Submitted:
30 September 2024
Posted:
30 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
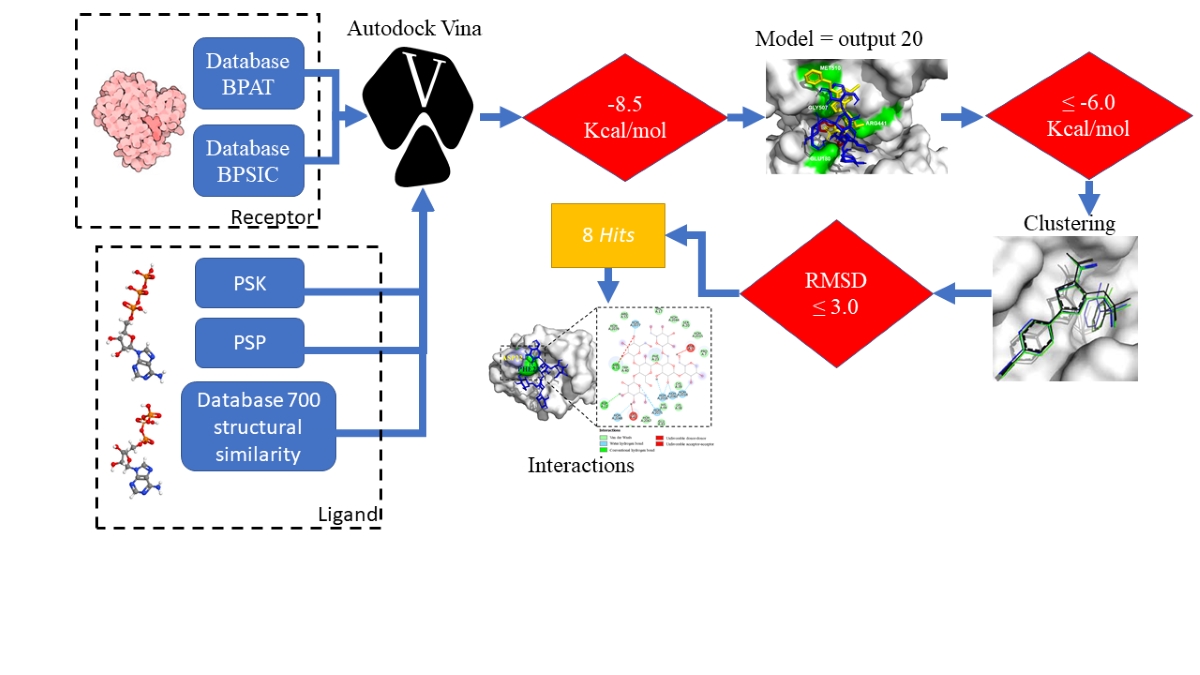
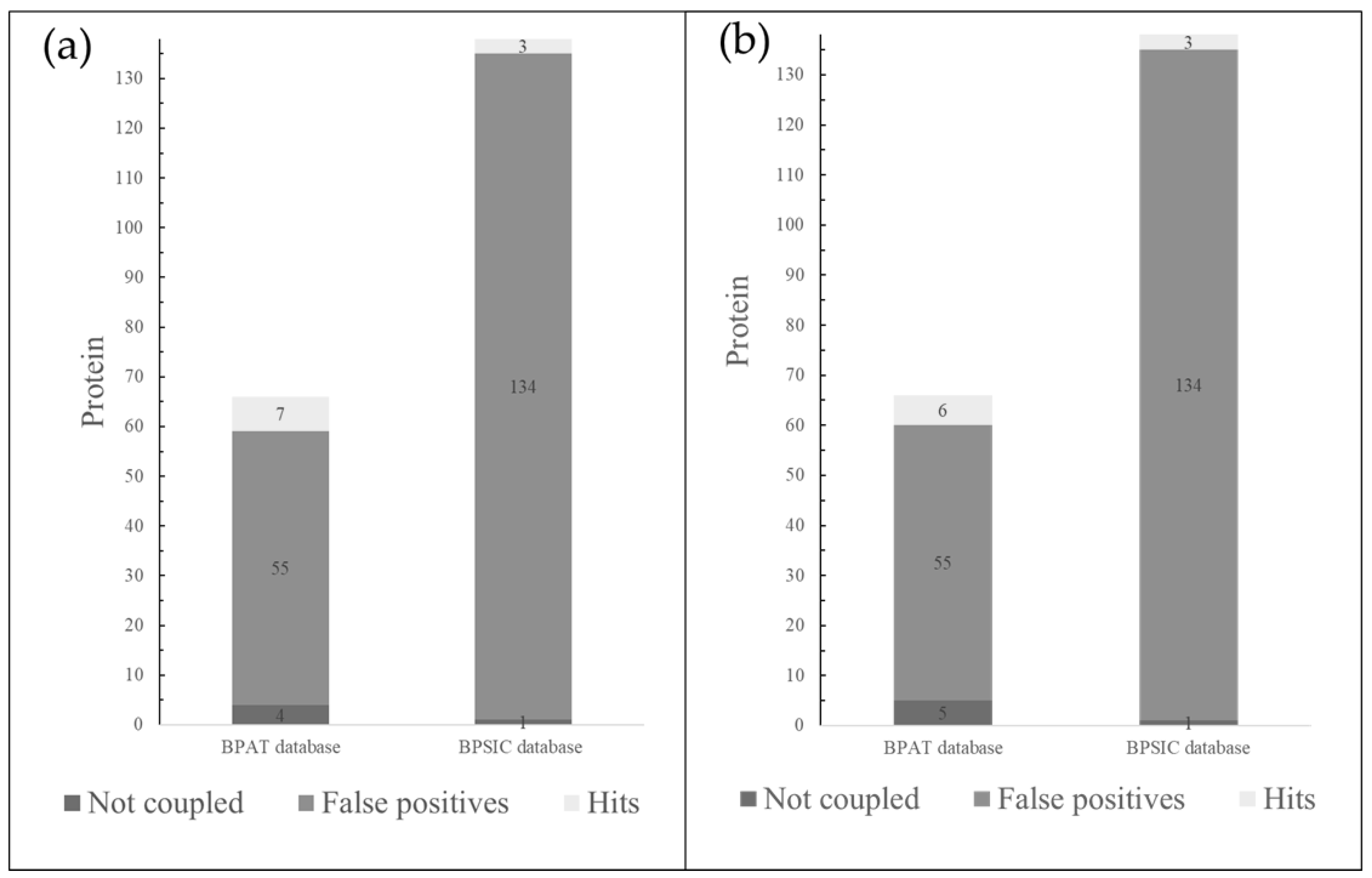
Inverse Molecular Docking
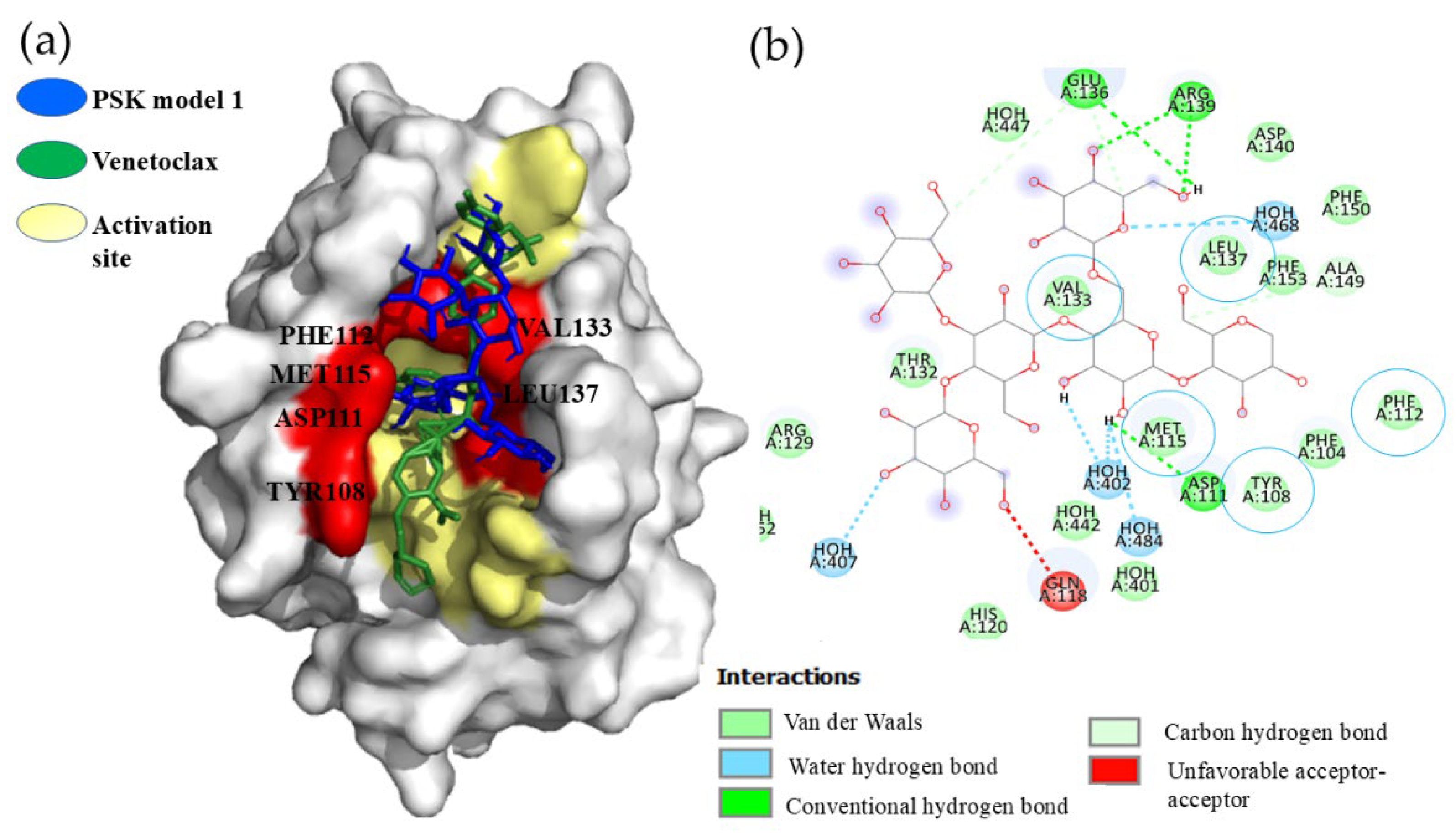
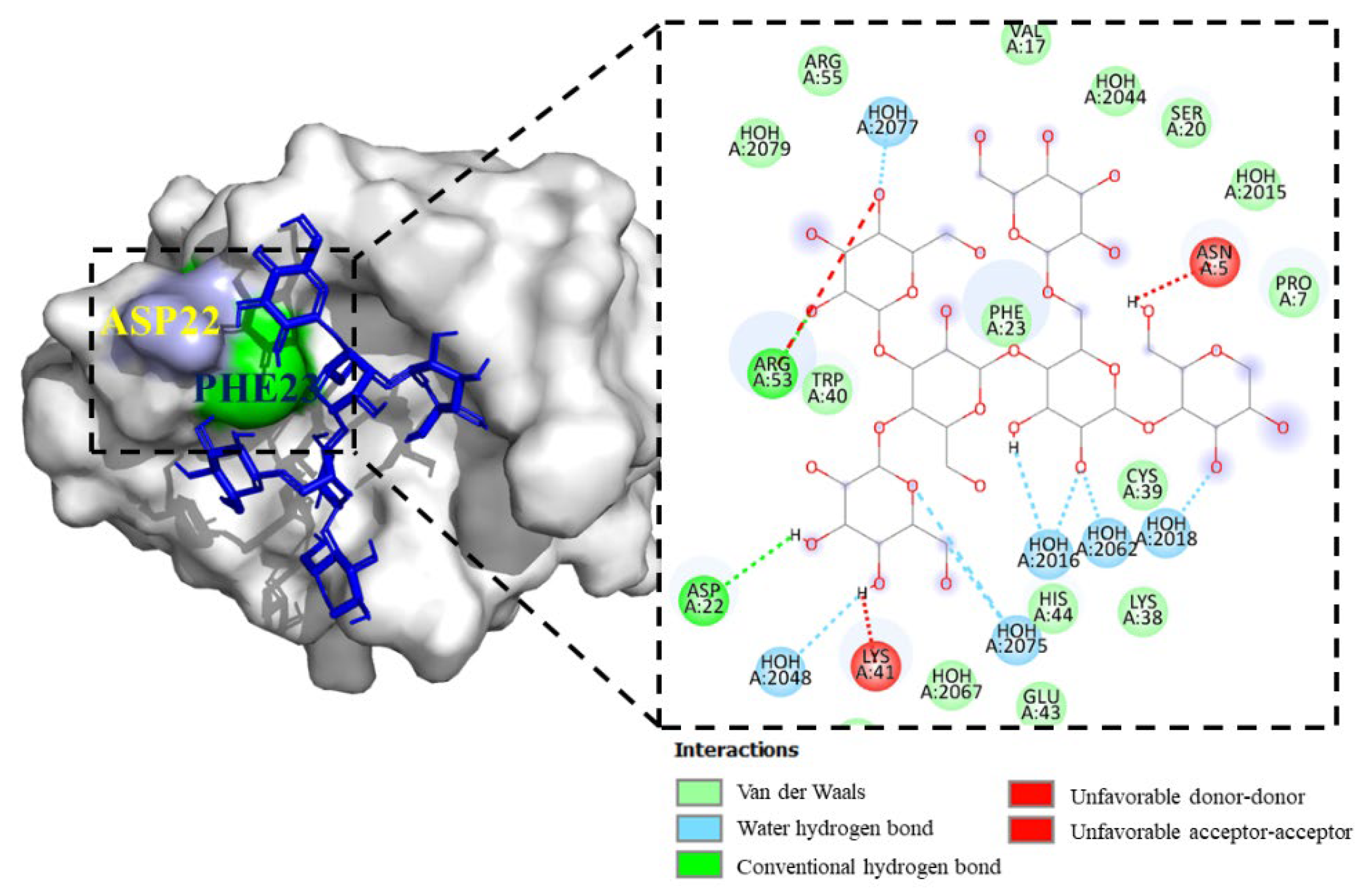
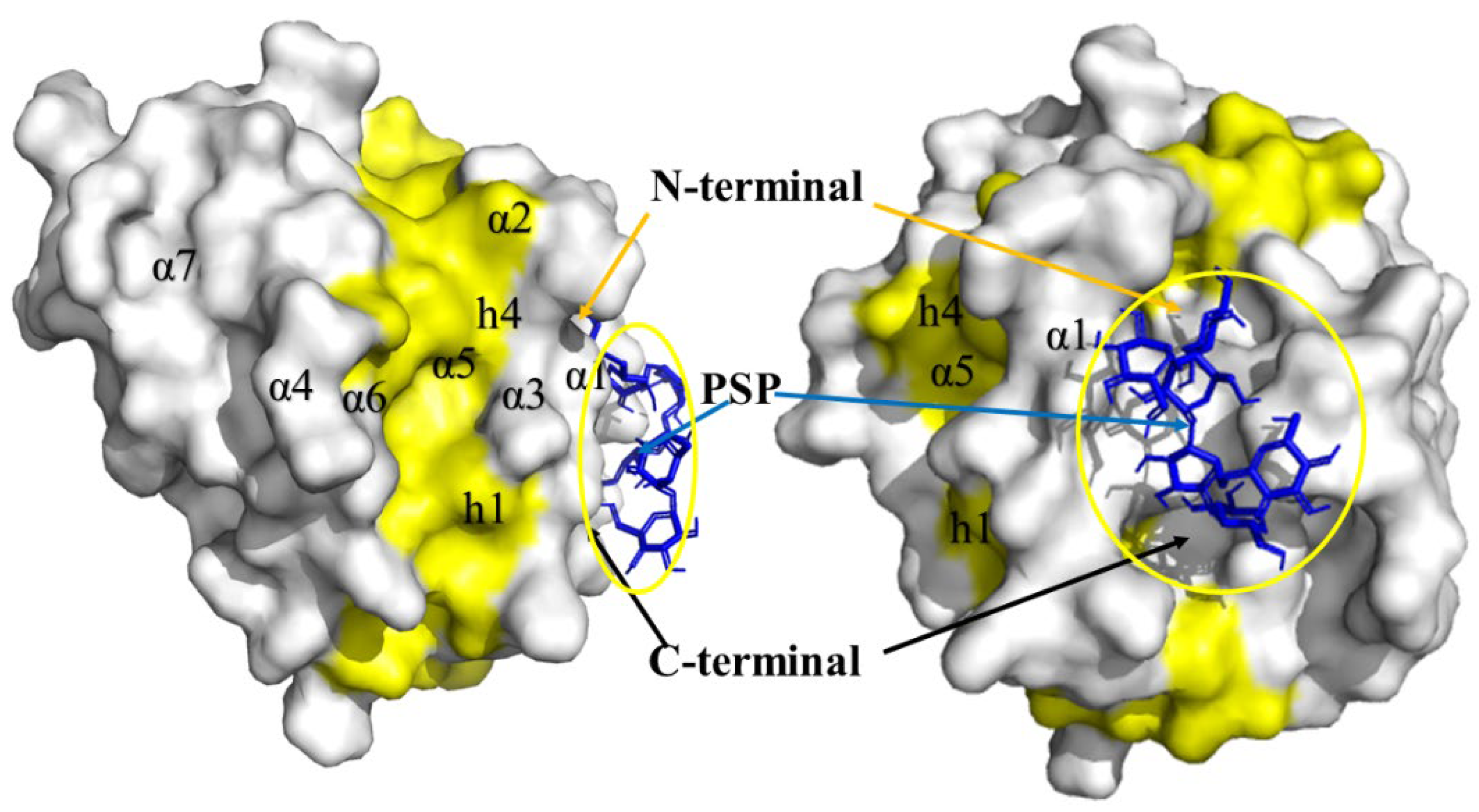
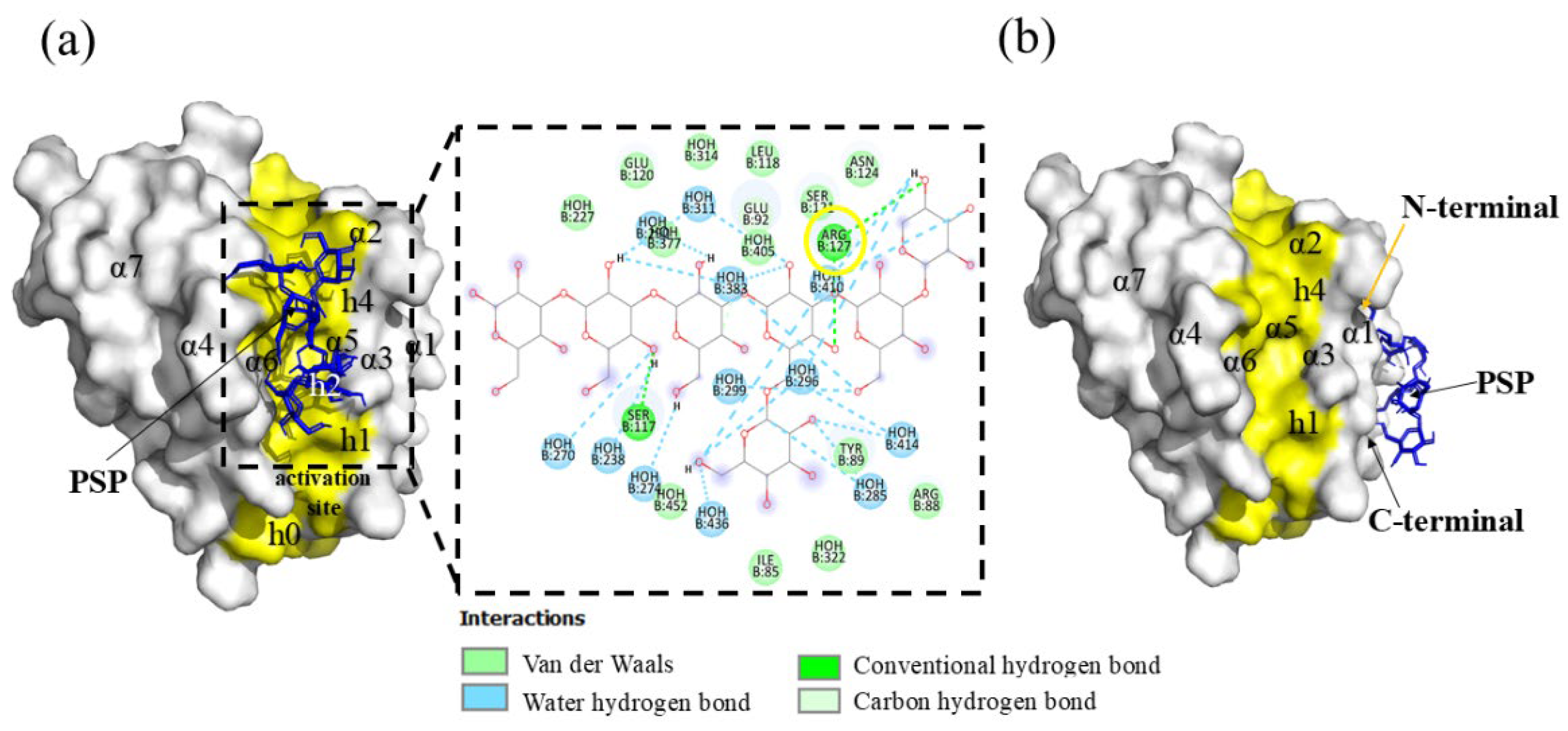
Hit 1, Bcl-2 Apoptosis Regulatory Protein (6O0K)
PSK-Bcl-2
PSP-Bcl-2
Hit 2, CD59 Human Antigen (2J8B)
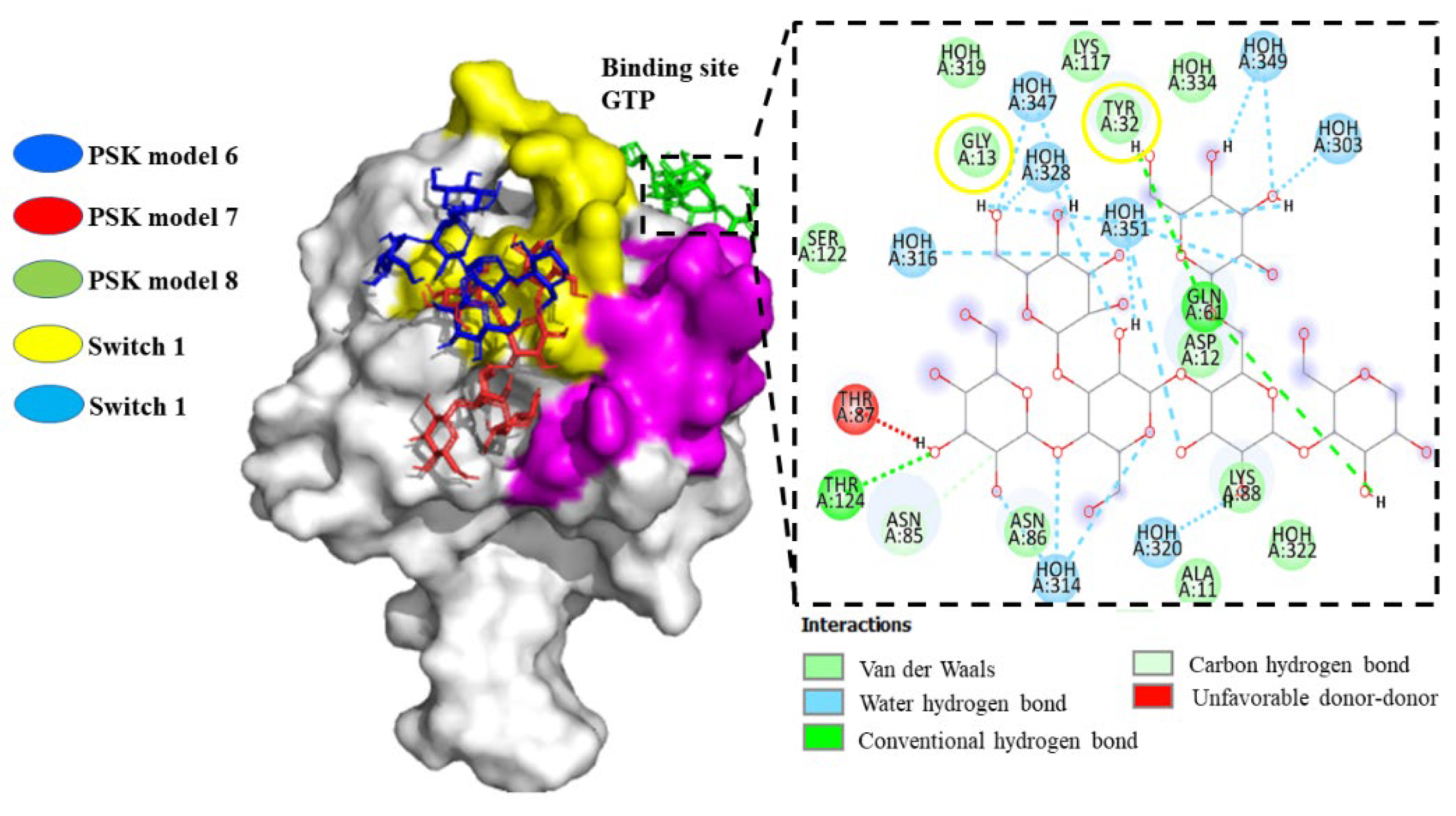
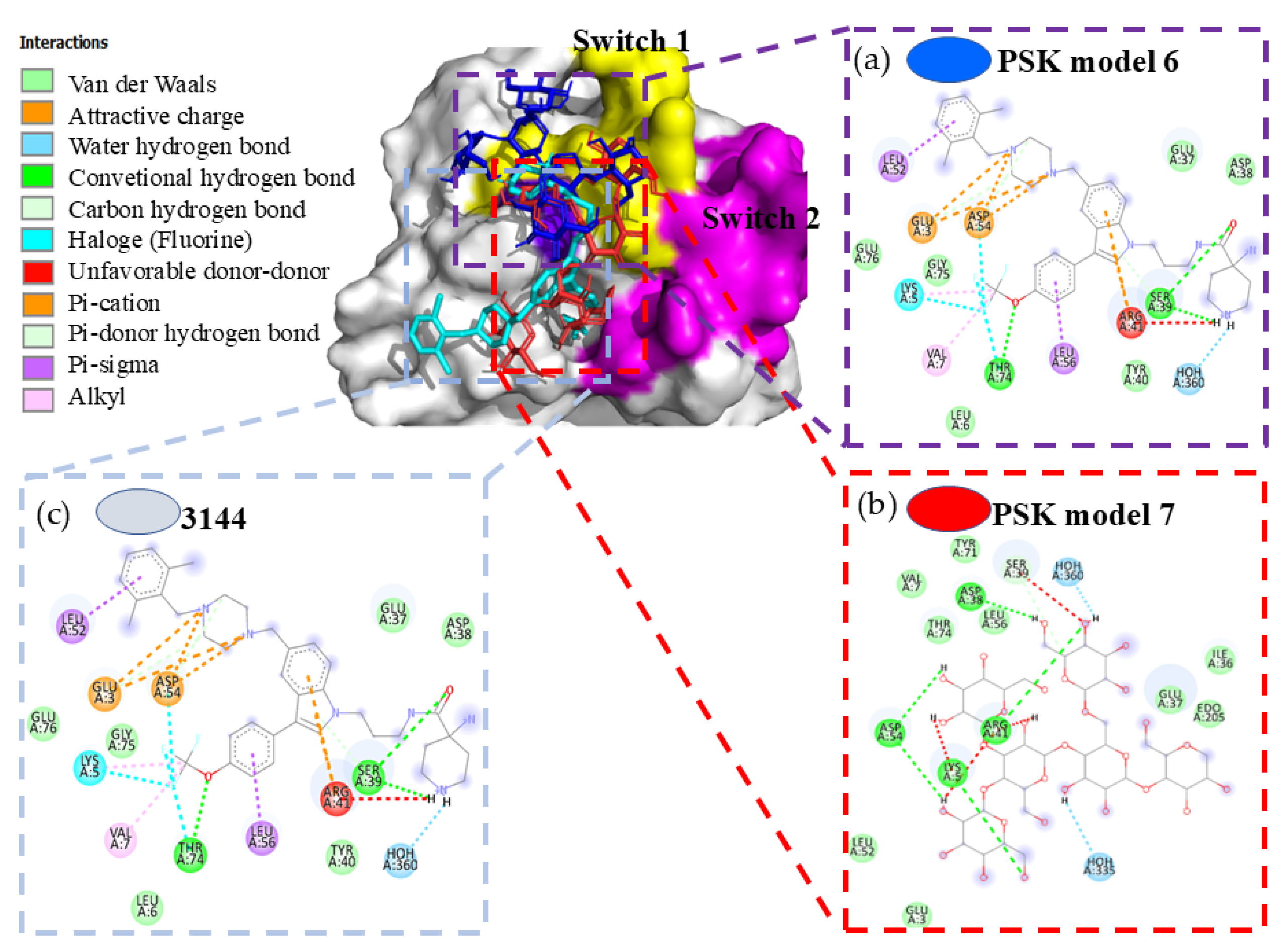
Hit 3, K-RAS Isoform (5USJ)
Hit 4, Ribonucleotide Reductase Enzymes (6L3R)
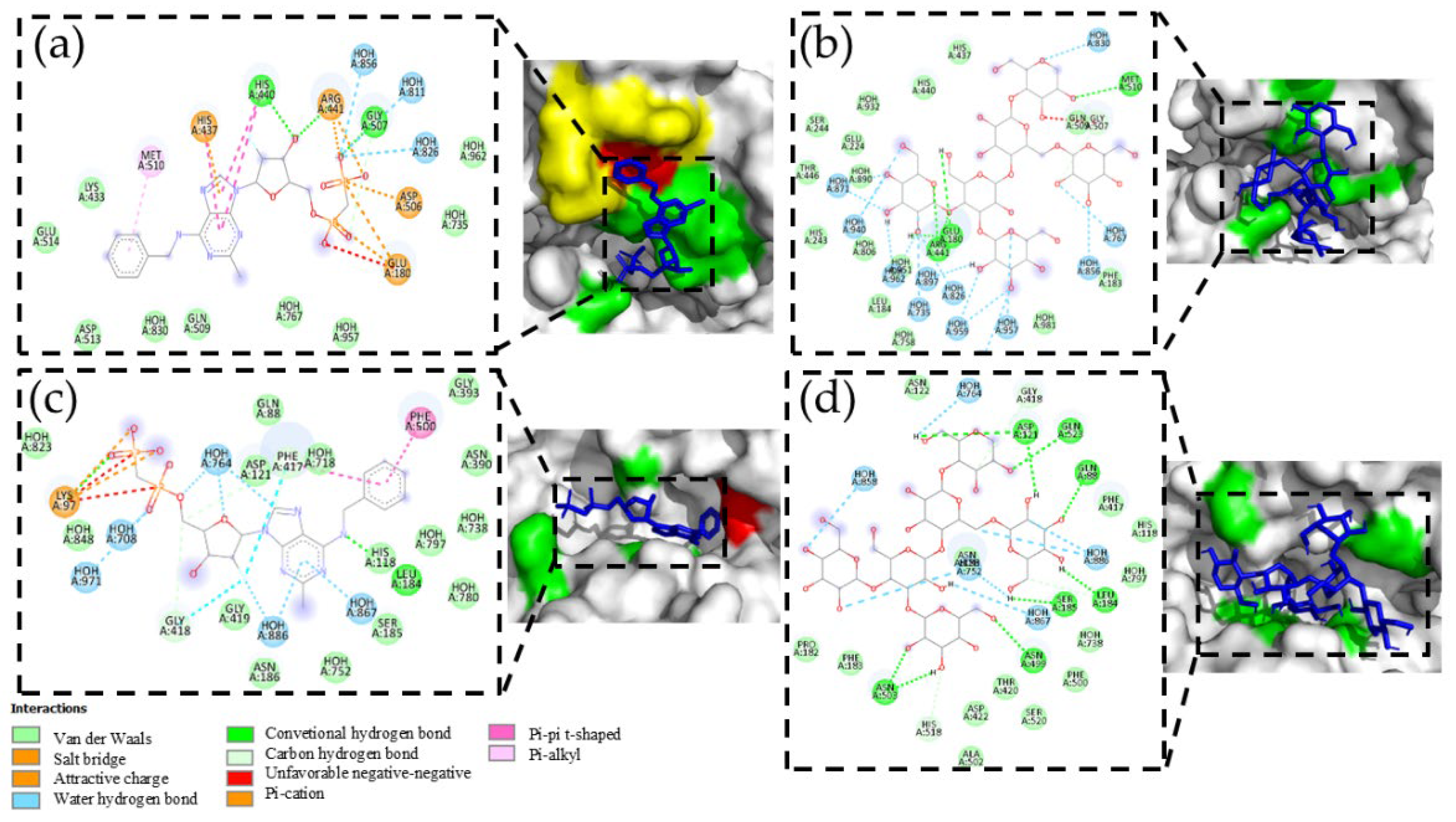
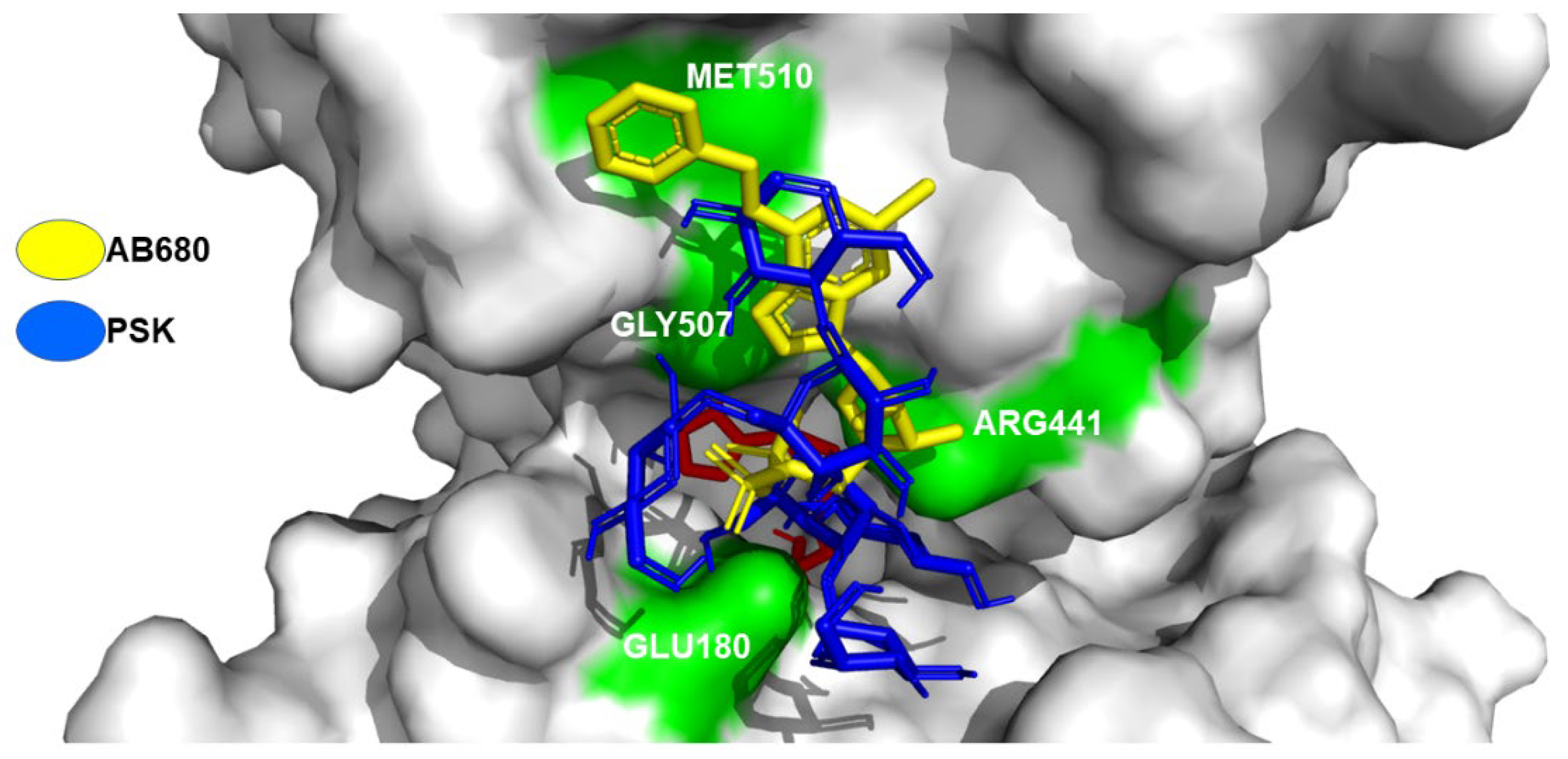
Hit 5, Bak Protein (5VX1)
PSK-Bak
PSP-Bak
Hit 6, the CD73 Protein (6Z9B)
3. Discussion
4. Materials and Methods
Computer Methodology
Specifications of the Computer Equipment
Preparation of the Ligands
Protein Database
Reverse Molecular Docking
Molecular Docking with Vina-Carb
Docking Analysis
Virtual Screening
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carlson, A.; Justo, A.; Hibbett, D.S. Species delimitation in Trametes : a comparison of ITS, RPB1, RPB2 and TEF1 gene phylogenies. Mycologia 2014, 106, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Justo, A.; Hibbett, D.S. Phylogenetic classification of Trametes (Basidiomycota, Polyporales) based on a five–marker dataset. Taxon 2011, 60, 1567–1583. [Google Scholar] [CrossRef]
- Ng, T.B. A review of research on the protein-bound polysaccharide (polysaccharopeptide, PSP) from the mushroom Coriolus versicolor (Basidiomycetes: Polyporaceae). General pharmacology 1998, 30, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.K.; Ng, T.B.; Sze, S.F.; Tsui, K.W. Activation of peritoneal macrophages by polysaccharopeptide from the mushroom, Coriolus versicolor. Immunopharmacology 1993, 26, 139–146. [Google Scholar] [CrossRef]
- Yeung, J.H.; Or, P.M. Polysaccharide peptides from Coriolus versicolor competitively inhibit model cytochrome P450 enzyme probe substrates metabolism in human liver microsomes. Phytomedicine 2012, 19, 457–463. [Google Scholar] [CrossRef]
- Awadasseid, A.; Hou, J.; Gamallat, Y.; Xueqi, S.; Eugene, K.D.; Musa Hago, A.; Bamba, D.; Meyiah, A.; Gift, C.; Xin, Y. Purification, characterization, and antitumor activity of a novel glucan from the fruiting bodies of Coriolus versicolor. PloS ONE 2017, 2017 12, e0171270. [Google Scholar] [CrossRef]
- Zhang, X.; Cheng, Y.; Zhou, Q.; Huang, H.; Dong, Y.; Yang, Y.; Zhao, M.; He, Q. The Effect of Chinese Traditional Medicine Huaiqihuang (HQH) on the Protection of Nephropathy. Oxid. Med. Cell. Longev. 2020, 1-10, 2153912. [Google Scholar] [CrossRef]
- Moazzem Hossen, S.M.; Akramul Hoque Tanim, M.; Shahadat Hossain, M.; Ahmed Sami, S.; Uddin Emon, N. Deciphering the CNS anti-depressant, antioxidant and cytotoxic profiling of methanol and aqueous extracts of Trametes versicolor and molecular interactions of its phenolic compounds. Saudi J. Biol. Sci. 2021, 28, 6375–6383. [Google Scholar] [CrossRef]
- Ebina, T.; Murata, K. Antitumor effect of PSK at a distant site: inductions of interleukin-8-like factor and macrophage chemotactic factor in murine tumor. Jpn. J. Cancer Res. 1990, 81, 1307–1313. [Google Scholar] [CrossRef]
- Kobayashi, H.; Matsunaga, K.; Fujii, M. PSK as a chemopreventive agent. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research. ASPO 1993, 2, 271–276. [Google Scholar]
- Kobayashi, H.; Matsunaga, K.; Oguchi, Y. Antimetastatic effects of PSK (Krestin), a protein-bound polysaccharide obtained from basidiomycetes: an overview. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research. ASPO 1995, 4, 275–281. [Google Scholar]
- Jiménez-Medina, E.; Berruguilla, E.; Romero, I.; Algarra, I.; Collado, A.; Garrido, F.; Garcia-Lora, A. The immunomodulator PSK induces in vitro cytotoxic activity in tumour cell lines via arrest of cell cycle and induction of apoptosis. BMC cancer 2008, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Fritz, H.; Kennedy, D.A.; Ishii, M.; Fergusson, D.; Fernandes, R.; Cooley, K.; Seely, D. Polysaccharide K and Coriolus versicolor extracts for lung cancer: a systematic review. Integr. Cancer Ther. 2015, 14, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.F.; Lin, M.C.; Tang, G. A Phytochemical-Based Copolymer Derived from Coriolus versicolor Polysaccharopeptides for Gene Delivery. Molecules (Basel, Switzerland) 2018, 23, 2273. [Google Scholar] [CrossRef]
- Pawlikowska, M.; Jędrzejewski, T.; Slominski, A.T.; Brożyna, A.A.; Wrotek, S. Pigmentation Levels Affect Melanoma Responses to Coriolus versicolor Extract and Play a Crucial Role in Melanoma-Mononuclear Cell Crosstalk. Int. J. Mol. Sci. 2021, 22, 5735. [Google Scholar] [CrossRef]
- Zhang, X.; Cai, Z.; Mao, H.; Hu, P.; Li, X. Isolation and structure elucidation of polysaccharides from fruiting bodies of mushroom Coriolus versicolor and evaluation of their immunomodulatory effects. Int. J. Biol. Macromol. 2021, 166, 1387–1395. [Google Scholar] [CrossRef]
- Yang, X.; Wei, S.; Lu, X.; Qiao, X.; Simal-Gandara, J.; Capanoglu, E.; Woźniak, L.; Zou, L.; Cao, H.; Xiao, J.; Tang, X.; Li, N. A neutral polysaccharide with a triple helix structure from ginger: Characterization and immunomodulatory activity. Food Chem. 2021, 350, 129261. [Google Scholar] [CrossRef]
- Liu, D.; Tang, W.; Yin, J.Y.; Nie, S.P.; Xie, M.Y. Monosaccharide composition analysis of polysaccharides from natural sources: Hydrolysis condition and detection method development. Food Hydrocoll 2021, 116, 106641. [Google Scholar] [CrossRef]
- Pacheco-Gómez, V.; Caballero-Zamora, A.; Martínez-González, S.; Prado-Rebolledo, O.; García-Casillas, A. Bioquímica y vías metabólicas de polisacáridos, lípidos y proteínas. Abanico Veterinario 2021, 11, NA. [Google Scholar] [CrossRef]
- Capecchi, A.; Probst, D.; Reymond, J.L. One molecular fingerprint to rule them all: drugs, biomolecules, and the metabolome. J. Cheminform. 2020, 12, 43. [Google Scholar] [CrossRef]
- Saldívar-González, F.; Prieto-Martínez, F.D.; Medina-Franco, J.L. Descubrimiento y desarrollo de fármacos: un enfoque computacional. Educación Química 2017, 28, 51–58. [Google Scholar] [CrossRef]
- Meng, Y.; Lyu, F.; Xu, X.; Zhang, L. Recent advances in chain conformation and bioactivities of triple-helix polysaccharides. Biomacromolecules 2020, 21, 1653–1677. [Google Scholar] [CrossRef] [PubMed]
- Awadasseid, A.; Hou, J.; Gamallat, Y.; Xueqi, S.; Eugene, K.D.; Musa Hago, A.; Bamba, D.; Meyiah, A.; Gift, C.; Xin, Y. Purification, characterization, and antitumor activity of a novel glucan from the fruiting bodies of Coriolus versicolor. PloS ONE 2017, 12, e0171270. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, Y.; Miao, C.; Liu, Y.; Liu, R. Coriolus versicolor polysaccharides (CVP) regulates neuronal apoptosis in cerebral ischemia-reperfusion injury via the p38MAPK signaling pathway. Ann. Transl. Med. 2020, 8, 1168. [Google Scholar] [CrossRef]
- Bae, H.; Song, G.; Lee, J.Y.; Hong, T.; Chang, M.J.; Lim, W. Laminarin-Derived from brown algae suppresses the growth of ovarian cancer cells via mitochondrial dysfunction and ER stress. Mar. Drugs 2020, 18, 152. [Google Scholar] [CrossRef]
- Chaichian, S.; Moazzami, B.; Sadoughi, F.; Haddad K., H.; Zaroudi, M.; Asemi, Z. Functional activities of beta-glucans in the prevention or treatment of cervical cancer. Ovarian Res. 2020, 13, 24. [Google Scholar] [CrossRef]
- Rosendahl, A.H.; Sun, C.; Wu, D.; Andersson, R. Polysaccharide-K (PSK) increases p21(WAF/Cip1) and promotes apoptosis in pancreatic cancer cells. Pancreatology 2012, 12, 467–474. [Google Scholar] [CrossRef]
- Hirai, R.; Oguchi, Y.; Sugita, N.; Matsunaga, K.; Nomoto, K. Enhancement of T-cell proliferation by PSK. Int. J. Immunopharmacol. 1993, 15, 745–750. [Google Scholar] [CrossRef]
- Kato, M.; Hirose, K.; Hakozaki, M.; Ohno, M.; Saito, Y.; Izutani, R.; Noguchi, J.; Hori, Y.; Okumoto, S.; Kuroda, S.; Nomura, H.; Nishimatsu, S.; Ohoyanagi, H. Induction of gene expression for immunomodulating cytokines in peripheral blood mononuclear cells in response to orally administered PSK, an immunomodulating protein-bound polysaccharide. CII 1995, 40, 152–156. [Google Scholar] [CrossRef]
- Harada, M.; Matsunaga, K.; Oguchi, Y.; Iijima, H.; Tamada, K.; Abe, K.; Takenoyama, M.; Ito, O.; Kimura, G.; Nomoto, K. Oral administration of PSK can improve the impaired anti-tumor CD4+ T-cell response in gut-associated lymphoid tissue (GALT) of specific-pathogen-free mice. Int. J. Cancer Res. 1997, 70, 362–372. [Google Scholar] [CrossRef]
- Lu, H.; Yang, Y.; Gad, E.; Wenner, C.A.; Chang, A.; Larson, E.R.; Dang, Y.; Martzen, M.; Standish, L.J.; Disis, M.L. Polysaccharide krestin is a novel TLR2 agonist that mediates inhibition of tumor growth via stimulation of CD8 T cells and NK cells. Clin Cancer Res. 2011, 17, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, K.; Mitsuhashi, N.; Saito, Y.; Takahashi, M.; Katano, S.; Shiojima, K.; Furuta, M.; Niibe, H. Effect of krestin (PSK) as adjuvant treatment on the prognosis after radical radiotherapy in patients with non-small cell lung cancer. Anticancer Res. 1993, 13, 1815–1820. [Google Scholar] [PubMed]
- Ohwada, S.; Ikeya, T.; Yokomori, T.; Kusaba, T.; Roppongi, T.; Takahashi, T.; Nakamura, S.; Kakinuma, S.; Iwazaki, S.; Ishikawa, H.; Kawate, S.; Nakajima, T.; Morishita, Y. Adjuvant immunochemotherapy with oral Tegafur/Uracil plus PSK in patients with stage II or III colorectal cancer: a randomised controlled study. Br. J. Cancer 2004, 90, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Oba, K.; Teramukai, S.; Kobayashi, M.; Matsui, T.; Kodera, Y.; Sakamoto, J. Efficacy of adjuvant immunochemotherapy with polysaccharide K for patients with curative resections of gastric cancer. CII 2007, 56, 905–911. [Google Scholar] [CrossRef] [PubMed]
- Luk, S.U.; Lee, T.K.; Liu, J.; Lee, D.T.; Chiu, Y.T.; Ma, S.; Ng, I.O.; Wong, Y.C.; Chan, F.L.; Ling, M.T. Chemopreventive effect of PSP through targeting of prostate cancer stem cell-like population. PloS ONE 2011, 6, e19804. [Google Scholar] [CrossRef]
- Ho, C.Y.; Lau, C.B.; Kim, C.F.; Leung, K.N.; Fung, K.P.; Tse, T.F.; Chan, H.H.; Chow, M.S. Differential effect of Coriolus versicolor (Yunzhi) extract on cytokine production by murine lymphocytes in vitro. Int. Immunopharmacol. 2004, 4, 1549–1557. [Google Scholar] [CrossRef]
- Yang, S.F.; Zhuang, T.F.; Si, Y.M.; Qi, K.Y.; Zhao, J. Coriolus versicolor mushroom polysaccharides exert immunoregulatory effects on mouse B cells via membrane Ig and TLR-4 to activate the MAPK and NF-κB signaling pathways. Mol. Immunol. 2015, 64, 144–151. [Google Scholar] [CrossRef]
- Sekhon, B.K.; Sze, D.M.; Chan, W.K.; Fan, K.; Li, G.Q.; Moore, D.E.; Roubin, R.H. PSP activates monocytes in resting human peripheral blood mononuclear cells: immunomodulatory implications for cancer treatment. Food Chem. 2013, 138, 2201–2209. [Google Scholar] [CrossRef]
- Saleh, M.H.; Rashedi, I.; Keating, A. Immunomodulatory properties of Coriolus versicolor: The role of polysaccharopeptide. Frontiers in Immunology 2017, 8, 1087. [Google Scholar] [CrossRef]
- Zaidman, B.Z.; Yassin, M.; Mahajna, J.; Wasser, S.P. Medicinal mushroom modulators of molecular targets as cancer therapeutics. Appl. Microbiol. Biotechnol. 2005, 67, 453–468. [Google Scholar] [CrossRef]
- Hsieh, T.C.; Wu, J.M. Cell growth and gene modulatory activities of Yunzhi (Windsor Wunxi) from mushroom Trametes versicolor in androgen-dependent and androgen-insensitive human prostate cancer cells. Int. J. Oncol. 2001, 18, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Habtemariam, S. Trametes versicolor (Synn. Coriolus versicolor) Polysaccharides in cancer therapy: Targets and efficacy. Biomedicines 2020, 8, 135. [Google Scholar] [CrossRef] [PubMed]
- Vasseur, R.; Baud, S.; Steffenel, L.A.; Vigouroux, X.; Martiny, L.; Krajecki, M.; Dauchez, M. Inverse docking method for new proteins targets identification: A parallel approach. Parallel Comput. 2015, 42, 48–59. [Google Scholar] [CrossRef]
- Cui, J.; Chisti, Y. Polysaccharopeptides of Coriolus versicolor: physiological activity, uses, and production. Biotechnol. Adv. 2003, 21, 109–122. [Google Scholar] [CrossRef]
- Zhang, H.; Pan, J.; Wu, X.; Zuo, A.R.; Wei, Y.; Ji, Z.L. Large-scale target identification of herbal medicine using a reverse docking approach. ACS Omega 2019, 4, 9710–9719. [Google Scholar] [CrossRef]
- Bianchi, V.; Bulek, A.; Fuller, A.; Lloyd, A.; Attaf, M.; Rizkallah, P.J.; Dolton, G.; Sewell, A.K.; Cole, D.K. A molecular switch abrogates glycoprotein 100 (gp100) T-cell Receptor (TCR) targeting of a human melanoma antigen. JBC 2016, 291, 8951–8959. [Google Scholar] [CrossRef]
- Kargbo R., B. Dual Inhibition of KRAS G12C and G12D mutants as a potential treatment in cancer therapy. ACS Med. Chem. Lett. 2021, 12, 1512–1513. [Google Scholar] [CrossRef]
- McCarthy, M.J.; Pagba, C.V.; Prakash, P.; Naji, A.K.; van der Hoeven, D.; Liang, H.; Gupta, A.K.; Zhou, Y.; Cho, K.J.; Hancock, J.F.; Gorfe, A.A. Discovery of high-affinity noncovalent allosteric KRAS inhibitors that disrupt effector binding. ACS Omega 2019, 4, 2921–2930. [Google Scholar] [CrossRef]
- Welsch, M.E.; Kaplan, A.; Chambers, J.M.; Stokes, M.E.; Bos, P.H.; Zask, A.; Zhang, Y.; Sanchez-Martin, M.; Badgley, M.A.; Huang, C.S.; Tran, T.H.; Akkiraju, H.; Brown, L.M.; Nandakumar, R.; Cremers, S.; Yang, W.S.; Tong, L.; Olive, K.P.; Ferrando, A.; Stockwell, B.R. Multivalent small-molecule Pan-RAS inhibitors. Cell 2017, 168, 878–889.e29. [Google Scholar] [CrossRef]
- Elledge, S.J.; Zhou, Z.; Allen, J.B. Ribonucleotide reductase: regulation, regulation, regulation. TIBS 1992, 17, 119–123. [Google Scholar] [CrossRef]
- Park, D.; Anisuzzaman, A.; Magis, A.T.; Chen, G.; Xie, M.; Zhang, G.; Behera, M.; Sica, G.L.; Ramalingam, S.S.; Owonikoko, T.K.; Deng, X. Discovery of small molecule Bak activator for lung cancer therapy. Theranostics 2021, 11, 8500–8516. [Google Scholar] [CrossRef] [PubMed]
- Lawson, K.V.; Kalisiak, J.; Lindsey, E.A.; Newcomb, E.T.; Leleti, M.R.; Debien, L.; Rosen, B.R.; Miles, D.H.; Sharif, E.U.; Jeffrey, J.L.; Tan, J.; Chen, A.; Zhao, S.; Xu, G.; Fu, L.; Jin, L.; Park, T.W.; Berry, W.; Moschütz, S.; Scaletti, E.; Powers, J.P. Discovery of AB680: A potent and selective inhibitor of CD73. J. Med. Chem. 2020, 63, 11448–11468. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.J.; Tait, S. Targeting BCL-2 regulated apoptosis in cancer. Open Biol. 2018, 8, 180002. [Google Scholar] [CrossRef]
- Guerra, V.A.; DiNardo, C.; Konopleva, M. Venetoclax-based therapies for acute myeloid leukemia. Best Pract Res Clin Haematol. 2019, 32, 145–153. [Google Scholar] [CrossRef]
- Vogler, M.; Dinsdale, D.; Dyer, M.J.; Cohen, G.M. ABT-199 selectively inhibits BCL2 but not BCL2L1 and efficiently induces apoptosis of chronic lymphocytic leukaemic cells but not platelets. Br. J. Haematol. 2013, 163, 139–142. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, Q.; Liao, Q.; Zhao, Y. CD59: a promising target for tumor immunotherapy. Future Oncol. 2018, 14, 781–791. [Google Scholar] [CrossRef] [PubMed]
- You, T.; Hu, W.; Ge, X.; Shen, J.; Qin, X. Application of a novel inhibitor of human CD59 for the enhancement of complement-dependent cytolysis on cancer cells. Cell Mol. Immunol. 2011, 8, 157–163. [Google Scholar] [CrossRef]
- Cai, B.; Xie, S.; Liu, F.; Simone, L.C.; Caplan, S.; Qin, X.; Naslavsky, N. Rapid degradation of the complement regulator, CD59, by a novel inhibitor. JBC 2014, 289, 12109–12125. [Google Scholar] [CrossRef] [PubMed]
- Wickham, S.E.; Hotze, E.M.; Farrand, A.J.; Polekhina, G.; Nero, T.L.; Tomlinson, S.; Parker, M.W.; Tweten, R.K. Mapping the intermedilysin-human CD59 receptor interface reveals a deep correspondence with the binding site on CD59 for complement binding proteins C8 alpha and C9. JBC 2011, 286, 20952–20962. [Google Scholar] [CrossRef]
- Prior, I.A.; Lewis, P.D.; Mattos, C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012, 72, 2457–2467. [Google Scholar] [CrossRef]
- Uprety, D.; Adjei, A.A. KRAS: From undruggable to a druggable Cancer Target. Cancer Treat. Rev. 2020, 89, 102070. [Google Scholar] [CrossRef] [PubMed]
- Kargbo R., B. Targeting the KRAS G12D mutant as potential therapy in cancer. ACS Med. Chem. Lett. 2021, 12, 1212–1213. [Google Scholar] [CrossRef] [PubMed]
- Pantsar, T. The current understanding of KRAS protein structure and dynamics. Comput Struct Biotechnol J. 2019, 18, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Aye, Y.; Li, M.; Long, M.J.; Weiss, R.S. Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene 2015, 34, 2011–2021. [Google Scholar] [CrossRef] [PubMed]
- Greene, B. L; Kang, G.; Cui, C.; Bennati, M.; Nocera, D.G.; Drennan, C.L.; Stubbe, J. Ribonucleotide reductases: Structure, chemistry, and metabolism suggest new therapeutic targets. Annu. Rev. Biochem. 2020, 89, 45–75. [Google Scholar] [CrossRef]
- Fairman, J.W.; Wijerathna, S.R.; Ahmad, M.F.; Xu, H.; Nakano, R.; Jha, S.; Prendergast, J.; Welin, R.M.; Flodin, S.; Roos, A.; Nordlund, P.; Li, Z.; Walz, T.; Dealwis, C.G. Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat. Struct. Mol. Biol. 2011, 18, 316–322. [Google Scholar] [CrossRef]
- Ahmad, M.F.; Dealwis, C.G. The structural basis for the allosteric regulation of ribonucleotide reductase. Prog. Mol. Biol. Transl. Sci. 2013, 117, 389–410. [Google Scholar] [CrossRef]
- Edlich, F. BCL-2 proteins and apoptosis: Recent insights and unknowns. Biochem. Biophys. Res. Commun. 2018, 500, 26–34. [Google Scholar] [CrossRef]
- Brouwer, J.M.; Lan, P.; Cowan, A.D.; Bernardini, J.P.; Birkinshaw, R.W.; van Delft, M.F.; Sleebs, B.E.; Robin, A.Y.; Wardak, A.; Tan, I.K.; Reljic, B.; Lee, E.F.; Fairlie, W.D.; Call, M.J.; Smith, B.J.; Dewson, G.; Lessene, G.; Colman, P.M.; Czabotar, P.E. Conversion of Bim-BH3 from activator to inhibitor of Bak through structure-based design. Mol. Cell 2017, 68, 659–672.e9. [Google Scholar] [CrossRef]
- Czabotar, P.E.; Westphal, D.; Dewson, G.; Ma, S.; Hockings, C.; Fairlie, W.D.; Lee, E.F.; Yao, S.; Robin, A.Y.; Smith, B.J.; Huang, D.C.; Kluck, R.M.; Adams, J.M.; Colman, P.M. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell 2013, 152, 519–531. [Google Scholar] [CrossRef]
- Kalkavan, H.; Green, D.R. MOMP, cell suicide as a BCL-2 family business. Cell Death Differ. 2018, 25, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Upton, J.P.; Valentijn, A.J.; Zhang, L.; Gilmore, A.P. The N-terminal conformation of Bax regulates cell commitment to apoptosis. Cell Death Differ. 2007, 14, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.; Bell, F.; Westphal, D.; Anwari, K.; Gulbis, J.; Smith, B.J.; Dewson, G.; Kluck, R.M. Bak apoptotic pores involve a flexible C-terminal region and juxtaposition of the C-terminal transmembrane domains. Cell Death Differ. 2015, 22, 1665–1675. [Google Scholar] [CrossRef]
- Antonioli, L.; Pacher, P.; Vizi, E.S.; Haskó, G. CD39 and CD73 in immunity and inflammation. Trends Mol. Med. 2013, 19, 355–367. [Google Scholar] [CrossRef]
- Schäkel, L.; Schmies, C.C.; Idris, R.M.; Luo, X.; Lee, S.Y.; Lopez, V.; Mirza, S.; Vu, T.H.; Pelletier, J.; Sévigny, J.; Namasivayam, V.; Müller, C.E. Nucleotide analog ARL67156 as a lead structure for the development of CD39 and dual CD39/CD73 ectonucleotidase inhibitors. Front. Pharmacol. 2020, 11, 1294. [Google Scholar] [CrossRef]
- Lawson, K.V.; Kalisiak, J.; Lindsey, E.A.; Newcomb, E.T.; Leleti, M.R.; Debien, L.; Rosen, B.R.; Miles, D.H.; Sharif, E.U.; Jeffrey, J.L.; Tan, J.; Chen, A.; Zhao, S.; Xu, G.; Fu, L.; Jin, L.; Park, T.W.; Berry, W.; Moschütz, S.; Scaletti, E.; Powers, J.P. Discovery of AB680: A potent and selective inhibitor of CD73. J. Med. Chem. 2020, 63, 11448–11468. [Google Scholar] [CrossRef] [PubMed]
- Novak, M.; Vetvicka, V. Beta-glucans, history, and the present: immunomodulatory aspects and mechanisms of action. J. Immunotoxicol. 2008, 5, 47–57. [Google Scholar] [CrossRef]
- Kojima, T.; Tabata, K.; Itoh, W.; Yanaki, T. Molecular weight dependence of the antitumor activity of Schizophyllan. Agric. Biol. Chem. 1986, 50, 231–232. [Google Scholar] [CrossRef]
- Young, S.H.; Jacobs, R.R. Sodium hydroxide induced conformational change in schizophyllan detected by the fluorescence dye, aniline blue. Carbohydr. Res. 1998, 310, 91–99. [Google Scholar] [CrossRef]
- Saitô, H.; Yamada, J.; Yoshioka, Y.; Shibata, Y.; Erata, T. Evidence of three distinct conformations-single chain, single helix, and triple helix-of (1 → 3)-β-D-xylan in the solid and intact frond of green algae as studied by 13C-nmr spectroscopy. Biopolymers 1991, 31, 933–940. [Google Scholar] [CrossRef]
- Ohno, N.; Miura, N.N.; Chiba, N.; Adachi, Y.; Yadomae, T. Comparison of the immunopharmacological activities of triple and single-helical Schizophyllan in mice. Biol. Pharm. Bull. 1995, 18, 1242–1247. [Google Scholar] [CrossRef] [PubMed]
- Kabat, E.A.; Wu, T.T.; Bilofsky, H. Attempts to locate residues in complementarity-determining regions of antibody combining sites that make contact with antigen. Proc. Natl. Acad. Sci. USA 1976, 73, 617–619. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.; Zhao, M.; Su, Q.; Zhang, M.; Lv, W.; Chen, Q.; Chen, F.; Chu, D.; Du, D.; Zhang, Y. 2D-SAR and 3D-QSAR analyses for acetylcholinesterase inhibitors. Mol. Divers. 2017, 21, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A. MMFF VI. MMFF94s option for energy minimization studies. J. Comput. Chem. 1999, 20, 720–729. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Lešnik, S.; Štular, T.; Brus, B.; Knez, D.; Gobec, S.; Janežič, D.; Konc, J. LiSiCA: A software for ligand-based virtual screening and its application for the discovery of Butyrylcholinesterase inhibitors. J. Chem. Inf. Model. 2015, 55, 1521–1528. [Google Scholar] [CrossRef]
- Nivedha, A.K.; Thieker, D.F.; Makeneni, S.; Hu, H.; Woods, R.J. Vina-Carb: improving glycosidic angles during carbohydrate docking. J. Chem. Theory Comput. 2016, 12, 892–901. [Google Scholar] [CrossRef]
- Nivedha, A.K.; Makeneni, S.; Foley, B.L.; Tessier, M.B.; Woods, R.J. Importance of ligand conformational energies in carbohydrate docking: Sorting the wheat from the chaff. J. Comput. Chem. 2014, 35, 526–539. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G. Open Babel: An open chemical toolbox. J. Cheminf. 2011, 3, 33. [Google Scholar] [CrossRef]
- Cereto-Massagué, A.; Ojeda, M.J.; Valls, C.; Mulero, M.; Garcia-Vallvé, S.; Pujadas, G. Molecular fingerprint similarity search in virtual screening. Methods 2015, 71, 58–63. [Google Scholar] [CrossRef]
- Baldi, P.; Nasr, R. When is chemical similarity significant? The statistical distribution of chemical similarity scores and its extreme values. J. Chem. Inf. Model 2010, 50, 1205–1222. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Wang, D.; Shen, Z.; Li, S.; Li, H. Multi-Body Interactions in Molecular Docking Program Devised with Key Water Molecules in Protein Binding Sites. Molecules (Basel, Switzerland) 2018, 23, 2321. [Google Scholar] [CrossRef] [PubMed]
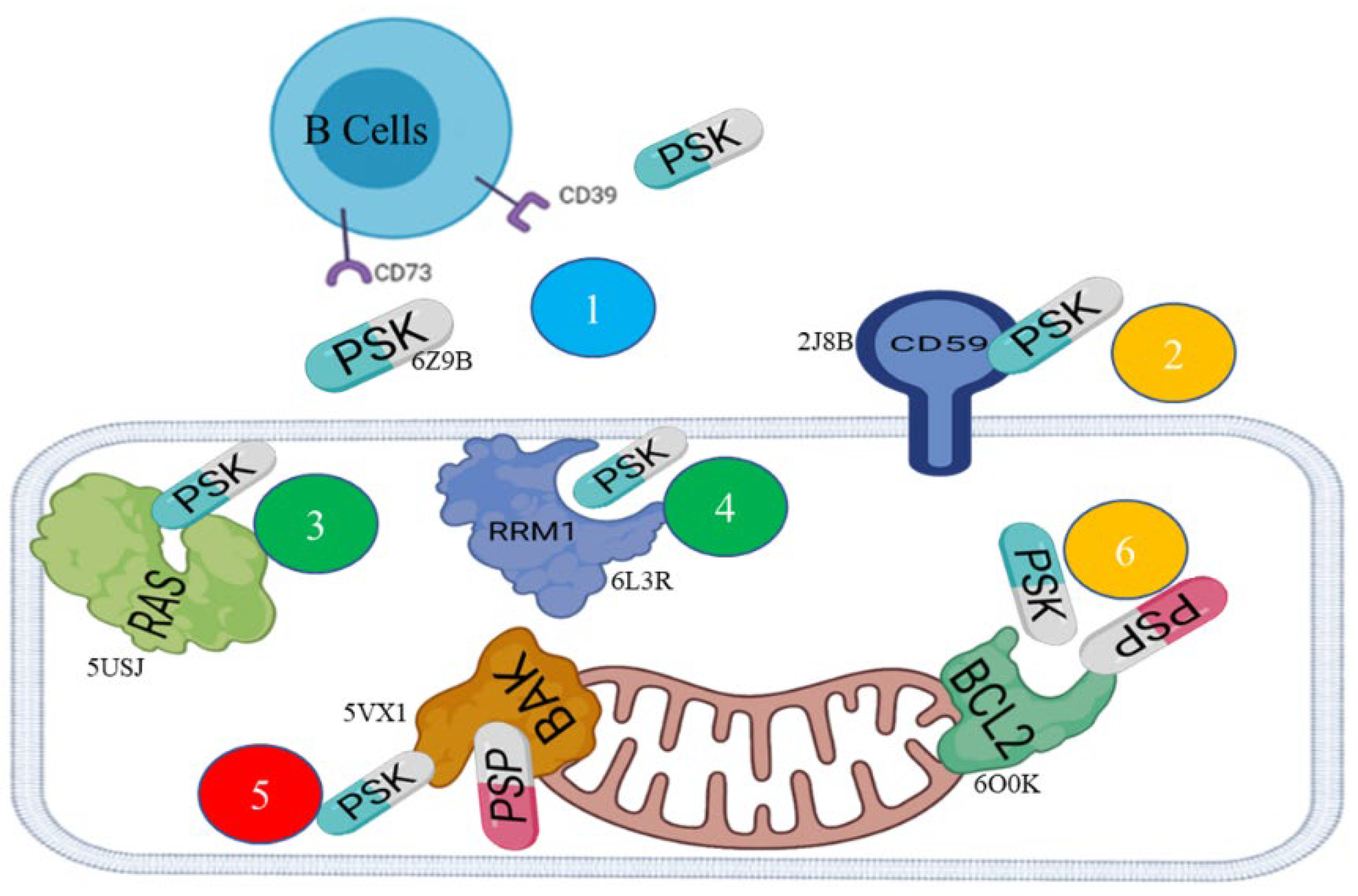
| Hits | Codes | Scoring (Kcal/mol) | Protein type | Mechanism | |
| PSK | PSP | ||||
| 1 | 6O0K | -6.5 | -6.8 | Bcl-2 apoptosis regulator | Promotes anti-apoptosis |
| 2 | 2J8B | -6.3 | Membrane-bound glycoprotein | Protects host cells from lysis | |
| 3 | 5USJ | -6.3 | Mutant KRAS G12D | Active molecular switch-regulators that increase the capacity for invasion and metastasis, and decrease apoptosis | |
| 4 | 6L3R | -7.2 | RRM1: large subunit of ribonucleoside-diphosphate reductase | RRM1 participates in regulating cell proliferation | |
| 5 | 5VX1 | -7.6 | -7.8 | BAK | Initiates oligomerization and permeabilization of the outer mitochondrial membrane |
| 6 | 6Z9B | -6.3 | Hydrolases | Hydrolyzes ATP and AMP to generate adenosines, which inhibit the immune response | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





