virgin (c, lanes 3-4, 9-10), refined (a, lane 4-5, B. 3) extracted by modified CTAB (a, lane 1-5, B, 1-3); NucleoSpin (c, lanes 1-5, 7-11). Water – negative control (a, lane 6, b. lane 4, c, lanes 6, 12). M. Molecular weight markers: Qiagen GelPilot 50 bp ladder).
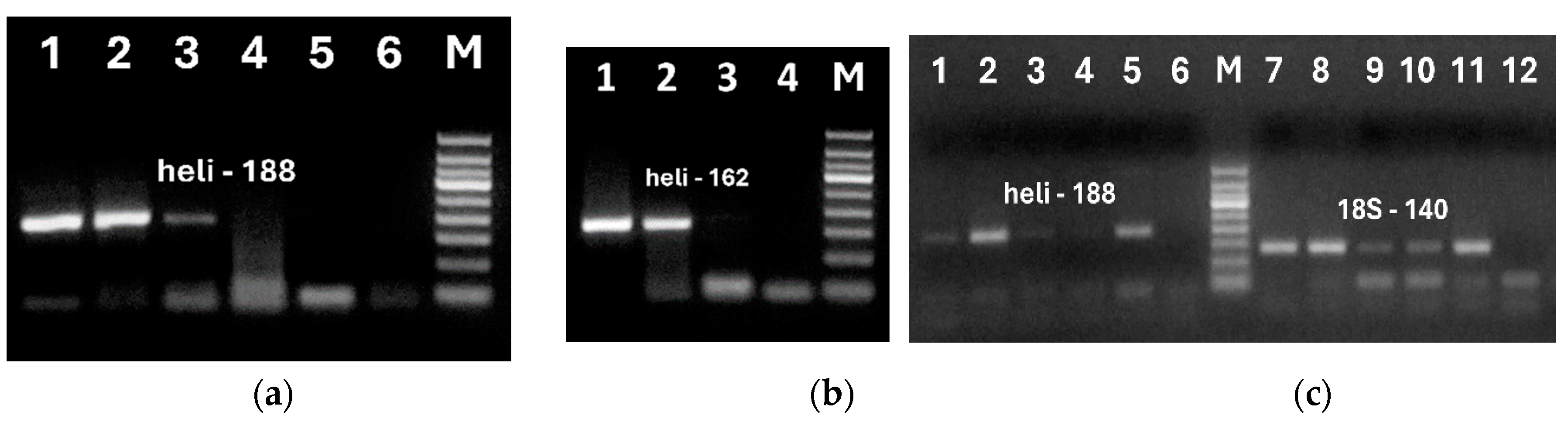
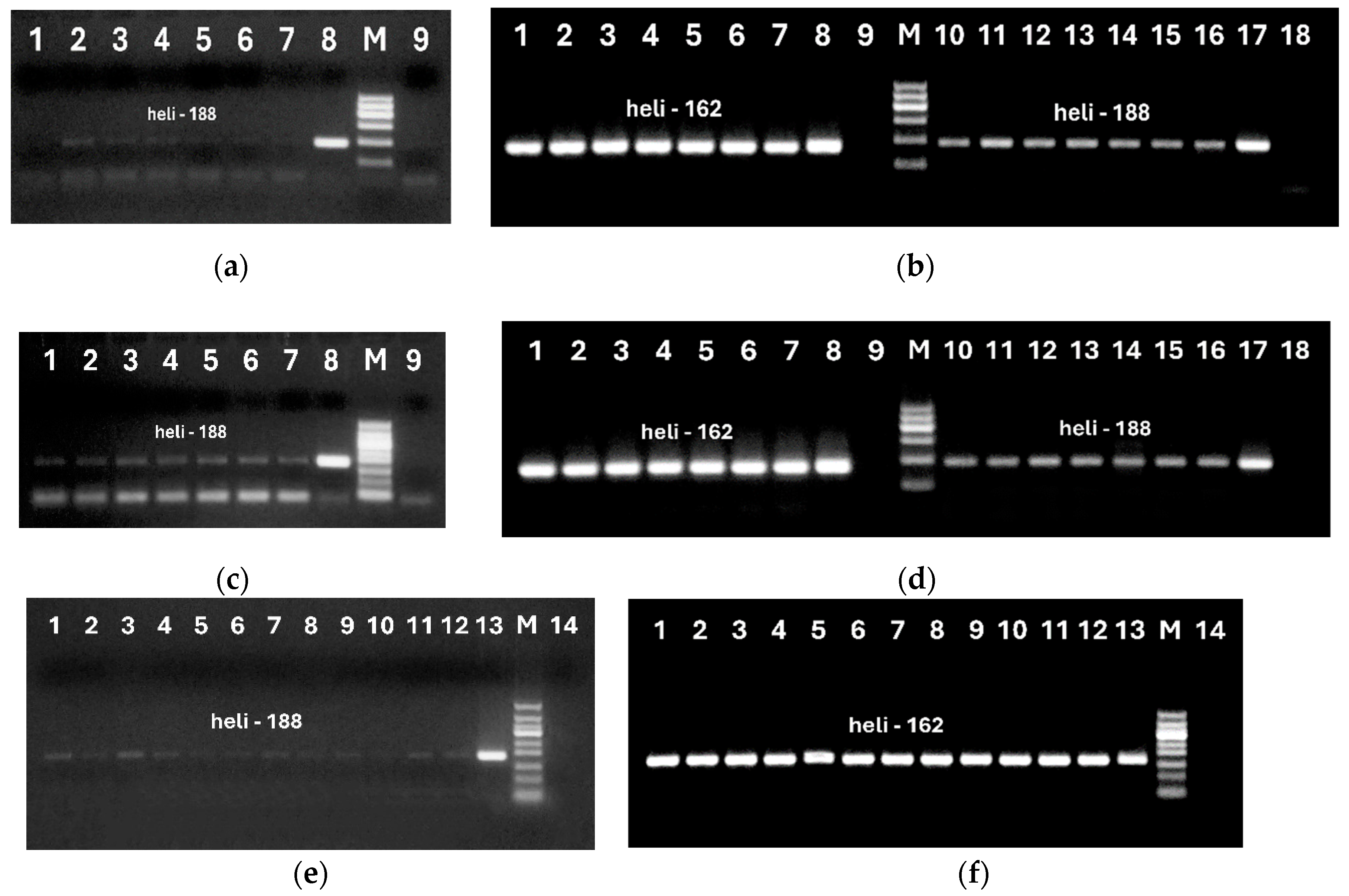
Figure 5.
Primary (a, c, e), Nested (b, d, lanes 1-9; f, lanes 1-14)) and Double (b, d, lanes 10-18) PCRs using primers heli188f/heli188r (a, c, e, and b, d, lanes 10-18) and heli162f/ heli162r (b, d, lanes 1-9, f, lanes 1-14) of sunflower DNA extracted from oils by NucleoSpin (a, lanes 1-9, b, lanes 1-18, e, f, lanes 1-4), oil kit (c, lanes 1-9, d, lanes 1-18, e, f, lanes 5-8); Qiagen Food (e, f, lanes 9-12). Samples: refined (a, c, lanes1-2; b, d, lanes 1-2, 10-11; e, f, lanes 1, 5, 9); extra virgin (a, c, lanes 3-4, b, d, lanes 3-4, 12-13; e, f, lanes 2, 6, 10); cold pressed (a, c, lanes 5-6, b, d, lanes 5-6, 14-15; e, f, lanes 3, 7, 11); crude (a, c, lane 7, b, d, lanes 7, 16; e, f, lanes 4, 8, 12); seeds (a, c, lane 8; b, d, lanes 8, 17; e, f, lane 13); water – negative control (a, c, lane 9; b, d, lanes 9, 18; e, f, lane 14); M. Molecular weight markers: Qiagen GelPilot 100 bp ladder (a, b, d) and Qiagen GelPilot 50 bp ladder) (c, e, f).
Figure 5.
Primary (a, c, e), Nested (b, d, lanes 1-9; f, lanes 1-14)) and Double (b, d, lanes 10-18) PCRs using primers heli188f/heli188r (a, c, e, and b, d, lanes 10-18) and heli162f/ heli162r (b, d, lanes 1-9, f, lanes 1-14) of sunflower DNA extracted from oils by NucleoSpin (a, lanes 1-9, b, lanes 1-18, e, f, lanes 1-4), oil kit (c, lanes 1-9, d, lanes 1-18, e, f, lanes 5-8); Qiagen Food (e, f, lanes 9-12). Samples: refined (a, c, lanes1-2; b, d, lanes 1-2, 10-11; e, f, lanes 1, 5, 9); extra virgin (a, c, lanes 3-4, b, d, lanes 3-4, 12-13; e, f, lanes 2, 6, 10); cold pressed (a, c, lanes 5-6, b, d, lanes 5-6, 14-15; e, f, lanes 3, 7, 11); crude (a, c, lane 7, b, d, lanes 7, 16; e, f, lanes 4, 8, 12); seeds (a, c, lane 8; b, d, lanes 8, 17; e, f, lane 13); water – negative control (a, c, lane 9; b, d, lanes 9, 18; e, f, lane 14); M. Molecular weight markers: Qiagen GelPilot 100 bp ladder (a, b, d) and Qiagen GelPilot 50 bp ladder) (c, e, f).
Figure 6.
Primary (a) and nested (b) PCRs with primers heli188f/heli188r (a) and heli162f/ heli162r (b) of DNAs from different oils. Samples: sunflower oils (a, b, lanes 1-3), soybean oil (a, b, lane 4) and maize oil (a, b, lane 5), sunflower seeds (a, b, lane 6); water (a, lane 7, b, lanes 7-8).
Figure 6.
Primary (a) and nested (b) PCRs with primers heli188f/heli188r (a) and heli162f/ heli162r (b) of DNAs from different oils. Samples: sunflower oils (a, b, lanes 1-3), soybean oil (a, b, lane 4) and maize oil (a, b, lane 5), sunflower seeds (a, b, lane 6); water (a, lane 7, b, lanes 7-8).
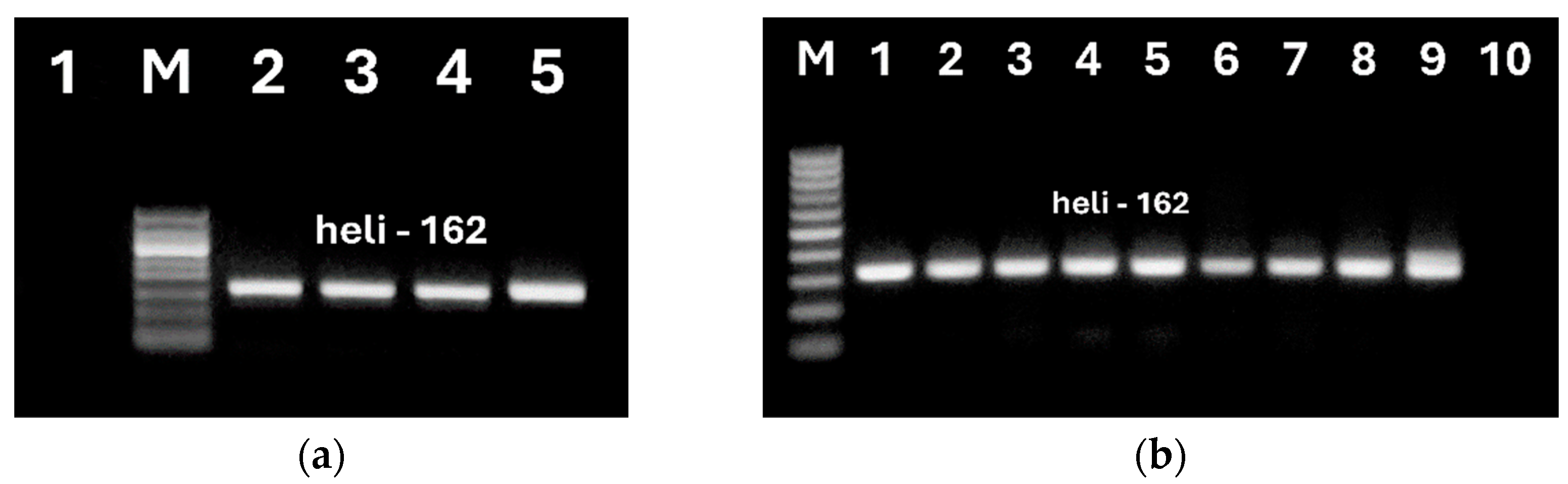
Figure 7.
Second PCRs using primers heli162f/ heli162r of DNAs from sunflower oils and UCO extracted by NucleoSpin kit (a, lanes 2-5, b, lanes 6-8) and modified CTAB (b, lanes 1-5) methods. Samples: refined oil (a, lanes 2, b. lanes 3-4, 6-7); extra virgin oil (b, lanes 5, 8); UCO (a, lanes 3-4, b, lanes 1-2); seeds (a, lane 5, b, lane 9), water (a, lane 1, b, lane 10).
Figure 7.
Second PCRs using primers heli162f/ heli162r of DNAs from sunflower oils and UCO extracted by NucleoSpin kit (a, lanes 2-5, b, lanes 6-8) and modified CTAB (b, lanes 1-5) methods. Samples: refined oil (a, lanes 2, b. lanes 3-4, 6-7); extra virgin oil (b, lanes 5, 8); UCO (a, lanes 3-4, b, lanes 1-2); seeds (a, lane 5, b, lane 9), water (a, lane 1, b, lane 10).
Table 1.
Oligonucleotide primers used in PCR.
Table 1.
Oligonucleotide primers used in PCR.
| Primer |
Sequence 5'→3' |
Target gene |
Amplicon size (bp) |
Reference |
| heli160f |
TCAACGCCCACAATCTTCTC |
helianthinin |
160 |
This study |
| heli160r |
CTTCCTTGTTCATTGGCTCTCT |
|
|
|
| |
|
|
|
|
| heli162f |
CTTCCCAGGCTGACTTTGTAA |
helianthinin |
162 |
This study |
| heli162r |
GAAGATTGTGGGCGTTGATTG |
|
|
|
| |
|
|
|
|
| heli177f |
CCTTCCTACGTCAACACCCC |
helianthinin |
177 |
This study |
| heli177r |
TCATAGGTTCTGCGGCATCC |
|
|
|
| |
|
|
|
|
| heli188f |
CCTTCCCAGGCTGACTTTGT |
helianthinin |
188 |
This study |
| heli188r |
CTCAAGGCTCCCTCGGTTAC |
|
|
|
| |
|
|
|
|
| 18S-140f |
TCTGC-CCTATCAACTTTCGATGGTA |
18S rRNA |
140 |
Zhang , 2015 |
| 18S-140r |
AATTTGCGCGCCTGCTGCCTTCCTT |
|
|
|
| |
|
|
|
|
| 18S-167f |
GCAAGACCGAAACTCAAAGGA |
18S rRNA |
167 |
Duan, 2021 |
| 18S-167r |
ACGACAGCCATGCAGCACC |
|
|
|
Table 2.
DNA concentration and purity of the oil extracts obtained with Nucleospin, Qiagen Food, oil kit, and modified CTAB methods.
Table 2.
DNA concentration and purity of the oil extracts obtained with Nucleospin, Qiagen Food, oil kit, and modified CTAB methods.
| Extraction Method |
Oil Samples (0.7 ml) |
DNA (ng/µL) |
A260/A280
|
A260/A230
|
| NucleSspin |
Crude |
2.39 ± 0.76 |
1.80 ± 0.90 |
0.53 ± 0.09 |
| Cold pressed |
1.51 ± 0.64 |
1.60 ± 0.50 |
0.42 ±0.07 |
| Extra virgin |
4.82 ± 2.07 |
1.49 ± 0.07 |
0.59 ± 0.04 |
| Refined |
4.25 ± 0.90 |
1.42 ± 0.01 |
0.62 ± 0.02 |
| UCO |
3.415 ± 0.70 |
2.09 ± 0.05 |
0.55 ± 0.07 |
| Qiagen Food kit |
Crude |
1.41 ± 0.02 |
1.14 ± 0.04 |
0.24 ± 0.01 |
| Cold pressed |
1.49 ± 0.55 |
1.00 ± 0.34 |
0.25 ± 0.03 |
| Extra virgin |
1.30 ± 0.14 |
2.14 ± 1.58 |
0.25 ± 0.02 |
| Refined |
1.40 ± 0.00 |
1.87 ± 0.20 |
0.25 ± 0.02 |
| Oil Kit |
Crude |
3.45 ± 0.10 |
2.03 ± 0.31 |
0.46 ± 0.00 |
| Cold pressed |
2.58 ± 0.55 |
2.03 ± 0.21 |
0.45 ± 0.05 |
| Extra virgin |
2.47 ± 1.75 |
1.73 ± 0.17 |
0.44 ± 0.16 |
| Refined |
1.72 ± 0.56 |
2.02 ± 0.10 |
0.36 ± 0.07 |
| Modified CTAB |
Crude |
5.15 ± 0.54 |
2.32 ± 0.13 |
0.24 ± 0.02 |
| Cold pressed |
5.41 ± 0.31 |
2.62 ± 0.80 |
0.25 ± 0.10 |
| Extra virgin |
6.58 ± 1.75 |
2.11 ± 0.31 |
0.20 ± 0.01 |
| Refined |
5.12 ± 0.71 |
1.91 ± 0.01 |
0.18 ± 0.01 |
| UCO |
7.05 ± 1.68 |
2.09 ± 0.04 |
0.24 ± 0.02 |