1. Introduction
The California Genetic Disease Screening Program (GDSP) identifies children diagnosed with Cystic Fibrosis (CF) and CF-Related Metabolic Syndrome (CRMS) using a 3-tier IRT-DNA-DNA testing method. When our 1st tier assay of immunoreactive trypsinogen (IRT) is higher than a cutoff, we send the dried bloodspots (DBS) for analysis with panel of disease-causing genetic mutations in the CFTR gene; if only one panel mutation is detected, we sequence the CFTR gene. The IRT cutoff is an important gateway to further testing for CF.
Our fixed IRT cutoff has changed periodically to anticipate a nominal population-based [
1] yearly IRT screen positive rate after a new reagent kit has been introduced. Observed seasonal variation in our population IRT percentiles led us to develop a seasonal model to help set initial kit-based cutoffs to run throughout the duration of each kit which can exceed 6 months. Upon introduction of a new IRT reagent kit, we adjust our IRT cutoff to approximate a 1.6% yearly IRT screen-positive rate, to avoid unnecessary genetic testing.
It is known that IRT levels in newborns differ by race, gestational age, and gestational weight. [
2,
3] IRT population means can also change by season and can differ between laboratory reagent kits [
3,
4,
5]. Some state screening programs establish cutoffs based on floating daily percentiles; however those programs call out a much higher percentage (4-5% vs. 1.6%) of the newborn population for molecular testing than does California [
3,
6]. We cannot predict the diverse mix of races and birth conditions among the newborns screened in California laboratories on any given day, but we can leverage statewide population data to take regular seasonal variability into account, to set fixed cutoffs, and still identify CF cases efficiently. Seasonal variation must be monitored to maintain an effective fixed cutoff, whereas variation is automatically built into a floating cutoff. We wanted to evaluate the trade-offs for each method. There is another approach, which is a repeat IRT after a positive IRT screening test followed by molecular testing (IRT-IRT-DNA) [
7,
8]. IRT-IRT-DNA shows promise in lowering the false-positive IRT results, but we do not do repeat testing in California and cannot emulate the method.
We reevaluated our fixed cutoffs in the summer of 2017 after we missed two CF cases close to the IRT cutoff boundary. In May of 2017 a new reagent kit was introduced, the number of IRT positives sent for molecular testing dropped and we missed two cases over the following two months of summer. This was a perfect storm where a new kit required a change to a lower fixed cutoff moving into the summer when population IRT positive percentiles are the lowest of the year. There are many reasons a case of CF can be missed [
9]; for our investigation we address false-negative CF cases due to a low IRT result below the cutoff.
We provide results of our seasonal analysis of population data which establishes initial IRT cutoffs for new reagent kits that anticipate variation throughout the life of a kit. We also compare GDSP cutoffs with the alternative method of floating laboratory-based cutoffs to see which method would correctly identify more CF cases near the IRT detection boundary. It is also important to compare which method may increase or reduce the number of costly molecular testing among newborns without CF, testing that can identify variants of unknown significance (VUS) creating distress for families of newborns thus identified.
2. Methods
2.1. Study Population
Initial IRT screening is performed by five laboratories contracted by GDSP that send results and specimens to the central GDSP Genetic Disease Laboratory. IRT values (ng/mL) are measured using the AutoDELFIA Neonatal IRT Kit (Revvity, Waltham, MA) on newborn dried bloodspot (DBS) cards. Cards with IRT values that exceed our fixed cutoff are sent to the Stanford University Molecular Pathology Laboratory, which performs the California 75 mutation panel followed by Sanger sequencing of the CFTR gene when a panel mutation is identified [
10,
11]. California moved from a 35 to a 75 mutation panel in 2020. Once genetic testing is complete, results of screening are provided by our Screening Information System (SIS) to the ordering physician and one of our Area Service Centers (ASCs) that notify the pediatrician to arrange a referral to one of the five contracted CF Special Care Centers for clinical management, diagnosis, and treatment. The Centers work with specialists and our ASC coordinators to report a diagnosis of CF shortly after birth, including cases missed by NBS.
We established a 15-year study cohort from July 16, 2007 through December 31, 2022 and identified IRT values and cutoffs on a given day for all newborns screened by GDSP in California. All missed cases identified by July of 2024 and tested by NBS initially within the cohort window were included providing at least 1.5 years of follow-up after the end of the cohort. We excluded cases initially tested by NBS who were born out of state.
2.2. Statistical Analysis
We utilized an ARIMA (autoregressive integrated moving average) model to fit monthly IRT screen-positive percentiles. Model results were used to test for seasonality and to estimate regular seasonal expectations for monthly screen-positive percentiles. The final seasonal model results were used to estimate IRT cutoffs as follows:
Monthly target percentile = Monthly seasonal % * percent positive IRT target (1.6%).
Monthly IRT cutoff value = IRT value calculated at the target percentile.
To emulate a floating cutoff, we used existing data from the study period, and calculated daily IRT distributions per contract laboratory at different cutoff levels determined by the top 1.6, 2.0, 3.0, 4.0, and 5.0 percentiles of each of the contact labs distributions. For each cutoff percentile, the frequency of known missed cases that would now be IRT positive or remain IRT negative was calculated and summed over the labs. The frequency of new IRT negative missed CF cases was also calculated. Cutoff levels and screening result frequencies were also calculated using the total previous seven-day IRT weekly distribution.
We used SAS for tests of significance and graphics [SAS Institute, 2023]. Results are presented as odds ratios (OR) and 95% confidence intervals (95% CI). The statistical significance threshold was set at p < 0.05.
3. Results
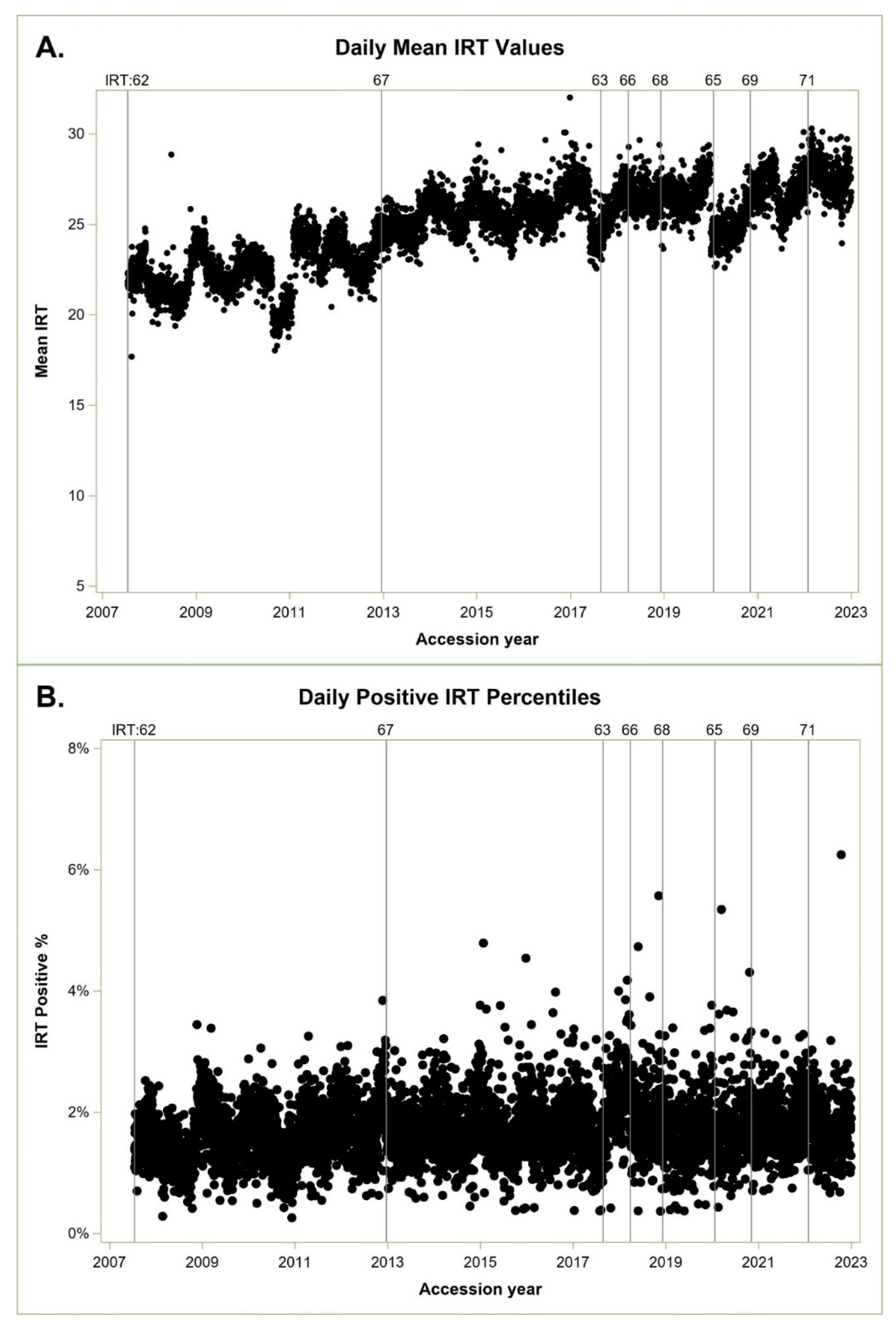
From July 16, 2007 through December 31,2022, we screened 7,410,016newborns with 123,313 IRT screen positive results for an overall IRT screen positive rate of 1.66%. We missed 36 cases below the IRT cutoff; five were within 3 ng/mL of the cutoff and considered near the IRT detection boundary. The ARIMA model indicated a statistically significant seasonal affect (Stable Seasonality F-test p<= 0.0001), no evidence of moving seasonality for the positive percentile (Moving Seasonality F-test P<=0.22), but some evidence of moving seasonality for mean monthly IRT values (Moving Seasonality F-test P<=0.07), which relates to the observed increase in mean IRT values over time in
Figure 1A.
Figure 1A shows statewide daily IRT means over time, and
Figure 1B shows daily IRT positive percentiles over time.
These are difficult to compare until we perform separate ARIMA models and extract monthly seasonal factors indicating a consistent cyclic monthly percentage above and below the IRT screen-positive population target, represented as 100% in
Figure 2A.
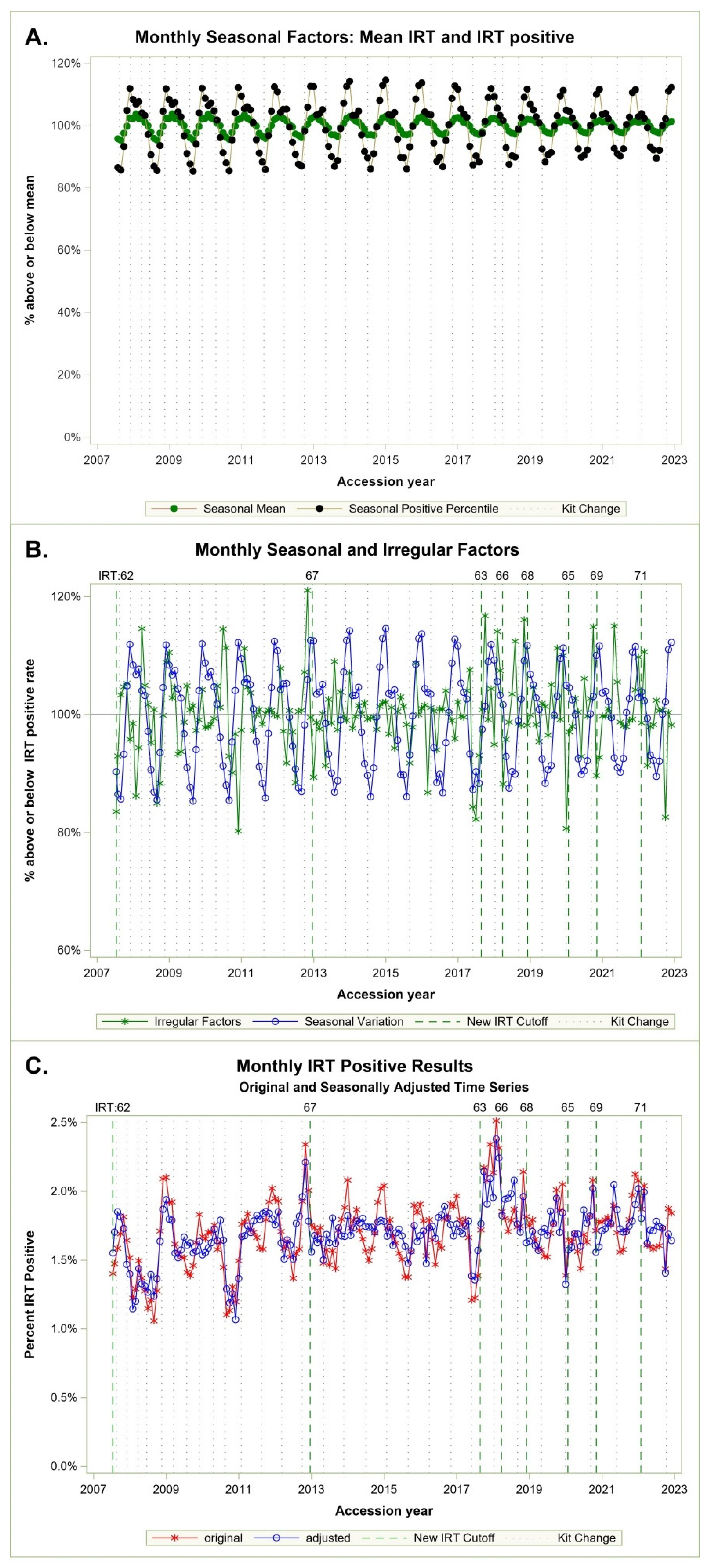
Figure 2A shows seasonal cycles of percentiles in black and IRT mean in green. Cycles are more pronounced for positive IRT percentiles, which vary in a range of 82% to 112%, compared with the IRT means, which vary between 95% to 104%. Both cycles are consistent and aligned, peaking in winter, and dropping in summer. IRT positive percentiles are our concern as these cutoffs determine which specimens are sent for molecular analysis.
Figure 2B shows both the seasonal and irregular monthly factor for the positive percentiles. Here the larger irregular factors appear to coincide with new kits and a subsequent change in cutoffs.
Figure 2C indicates original monthly IRT positive percent and time series adjusted by seasonal factors. The adjusted series can be interpreted as what we may have seen if NBS had made cutoff corrections each month based on the model vs. the IRT percent observed in our real data.
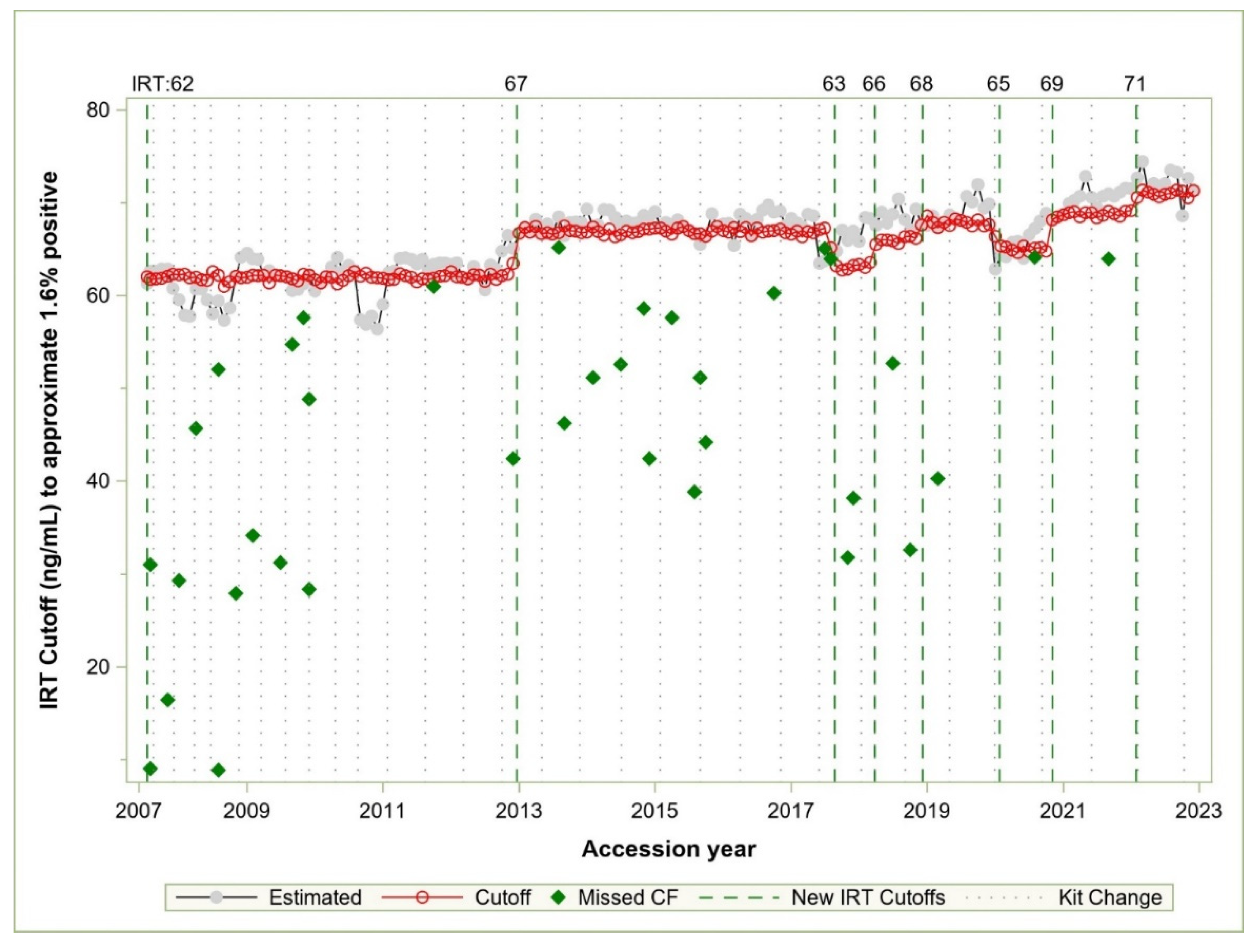
Figure 3 compares IRT values of each cutoff vs IRT values estimated by the model results. Between July 2007 and 2017, the cutoff was changed once, from 62 to 67 ng/mL in late 2012. There was one missed case within 3 ng/mL of the cutoff in 2011, and there were kits in 2008 and 2010 that appeared to require a cutoff far lower than the 62 in use at that time, though no known cases were missed. In the summer of 2017, we missed two cases of CF shortly after a kit change in May. The kit lot change led to low IRT percentiles due to continuation of the IRT cutoff of 67. We subsequently made a cutoff change to 63 in the summer based on the ARIMA model that we developed at that time.
Table 1 shows the results of a multivariate logistic regression with seasonal and regional effects comparing missed and identified CF cases. There was no detectable overall regional effect (Regional Chi-Square = 0.89, p < 0.826) compared with the Northern California Mountain regions. There was a strong statistically significant seasonal effect (Season Chi-Square = 9.96, p < 0.019), with OR of 6.51 (95% CI = 1.46, 28.98) in Fall and 6.32 (95% CI = 1.44, 27.82) in Summer compared with missed cases found below the IRT cutoff in the Spring. Winter was not significantly different than Spring 2.05 (95% CI = 0.37, 11.34).
The IRT percent screen positive targets listed in
Table 2 were created by extracting monthly seasonal percentiles and multiplying by desired yearly IRT population percentile. We show three estimates using 1.6%, 1.7% and 1.8% population percentiles. In California, percentiles are highest in December and January and lowest during the summer months of June through August.
In
Table 3, we estimated daily and weekly floating cutoffs using data summed from each of the five contract labs and our known missed and confirmed CF cases. We found a daily floating cutoff was able to identify many of the current low IRT missed cases at 4-5% screen positive rates and did not create new false-negative, missed cases. These cutoffs would require an additional 197,784 to 276,457 more genetic tests to be performed, with over twice as many than we currently conduct for the 5% daily floating cutoff. The weekly floating estimate did not add new false-positive cases starting at 3% and captured half of the current IRT false negative missed cases at a 5% cutoff percentile.
4. Discussion
ARIMA analysis allowed us to examine the regular seasonal pattern of IRT screen positive percentiles which has been observed in other states. Logistic analysis confirmed that low-IRT missed cases were more likely to occur during the summer and fall compared with spring months, even after we included region in the multivariate model. This remained true even after we removed the five results that were within 3 ug/mL of the IRT cutoff (not shown). Summer and fall comprise the hottest months when specimens are transferred at room temperature and IRT enzyme in specimens can degrade.
We used the analysis to define stable cutoffs based on the first two weeks of kit introduction. If we had used the adjustment factor derived from this model in 2017, we would not have missed CF cases near the IRT detection boundary; instead, we missed two cases in practice due to a delay in making a cutoff change (
Figure 2). After many years without a cutoff change, the team was caught off guard when a kit changed dramatically in 2017. We have since monitored IRT screen positive levels monthly with an emphasis on summer and fall.
We summarized the output of the ARIMA model in
Table 1 with monthly percentile targets, which can be used to determine the percentile we want to achieve when we have a new kit. We estimated the IRT value that falls within the population percentile for the new kit. The new cutoffs we estimated have been sufficient to set once throughout the life of a kit. The table can also be used to monitor and change cutoffs monthly, but we haven’t needed to change cutoffs once they were set. The most important set points are in the summer, and we have observed that monthly percentiles that drop below 1.4% to 1.5% raise the potential of a missed case close to the cutoff boundary. However, the bulk of missed CF cases due to low IRT values are well below the cutoffs in summer and fall and difficult to detect using IRT alone.
Results from California may not be generalizable to other states or countries due to different temperature gradients. The key minimum set point for any program is during the months when temperatures are highest. The other maximum IRT percentiles can then be set based on the desired yearly population percentiles.
When we went live with CF screening in 2007, we determined a cutoff of 62 based on population projections and historical test data, which led us to target a yearly percentile of 1.6%. The program has maintained a positive percentile between 1.6% and 1.7% yearly with rare changes in cutoff until 2017 when we dropped the cutoff from 67 to 63. We have been monitoring percentiles since then and results of our analysis suggest that we can move our percentile to 1.7% yearly, assuring that the lowest monthly percentile is 1.5% during the summer months.
Table 3 suggests daily variability in the screening population and in IRT results was great enough to lower screening efficacy for CF, and a weekly floating cutoff was more effective. If we went to a laboratory-based daily percentile, we would need to set a 4%-5% daily floating cutoff to lower missed cases at a cost of sending 152-212% more specimens for molecular testing. If our five contract laboratories were able to calculate a floating cutoff derived weekly, we could use a lower 3%-4% cutoff level, but we would still increase or molecular testing by 82%-142% respectively (
Table 3). This analysis, though limited to California, can help explain why states that use floating cutoffs may require a 4-5% IRT positive population rate in contrast to our lower 1.6%% rate. More than half of CF cases we miss in California are identified by high IRT where molecular testing only identifies none or one CF-causing variant.
4.1. Limitations
The study’s main limitation is the incomplete identification of missed cases. There may be a long lead time before a missed CF case is identified. Missed cases may be identified in other states or counties and never make it into our surveillance system. Missed cases in California may never be entered into our surveillance system, even though we maintain good relationships with our CF Special Care Centers who report such cases to us. Lack of complete case ascertainment may hamper the examination of floating cutoffs we presented using California data. If we had doubled the number of molecular tests our program conducted at lower IRT levels, we may have captured more confirmed CF cases. We suspect that number would be small given the episodic nature of low IRT false positive results, but we don’t know for certain.
5. Conclusions
Regular seasonal variation in IRT screen-positive rates can be leveraged to establish initial fixed cutoffs for a new reagent kit that can be monitored throughout the year. Daily and weekly population variability is large enough that IRT percentiles may reduce screening efficacy compared with a fixed cutoff as long as the fixed cutoff is monitored for monthly IRT-positive percentiles especially during the summer and fall. A population-based model and fixed cutoffs have been more efficacious, reduced missed cases, and minimized molecular testing for California. We try to reduce the burden of stress and uncertainty for families with newborns identified VUSs and with CRMS, a “watchful waiting” symptomless state at heightened risk for CF who we may identify at the low end of the screening threshold.
Institutional Review Board Statement
The California Health and Human Services Agency’s Committee for the Protection of Human Subjects has determined that the program evaluation and surveillance activities conducted by the California Newborn Screening Program represent exempt research per federal guidelines, Section 46.101(b)(4)(ii)). The data provided in this study have been de-identified in compliance with federal HIPAA standards.
Informed Consent Statement
Informed parental consent is not required for newborn screening in California, though parents may opt out of screening for religious reasons. Parents can request that the newborn blood spot be destroyed and not used for research when their newborn has been screened.
Data Availability Statement
Participant data cannot be made available due to legal and ethical requirements restricting access to individual level data from the California newborn screening program.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| ARIMA |
Autoregressive integrated moving average |
| CF |
Cystic fibrosis |
| GDSP |
Genetic Disease Screening Program |
| IRT |
Immunoreactive trypsinogen |
| NBS |
Newborn screening |
References
- Kharrazi, M.; Yang, J.; Bishop, T.; Lessing, S.; Young, S.; Graham, S.; et al. Newborn Screening for Cystic Fibrosis in California. Pediatrics. 2015, 136, 1062–72. [Google Scholar] [CrossRef] [PubMed]
- Korzeniewski, S.J.; Young, W.I.; Hawkins, H.C.; Cavanagh, K.; Nasr, S.Z.; Langbo, C.; et al. Variation in immunoreactive trypsinogen concentrations among Michigan newborns and implications for cystic fibrosis newborn screening. Pediatr Pulmonol. 2011, 46, 125–30. [Google Scholar] [CrossRef] [PubMed]
- Kay, D.M.; Maloney, B.; Hamel, R.; Pearce, M.; DeMartino, L.; McMahon, R.; et al. Screening for cystic fibrosis in New York State: considerations for algorithm improvements. Eur J Pediatr. 2016, 175, 181–93. [Google Scholar] [CrossRef] [PubMed]
- Kloosterboer, M.; Hoffman, G.; Rock, M.; Gershan, W.; Laxova, A.; Li, Z.; et al. Clarification of laboratory and clinical variables that influence cystic fibrosis newborn screening with initial analysis of immunoreactive trypsinogen. Pediatrics. 2009, 123, e338–46. [Google Scholar] [CrossRef] [PubMed]
- Maier, P.; Jeyaweerasinkam, S.; Eberhard, J.; Soueidan, L.; Hämmerling, S.; Kohlmüller, D.; et al. Influence of Season, Storage Temperature and Time of Sample Collection in Pancreatitis-Associated Protein-Based Algorithms for Newborn Screening for Cystic Fibrosis. International Journal of Neonatal Screening. 2024, 10, 5. [Google Scholar] [CrossRef] [PubMed]
- Sicko, R.J.; Stevens, C.F.; Hughes, E.E.; Leisner, M.; Ling, H.; Saavedra-Matiz, C.A.; et al. Validation of a Custom Next-Generation Sequencing Assay for Cystic Fibrosis Newborn Screening. Int J Neonatal Screen. 2021, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Sontag, M.K.; Lee, R.; Wright, D.; Freedenberg, D.; Sagel, S.D. Improving the Sensitivity and Positive Predictive Value in a Cystic Fibrosis Newborn Screening Program Using a Repeat Immunoreactive Trypsinogen and Genetic Analysis. The Journal of pediatrics. 2016, 175, 150–8. [Google Scholar] [CrossRef] [PubMed]
- Martiniano, S.L.; Croak, K.; Bonn, G.; Sontag, M.K.; Sagel, S.D. Improving outcomes for Colorado’s IRT-IRT-DNA cystic fibrosis newborn screening algorithm by implementing floating cutoffs. Molecular genetics and metabolism. 2021, 134, 65–7. [Google Scholar] [CrossRef] [PubMed]
- Rock, M.J.; Levy, H.; Zaleski, C.; Farrell, P.M. Factors accounting for a missed diagnosis of cystic fibrosis after newborn screening. Pediatric Pulmonology. 2011, 46, 1166–74. [Google Scholar] [CrossRef] [PubMed]
- McGarry, M.E.; Sciortino, S.; Graham, S.; Bishop, T.; Gibb, E.R. Improved detection of cystic fibrosis by the California Newborn Screening Program for all races and ethnicities. Pediatr Pulmonol 2024, 28. [Google Scholar] [CrossRef] [PubMed]
- Matteson, J.; Sciortino, S.; Feuchtbaum, L.; Bishop, T.; Olney, R.S.; Tang, H. Adrenoleukodystrophy Newborn Screening in California Since 2016: Programmatic Outcomes and Follow-Up. Int J Neonatal Screen. 2021, 7, 2. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).