1. Introduction
Dengue virus (DENV) is one of the fastest-spreading mosquito-borne viruses worldwide. DENV infection is caused by any of the four DENV serotypes (DENV-1 to DENV-4) and presents a significant public health challenge in tropical and subtropical areas [
1]. Clinically, DENV infection varies from asymptomatic cases to mild fever, and in severe cases, it can become life-threatening. Early symptoms include high fever, severe headaches, pain behind the eyes, muscle and joint pain, nausea, vomiting, skin rash, and minor bleeding [
2]. In more severe cases, the disease can progress to a critical phase with increased capillary permeability and plasma leakage, leading to hypovolemic shock [
3]. Without timely treatment, complications such as organ failure, severe bleeding, and circulatory collapse can occur, often resulting in death [
4]. Dengue remains a leading cause of hospitalization and death in both children and adults in endemic regions [
5].
The virus is transmitted primarily by
Aedes aegypti and
Aedes albopictus mosquitoes [
6]. DENV transmission is expanding due to rising global temperatures and increased urbanization, leading to significant health, social, and economic impacts [
7]. The World Health Organization (WHO) estimates that there are approximately 390 million infections annually, with over 3.9 billion people in more than 100 countries at risk [
2]. By the end of April 2024, the WHO had recorded approximately 7.6 million cases of dengue fever thus far in 2024, with over 3.4 million clinically confirmed infections, 16,000 severe cases, and fatalities surpassing 3,000 [
8].
Latin American and Asian countries are experiencing hyperendemic transmission with frequent outbreaks [
9,
10]. In Africa, dengue has historically been underreported compared with other regions, but in the past two decades, cases have sharply increased, particularly in West and Central Africa, indicating that the true burden of the disease may be underestimated [
11]. In Ghana specifically, studies indicate established transmission, yet an accurate nationwide epidemiological profile remains unclear. Situated in the tropical coastal West Africa belt and experiencing intense urban growth, the country meets many requisite conditions for indigenous DENV circulation and outbreaks. While Ghana is on the fringes of the WHO's DENV endemic map, some regional hospital-based studies provide serological evidence of previous exposure in healthy individuals as well as acute febrile patients [
12,
13]. However, the nationwide seroprevalence is undefined, and the full spectrum of disease remains unclear because of the absence of systematic entomological and virological surveillance. The lack of detailed characterization poses challenges for effective prevention. This underscores the rationale for conducting an extensive review of all available evidence. This review aims to provide a comprehensive synthesis of the pooled prevalence of DENV infection markers—including DENV-specific IgG, IgM, and DENV RNA, as well as circulating serotypes—in Ghana to facilitate well-informed public health response planning.
2. Materials and Methods
This review was conducted following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [
14] to ensure a rigorous and transparent methodology.
2.1. Information sources and search strategy
Three databases (PubMed, Web of Science, and Scopus) were searched for relevant publications up to September 24, 2024. The following search terms were used in various combinations: ("Dengue virus" OR "DENV" OR "Dengue fever") AND ("Ghana") AND ("Epidemiology" OR "Seroprevalence" OR "Diagnosis" OR "Surveillance" OR "Case report" OR "Outbreak"). Additional studies were identified through manual searches of references from eligible articles and relevant reviews on dengue in Africa. No restrictions were placed on the study design or sample size. The relevant articles were imported into Mendeley Reference Manager Version 2.122.1 for processing and organization.
2.2. Study selection and data extraction
All records obtained from searches were imported into Rayyan software [
15]. Two reviewers independently screened the records according to predefined eligibility criteria. Specifically, the relevance of each study for investigating dengue virus epidemiology in Ghana through the detection of biological markers was assessed. Potentially eligible full texts were retrieved and evaluated. A standardized data extraction form was developed in Microsoft Excel and pilot tested on three randomly selected studies before finalizing. The following characteristics were extracted from the included studies by one reviewer and verified by another:
1. Author and year of publication
2. Study design (e.g., cross-sectional, case‒control)
3. Study setting and period
4. Study population and inclusion/exclusion criteria
5. Sample size
6. Dengue diagnostic tests used (e.g., IgM ELISA, IgG ELISA, RT‒PCR)
7. Main outcome variables (e.g., IgG seroprevalence, IgM seroprevalence, and DENV RNA detection)
8. Dengue virus serotypes identified
Any discrepancies in the extracted data between reviewers were resolved by reexamining the original text. Incomplete or unreported information was documented accordingly. The extracted data were compiled into evidence tables for analysis and visualization of key parameters across studies. This standardized methodology ensured accurate extraction of relevant details to address the review objectives.
2.3. Inclusion criteria
The following inclusion criteria were applied during the study selection process:
1. Primary research studies have been conducted among humans in Ghana.
2. Studies employing serological and/or molecular techniques to detect dengue virus immunoglobulins (IgG and IgM) and/or RNA.
3. No date restrictions were placed on the included studies.
4. Studies providing key details on methodology, study characteristics (design, location, population), diagnostic methods used and main findings.
5. Studies from all geographical regions and population groups across Ghana.
2.4. Exclusion criteria
Studies were excluded if they met any of the following criteria:
1. Review articles, systematic reviews, meta-analyses, case reports, opinion pieces, letters, and conference abstracts - as these do not report primary research.
2. Studies conducted outside Ghana or not among human populations.
3. Studies detecting antibodies or antigens of pathogens other than dengue virus.
4. Studies focusing only on vector surveillance or reporting vector indices without dengue virus detection.
5. Studies with incomplete/insufficient methodological information to assess quality and extract relevant data.
6. Studies published in languages other than English.
These clear inclusion and exclusion criteria helped obtain a focused set of studies relevant to the objectives of the review.
2.5. Quality assessment
Study quality was assessed using the Joanna Briggs Institute (JBI) Critical Appraisal tool relevant for cohort, cross-sectional and case‒control study designs [
16]. A customized nine-item checklist was used to examine methodological domains:
1. Was the sample frame appropriate to address the target population?
2. Were study participants sampled appropriately?
3. Was the sample size adequate?
4. Were the study subjects and setting described in detail?
5. Was data analysis sufficiently comprehensive of the identified sample?
6. Were valid methods used for condition identification?
7. Was the condition measured reliably for all participants?
8. Was statistical analysis appropriate?
9. Was the response rate adequate, and if not, was this managed appropriately?
Each item was scored "1" if adequately met and "0" if unreported or poorly described. The maximum quality score was 9. Studies scoring ≥7 points were considered "high" quality, those scoring 4--6 points were considered "moderate", and those scoring <4 points were considered "low". The quality assessment was independently conducted by two reviewers, with a third reviewer resolving any discrepancies.
2.6. Statistical analysis
The meta-analyses were performed using R software version 4.2.1 and applicable R packages. Specifically, the metafor package was used to conduct all the analyses. Random effects meta-analyses generated pooled proportions with 95% confidence intervals (CIs) for dengue IgM, IgG, and RNA detection and combined IgG/IgM results. Heterogeneity was assessed via the I2 statistic, with values greater than 50% indicating moderate to high heterogeneity. Forest plots graphically display prevalence outcomes with study-specific CIs and pooled estimates. Publication bias was evaluated using funnel plots.
3. Results
3.1. Search results
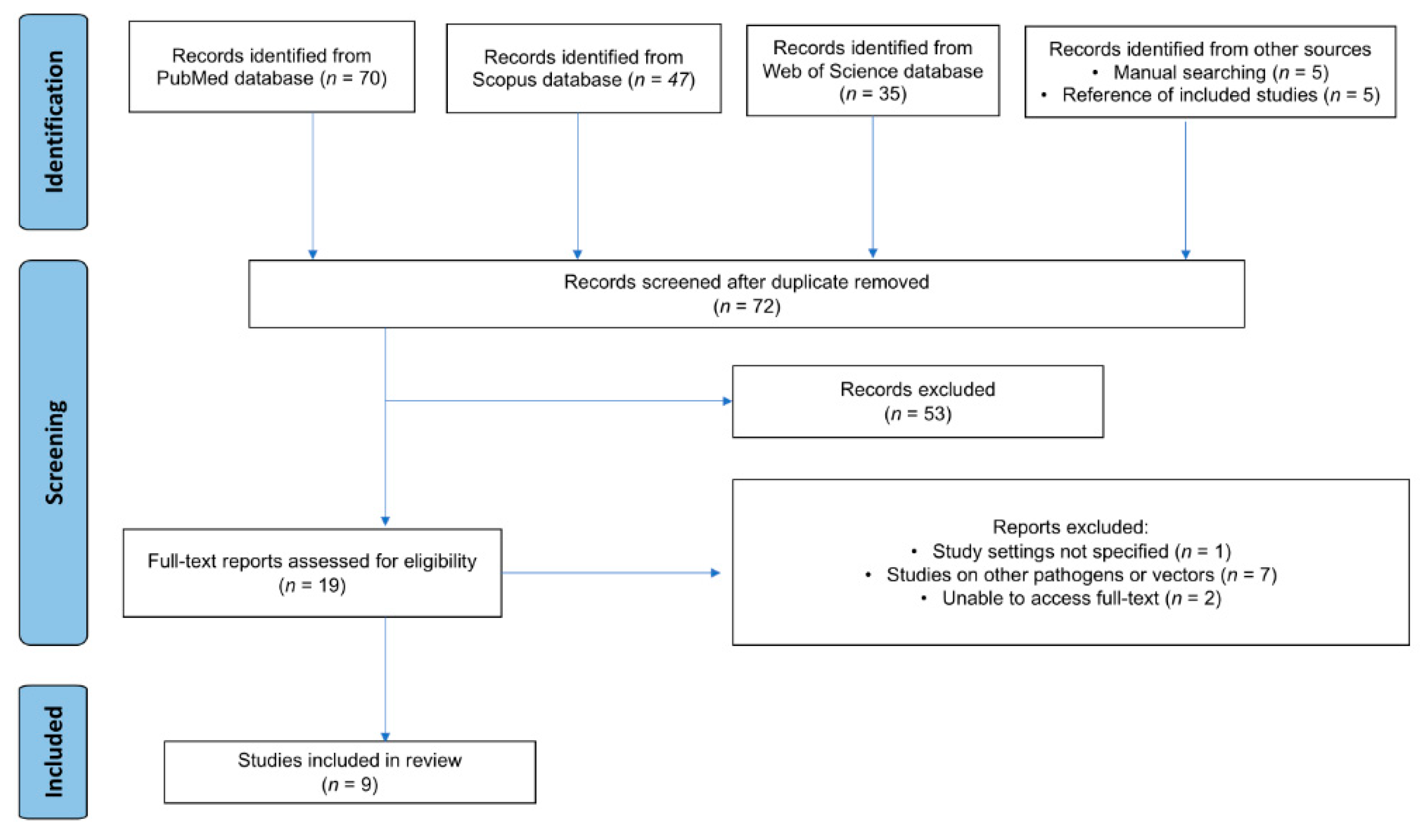
The database searches (PubMed, Scopus, and Web of Science) yielded 152 publications. An additional 10 studies were identified through manual techniques and reference screening, totaling 162. After 90 duplicate reports were removed, 72 studies underwent title/abstract review. Most (n=53) did not meet the criteria and were excluded at this stage. The remaining 19 patients underwent full-text assessment; 10 were excluded for the following reasons:
Study setting not specified (n=1)
Other pathogens/vectors studied (n=7)
Unable to access the full text (n=2)
Finally, 9 studies [
12,
13,
17,
18,
19,
20,
21,
22,
23] met all eligibility criteria and were included in the review. Any discrepancies in study selection between reviewers were resolved via discussion until consensus was reached. The overall selection process is summarized in a PRISMA flow diagram (
Figure 1).
3.2. Study characteristics
The detailed study characteristics are provided in
Table S1. Six studies utilized a cross-sectional design [
13,
17,
18,
21,
22,
23], one was a case study [
19], and two did not specify the design [
12,
20]. Two identified infections during outbreaks [
20,
22], whereas seven detected infections outside of outbreaks [
12,
13,
17,
18,
19,
21,
23]. All were clinical-based. Five studies used both RT‒PCR and ELISA [
12,
13,
20,
21,
23], one used both RDT and ELISA [
17], two used only RT‒PCR [
18,
19], and one used ELISA alone [
22]. Seven studies detected IgG [
12,
13,
17,
20,
21,
22,
23], five detected IgM [
12,
17,
20,
21,
23], three detected RNA [
18,
19,
20], and three detected IgG/IgM [
17,
20,
21]. Three studies identified DENV-2 [
18,
19,
20], and one identified DENV-3 [
20]. The target populations included patients aged 6 months to 82 years with suspected yellow fever [
23]; children aged 2–14 years with confirmed malaria [
12]; patients with fever and jaundice [
22]; healthy blood donors [
13]; suspected dengue and/or chikungunya fever patients [
21]; patients suspected of Ebola virus disease [
20]; suspected viral hemorrhagic fever cases [
19]; children aged 1–15 years presenting with fever [
18]; and adults aged ≥18 years with fever and three malaria-like symptoms [
17].
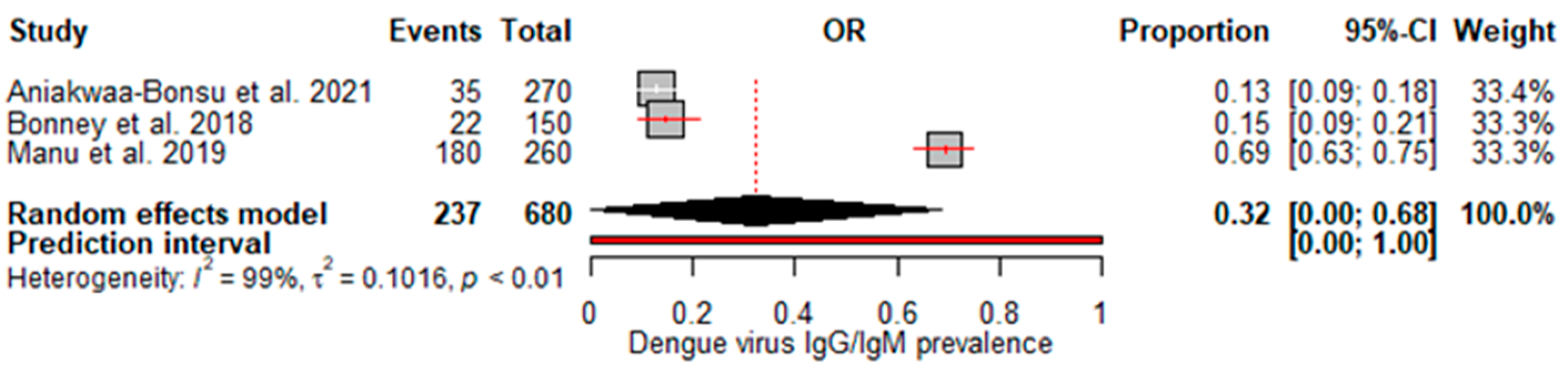
3.3. Pool prevalence of dengue virus in Ghana
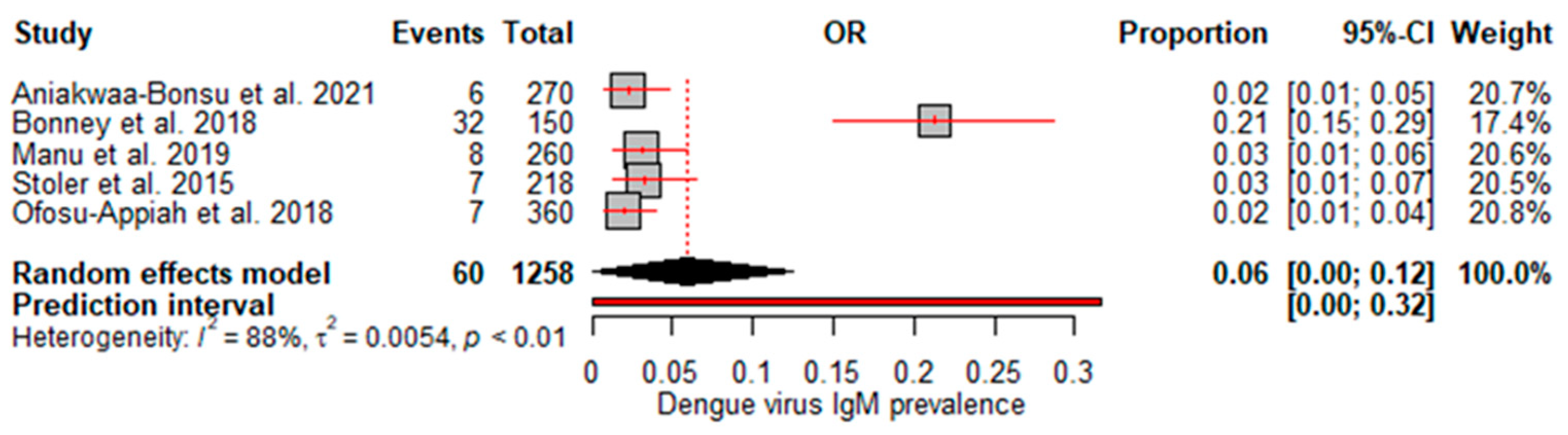
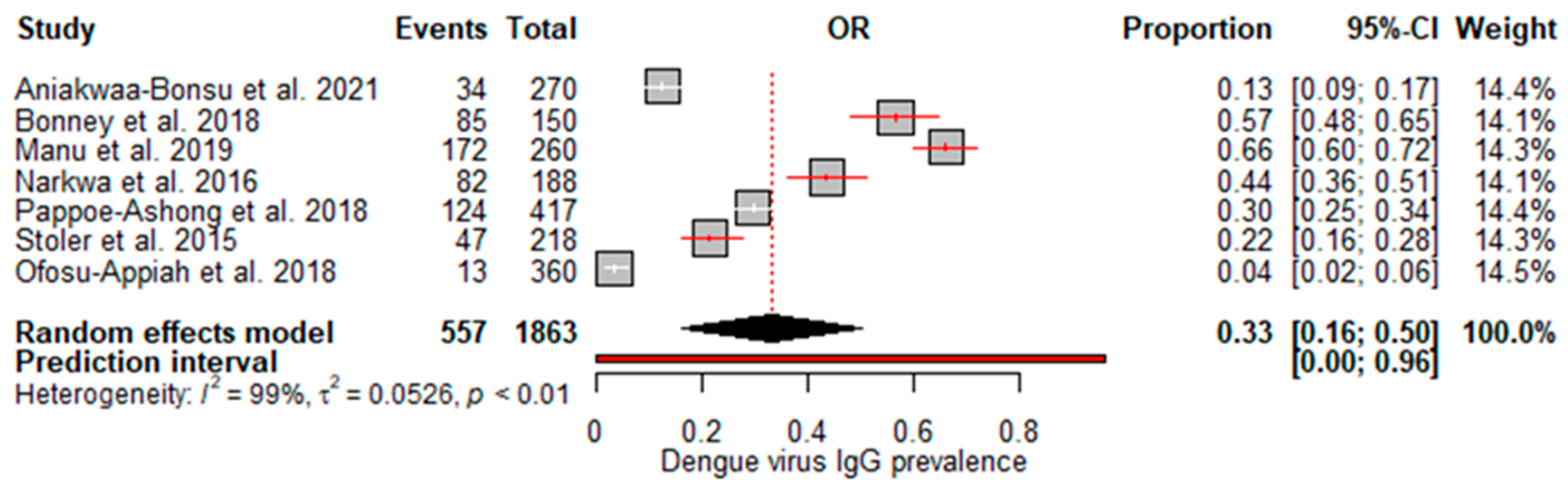
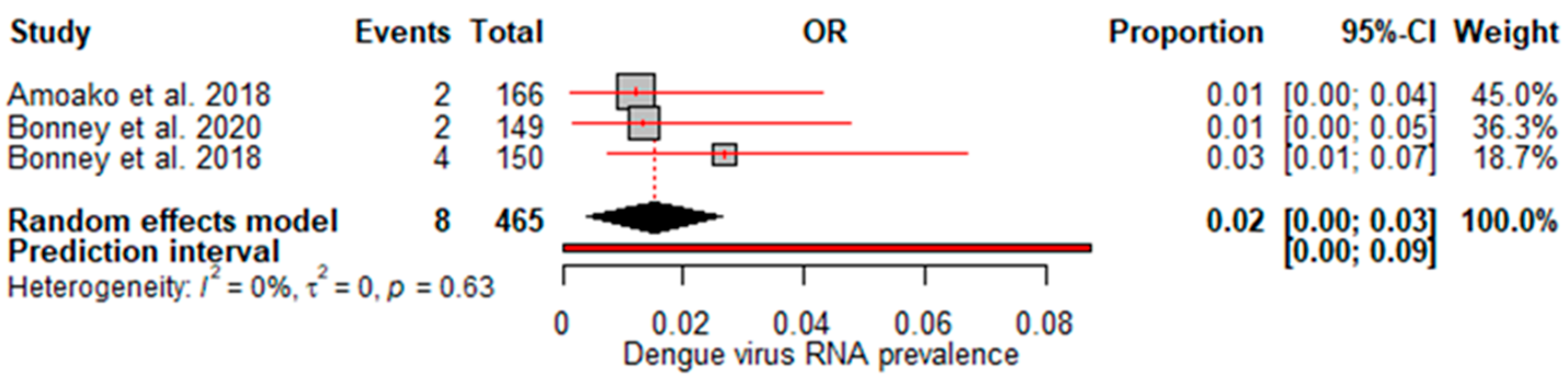
A random effects meta-analysis evaluated the pooled prevalence of dengue virus infection in Ghana. For IgM, the analysis estimated a pooled prevalence of 6% (95% CI: 0–12%), with high heterogeneity (I
2=88%, p<0.01) (
Figure 2), indicating relatively low recent infection rates at the population level. The pooled IgG incidence was greater at 33% (95% CI: 16–50%), again with significant heterogeneity (I
2=99%, p<0.01) (
Figure 3), revealing extensive past exposures in Ghana. The detection of DENV RNA by PCR revealed a pooled prevalence of 2% (95% CI: 0–3%), with no heterogeneity (
Figure 4), which is consistent with the low current viremic phase. Studies reporting combined IgG/IgM prevalence estimated a pooled 32% (95% CI: 0–68%), with substantial heterogeneity (
Figure 5).
3.4. Risk of bias
The quality of the included studies was assessed via the Joanna Briggs Institute (JBI) critical appraisal tool. The nine-item risk of bias checklist scores are presented in
Table S2.
Most studies (n=6) [
13,
17,
18,
19,
21,
23] achieved a '100%' rating, indicating a low risk of bias, as they adequately reported sample frames, sampling methods, sample sizes, settings/subjects, data analysis coverage, diagnostic validity, condition measurement, and statistical analysis. Two studies with scores of '88.9%' were considered to have a high risk of bias because of inadequate sampling and setting descriptions [
12,
20]. One study scored '88.9%' with an unclear risk of bias because of a lack of detail on the statistical methods used [
22]. Overall, the included studies were high quality, with clear reporting of methodological domains, enhancing confidence in their findings on dengue virus infection patterns in Ghana.
4. Discussion
This meta-analysis provides valuable insights into the epidemiology of dengue virus infection in Ghana. The estimated overall IgG seroprevalence was 33%, indicating widespread exposure to DENV in the population. However, there was significant heterogeneity (I2=99%), indicating that the prevalence differed depending on location, time period or other study characteristics. This underscores the need for further standardized surveillance across diverse regions. Our results are better than those of other meta-analyses. For example, our IgG seroprevalence is greater than the 21% IgG prevalence reported in Ethiopia [
24], the 15.6% reported in a 2019 meta-analysis in Africa [
25], a 2021 meta-analysis in Africa, which reported 14% [
26], the reported 25% in sub-Saharan Africa [
27], but lower than the 38.3% reported in a meta-analysis in India [
28].
We found a pooled IgM prevalence of only 6% and 2% DENV RNA detection, suggesting that current infection rates may be relatively low. The relatively low pooled estimate suggests that a small proportion of the population has experienced recent/acute infection at any given time. While RNA confirmation of acute cases is more standardized, low-test numbers may underestimate true burdens. However, the high heterogeneity (I
2=88%) indicates considerable variability between studies, which could be due to factors such as seasonality of outbreaks or regional differences in transmission intensity. Interestingly, no heterogeneity was observed for the RNA studies despite the limited amount of data. This suggests that molecular diagnostic methods may provide more consistent estimates than antibody assays, which can be subject to interassay variability [
29]. Our findings suggest that molecular diagnosis could offer a relatively consistent approach when optimized diagnostic algorithms are employed nationally. Notably, the meta-analysis by Simo et al. [
25] revealed a lower IgM prevalence of 3.5%, and no DENV RNA detection was identified in Africa. However, the meta-analysis by Eltom et al. [
27] reported a higher IgM prevalence and DENV RNA prevalence of 10% and 14%, respectively, in sub-Saharan Africa; similarly, the meta-analysis in Ethiopia revealed a higher IgM prevalence of 9% and a pooled DENV-RNA prevalence of 48% [
24].
The identification of DENV-2 and DENV-3 as the predominant serotypes circulating in Ghana provides some important insights but also leaves many unanswered questions. DENV-2 was consistently identified across multiple studies from 2011-2018, indicating that it is well established locally with ongoing endemic transmission. However, without genetic sequencing data, it is difficult to determine whether this represents a stable lineage or repeated reintroductions. The presence of DENV-3 confirms that it also maintains local transmission chains, although again, the phylogenetic context is lacking. An intriguing finding is that DENV-1 and DENV-4 were not detected, despite being common elsewhere, suggesting that vector or environmental factors may limit their spread in Ghana for unknown reasons. While DENV-2 and DENV-3 cause a substantial burden globally, their propensity to cause severe dengue versus milder outcomes varies substantially by region due to factors such as population-level immunity and socioeconomics [
26,
30]. Vietnam studies reported significant urban‒rural and regional differences, with prevalence gradients decreasing from densely populated south-central localities towards forested highlands, corroborating landscape influences on vector dynamics [
31,
32]. Similarly, a study in Brazil reported higher exposure burdens in Amazonian municipalities than in non-Amazonian municipalities, which was attributed to ecological factors affecting mosquito proliferation [
33]. To obtain a clearer picture of serotype dynamics, expanded sentinel surveillance with annual whole-genome sequencing is needed from multiple urban and rural sites. This could track whether strains are persistent over decades or periodically displaced by new introductions, with implications for developing adaptive control programs. Further serotyping of symptomatic versus asymptomatic infections may also provide clues about changes in virulence.
RT‒PCR, ELISA and combination methods are established approaches for dengue virus detection. RT‒PCR enables sensitive molecular confirmation of acute infection, but its availability is often limited to well-resourced laboratories. ELISA serology provides broader exposure assessment over the lifetime, but reliability depends heavily on the antigen/antibody kits employed between settings [
34]. Inconsistencies in standardizing methods, cut-offs and timing of specimen collection complicated efforts to meaningfully aggregate findings. Moving forward, national guidelines standardizing the serial use of both modalities according to accepted algorithms will optimize disease phase resolution critical for guiding clinical management and surveillance goals.
Limitations and recommendation
The studies included in our review targeted a range of population cohorts to gain insights into dengue virus infection in Ghana. As the majority of the included studies were hospital-based and involved acute fever patients, the prevalence estimates may overestimate the “true” community burden. Hospital-based studies provide a useful starting point for detecting clinical cases, but their populations are biased toward individuals able and willing to seek healthcare, precluding measuring the true prevalence across communities, particularly for mild/asymptomatic cases. The inclusion of apparently healthy blood donors offered an alternative perspective, but the generalizability of these convenience samples to the whole country is questionable. In the future, well-designed population-level serosurveys sampling all age groups are needed for more accurate risk profiling. This allows us to stratify analyses by demographics and local risk factors.
5. Conclusions
This review established the prevalence of DENV and the circulation of different serogroups in Ghana. The increased seropositivity and heterogeneity of DENV strains underscore the need for standardized, large-scale surveillance to optimize the characterization of the disease and guide appropriate control measures. Integrating dengue prevention strategies into existing vector control programs in Ghana could help reduce the risks posed by this disease.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Conceptualization, E.S.D., I.B., K.W.C.S; methodology, A.O., E.S.D., I.B., and K.W.C.S; validation, I.B., A.O., E.S.D., and K.W.C.S; formal analysis, A.O., I.B., E.S.D., and K.W.C.S; investigation, A.O., E.S.D., I.B., and K.W.C.S; resources, A.O., E.S.D., I.B., and K.W.C.S; data curation, A.O., E.S.D., I.B., and K.W.C.S; writing—original draft preparation, A.O., E.S.D., I.B., and K.W.C.S; writing—review and editing, A.O., E.S.D., I.B., and K.W.C.S; visualization, A.O., E.S.D., I.B., and K.W.C.S; supervision, E.S.D. and K.W.C.S.; project administration, E.S.D. and K.W.C.S.; funding acquisition, E.S.D. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Institutes of Health, USA, through the “Application of Data Science to Build Research Capacity in Zoonoses and Food-Borne Infections in West Africa (DS-ZOOFOOD) Training Programme” hosted at the Department of Medical Microbiology, University of Ghana Medical School (Grant Number: UE5TW012566-01). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Data Availability Statement
All the supporting data are presented in the manuscript and supplementary files.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Kularatne SA, Dalugama C. Dengue infection: Global importance, immunopathology and management. Clin Med (Lond) [Internet]. 2022 Jan;22(1):9–13. Available from: https://pubmed.ncbi.nlm.nih.gov/35078789. [CrossRef]
- Dengue and severe dengue [Internet]. [cited 2024 Sep 25]. Available from: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue.
- Singh RK, Tiwari A, Satone PD, Priya T, Meshram RJ. Updates in the Management of Dengue Shock Syndrome: A Comprehensive Review. Cureus [Internet]. 2023 Oct 9;15(10):e46713–e46713. Available from: https://pubmed.ncbi.nlm.nih.gov/38021722. [CrossRef]
- Uzair H, Waseem R, Kumar N, Hussain MS, Shah HH. Fatal outcome of dengue fever with multi-organ failure and hemorrhage: A case report. SAGE Open Med Case Rep [Internet]. 2023 Dec 25;11:2050313X231220808-2050313X231220808. Available from: https://pubmed.ncbi.nlm.nih.gov/38149117. [CrossRef]
- Ilic I, Ilic M. Global Patterns of Trends in Incidence and Mortality of Dengue, 1990-2019: An Analysis Based on the Global Burden of Disease Study. Medicina (Kaunas) [Internet]. 2024 Mar 1;60(3):425. Available from: https://pubmed.ncbi.nlm.nih.gov/38541151.
- Schaefer TJ, Panda PK, Wolford RW. Dengue Fever. BMJ Best Practice [Internet]. 2024 Mar 6 [cited 2024 Sep 25];5–6. Available from: https://www.ncbi.nlm.nih.gov/books/NBK430732/.
- Nakase T, Giovanetti M, Obolski U, Lourenço J. Population at risk of dengue virus transmission has increased due to coupled climate factors and population growth. Commun Earth Environ. 2024 Aug 30;5(1):475. [CrossRef]
- Dengue - Global situation [Internet]. [cited 2024 Sep 25]. Available from: https://www.who.int/emergencies/disease-outbreak-news/item/2024-DON518.
- Lessa CLS, Hodel KVS, Gonçalves M de S, Machado BAS. Dengue as a Disease Threatening Global Health: A Narrative Review Focusing on Latin America and Brazil. Trop Med Infect Dis [Internet]. 2023 Apr 23;8(5):241. Available from: https://pubmed.ncbi.nlm.nih.gov/37235289. [CrossRef]
- Hossain MS, Noman A Al, Mamun SMA Al, Mosabbir A Al. Twenty-two years of dengue outbreaks in Bangladesh: epidemiology, clinical spectrum, serotypes, and future disease risks. Trop Med Health [Internet]. 2023 Jul 11;51(1):37. Available from: https://pubmed.ncbi.nlm.nih.gov/37434247.
- Gainor EM, Harris E, LaBeaud AD. Uncovering the Burden of Dengue in Africa: Considerations on Magnitude, Misdiagnosis, and Ancestry. Viruses [Internet]. 2022 Jan 25;14(2):233. Available from: https://pubmed.ncbi.nlm.nih.gov/35215827. [CrossRef]
- Stoler J, Delimini RK, Bonney JHK, Oduro AR, Owusu-Agyei S, Fobil JN, et al. Evidence of recent dengue exposure among malaria parasite-positive children in three urban centers in Ghana. Am J Trop Med Hyg [Internet]. 2015;92(3):497–500. Available from: ["http://dx.doi.org/10.4269/ajtmh.14-0678", ‘https://www.ncbi.nlm.nih.gov/pubmed/25582693’, ‘https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4350537’]. [CrossRef]
- Narkwa PW, Mutocheluh M, Kwofie TB, Owusu M, Annan A, Ali I, et al. Dengue virus exposure among blood donors in Ghana. JOURNAL OF MEDICAL AND BIOMEDICAL SCIENCE. 2016;5(2):30–5. [CrossRef]
- Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. The BMJ. 2021 Mar 29;372.
- Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. Rayyan—a web and mobile app for systematic reviews. Syst Rev. 2016 Dec 5;5(1):210. [CrossRef]
- JBI Critical Appraisal Tools | JBI [Internet]. [cited 2024 Sep 25]. Available from: https://jbi.global/critical-appraisal-tools.
- Aniakwaa-Bonsu E, Amoako-Sakyi D, Dankwa K, Prah JK, Nuvor SV, Aniakwaa-Bonsu E, et al. Seroprevalence of Dengue Viral Infection among Adults Attending the University of Cape Coast Hospital. Adv Infect Dis [Internet]. 2021 Feb 1 [cited 2024 Sep 25];11(1):60–72. Available from: https://www.scirp.org/journal/paperinformation?paperid=107886. [CrossRef]
- Amoako N, Duodu S, Dennis FE, Bonney JHK, Asante KP, Ameh J, et al. Detection of dengue virus among children with suspected malaria, Accra, Ghana. Emerg Infect Dis [Internet]. 2018;24(8):1544–7. Available from: ["http://dx.doi.org/10.3201/eid2408.180341", ‘https://www.ncbi.nlm.nih.gov/pubmed/30015610’, ‘https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6056106’]. [CrossRef]
- Bonney JHK, Asigbee TW, Kotey E, Attiku K, Asiedu-Bekoe F, Mawuli G, et al. Molecular detection of viral pathogens from suspected viral hemorrhagic fever patients in Ghana. Health Sciences Investigations Journal [Internet]. 2020;1(1):31–5. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85170560500&doi=10.46829%2fhsijournal.2020.6.1.1.31-35&partnerID=40&md5=b2c5670be32c6a90f6a9384b9bd7e478. [CrossRef]
- Bonney JHK, Hayashi T, Dadzie S, Agbosu E, Pratt D, Nyarko S, et al. Molecular detection of dengue virus in patients suspected of Ebola virus disease in Ghana. PLoS One [Internet]. 2018;13(12):e0208907. Available from: ["http://dx.doi.org/10.1371/journal.pone.0208907", ‘https://www.ncbi.nlm.nih.gov/pubmed/30566466’, ‘https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6300295’]. [CrossRef]
- Manu SK, Bonney JHK, Pratt D, Abdulai FN, Agbosu EE, Frimpong PO, et al. Arbovirus circulation among febrile patients at the greater Accra Regional Hospital, Ghana. BMC Res Notes [Internet]. 2019;12(1):332. Available from: ["http://dx.doi.org/10.1186/s13104-019-4378-x", ‘https://www.ncbi.nlm.nih.gov/pubmed/31186058’, ‘https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6560752’]. [CrossRef]
- Pappoe-Ashong PJ, Ofosu-Appiah LH, Mingle JA, Jassoy C. Seroprevalence of dengue virus infections in Ghana. East Afr Med J [Internet]. 2018;95(11):2132–40. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85094914330&partnerID=40&md5=d6a1f176a34467c95092b60e34814c18.
- Ofosu-Appiah L, Kutame R, Ayensu B, Bonney J, Boateng G, Adade R, et al. Detection of Dengue Virus in Samples from Suspected Yellow Fever Cases in Ghana. Microbiol Res J Int. 2018 May 22;24(1):1–10. [CrossRef]
- Nigussie E, Atlaw D, Negash G, Gezahegn H, Baressa G, Tasew A, et al. A dengue virus infection in Ethiopia: a systematic review and meta-analysis. BMC Infect Dis [Internet]. 2024 Mar 6;24(1):297. Available from: https://pubmed.ncbi.nlm.nih.gov/38448847. [CrossRef]
- Simo FBN, Bigna JJ, Kenmoe S, Ndangang MS, Temfack E, Moundipa PF, et al. Dengue virus infection in people residing in Africa: a systematic review and meta-analysis of prevalence studies. Sci Rep [Internet]. 2019 Sep 20;9(1):13626. Available from: https://pubmed.ncbi.nlm.nih.gov/31541167. [CrossRef]
- Mwanyika GO, Mboera LEG, Rugarabamu S, Ngingo B, Sindato C, Lutwama JJ, et al. Dengue Virus Infection and Associated Risk Factors in Africa: A Systematic Review and Meta-Analysis. Viruses [Internet]. 2021 Mar 24;13(4):536. Available from: https://pubmed.ncbi.nlm.nih.gov/33804839. [CrossRef]
- Eltom K, Enan K, El Hussein ARM, Elkhidir IM. Dengue Virus Infection in Sub-Saharan Africa Between 2010 and 2020: A Systematic Review and Meta-Analysis. Front Cell Infect Microbiol [Internet]. 2021 May 25;11:678945. Available from: https://pubmed.ncbi.nlm.nih.gov/34113579. [CrossRef]
- Ganeshkumar P, Murhekar M V, Poornima V, Saravanakumar V, Sukumaran K, Anandaselvasankar A, et al. Dengue infection in India: A systematic review and meta-analysis. PLoS Negl Trop Dis [Internet]. 2018 Jul 16;12(7):e0006618–e0006618. Available from: https://pubmed.ncbi.nlm.nih.gov/30011275. [CrossRef]
- Raafat N, Blacksell SD, Maude RJ. A review of dengue diagnostics and implications for surveillance and control. Trans R Soc Trop Med Hyg. 2019 Nov 1;113(11):653–60. [CrossRef]
- Garcia-Bates TM, Cordeiro MT, Nascimento EJM, Smith AP, Soares de Melo KM, McBurney SP, et al. Association between Magnitude of the Virus-Specific Plasmablast Response and Disease Severity in Dengue Patients. The Journal of Immunology. 2013 Jan 1;190(1):80–7. [CrossRef]
- Huynh TTT, Minakawa N. A comparative study of dengue virus vectors in major parks and adjacent residential areas in Ho Chi Minh City, Vietnam. PLoS Negl Trop Dis. 2022 Jan 12;16(1):e0010119. [CrossRef]
- Pham H V, Doan HT, Phan TT, Tran Minh NN. Ecological factors associated with dengue fever in a central highlands Province, Vietnam. BMC Infect Dis. 2011 Dec 16;11(1):172.
- Lowe R, Lee S, Martins Lana R, Torres Codeço C, Castro MC, Pascual M. Emerging arboviruses in the urbanized Amazon rainforest. BMJ. 2020 Nov 13;m4385. [CrossRef]
- Kabir MA, Zilouchian H, Younas MA, Asghar W. Dengue Detection: Advances in Diagnostic Tools from Conventional Technology to Point of Care. Biosensors (Basel). 2021 Jun 23;11(7):206. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).