Submitted:
01 October 2024
Posted:
01 October 2024
You are already at the latest version
Abstract
Keywords:
Introduction
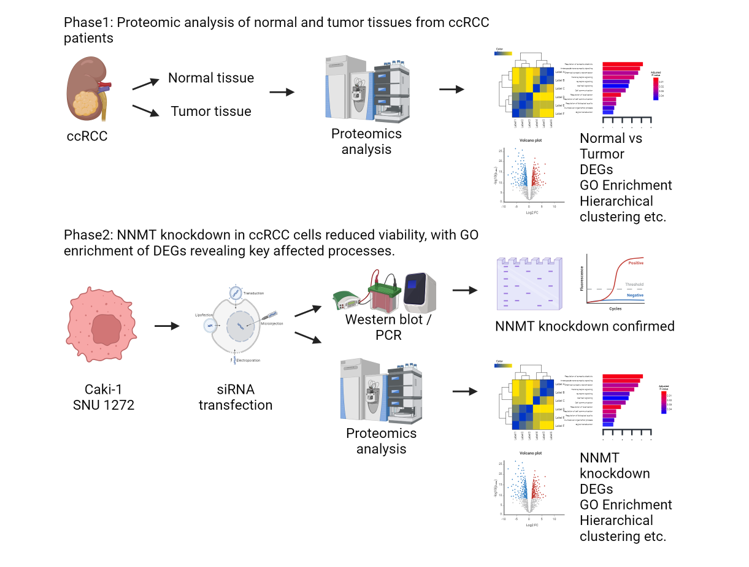
Material and Methods
Medical Samples
RNA Transcriptome Sequencing
GO Enrichment Analysis
Analysis of CIBERSORT Deconvolution
Renal Cell Carcinoma Tissue Reverse Transcription PCR
Real-Time Quantitative Reverse Transcription PCR
Cell Culture
siRNA Transfection
CCK-8 Assay
Protein Extraction and Western Blot
Statistical Analysis
Ethical Approval
Results
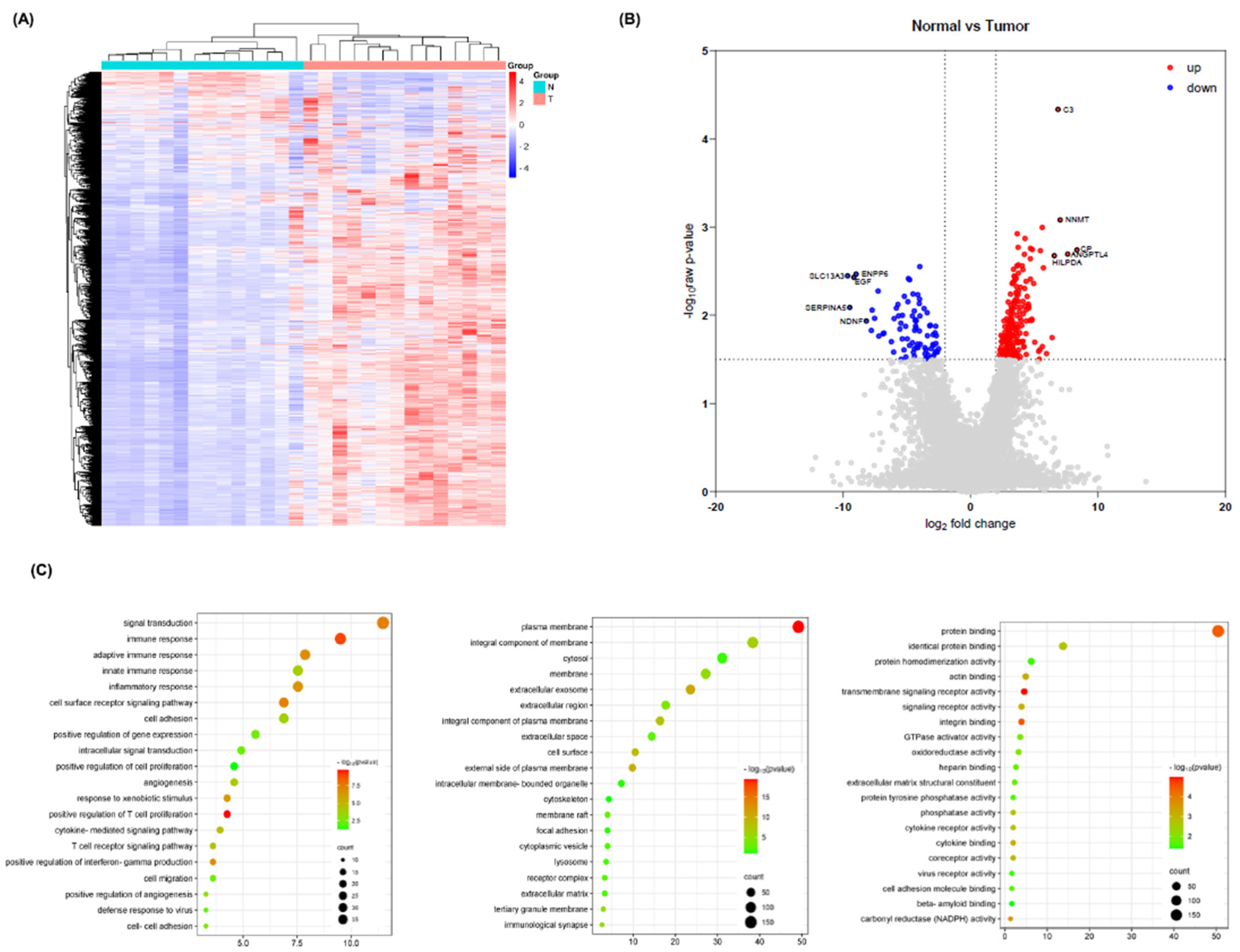
Identification of Common Differentially Expressed Genes
DAVID Enrichment Analysis for Common Differentially Expressed Genes
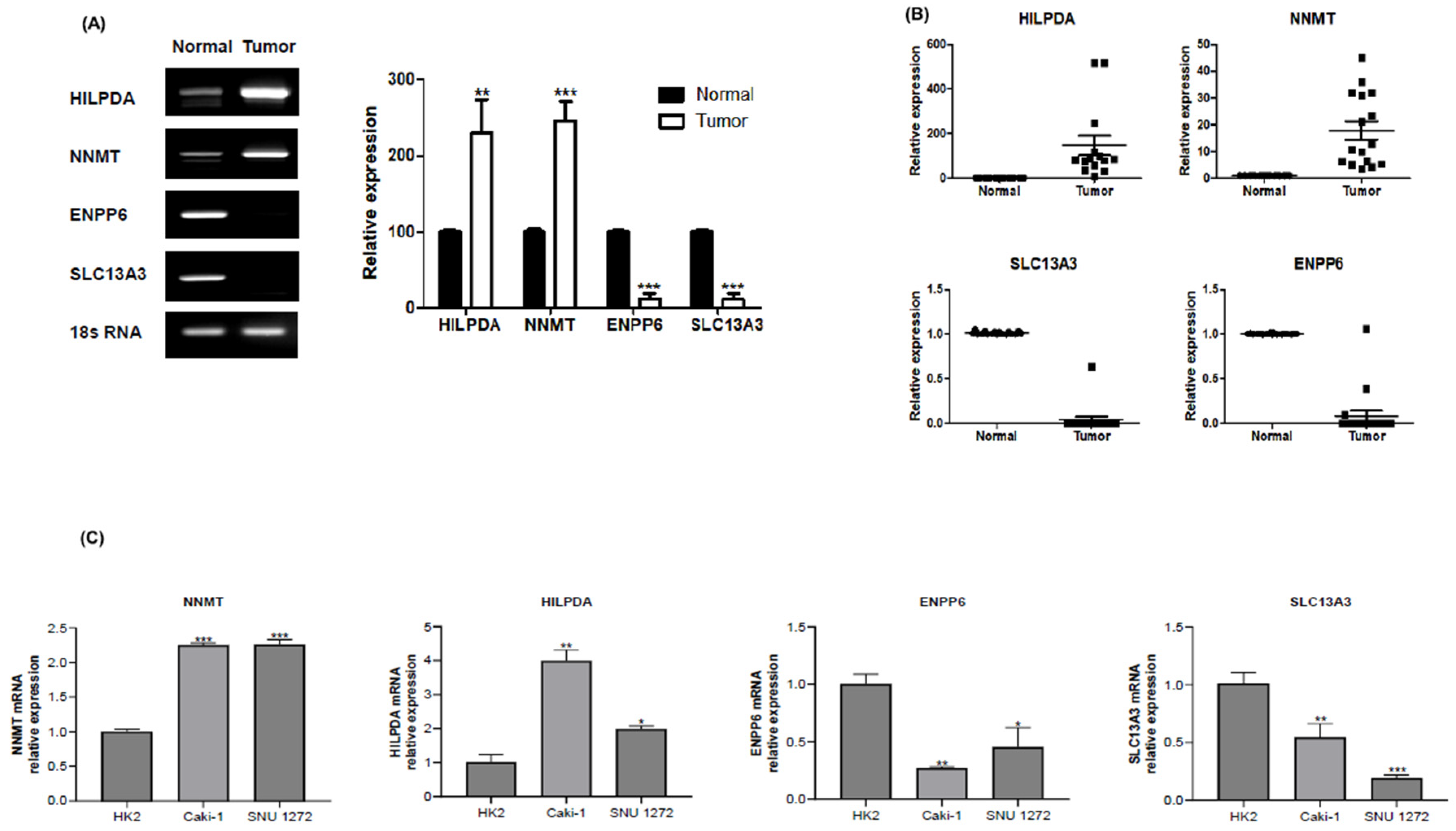
Gene Expression Alterations in Renal Carcinoma: A qPCR Validation Study
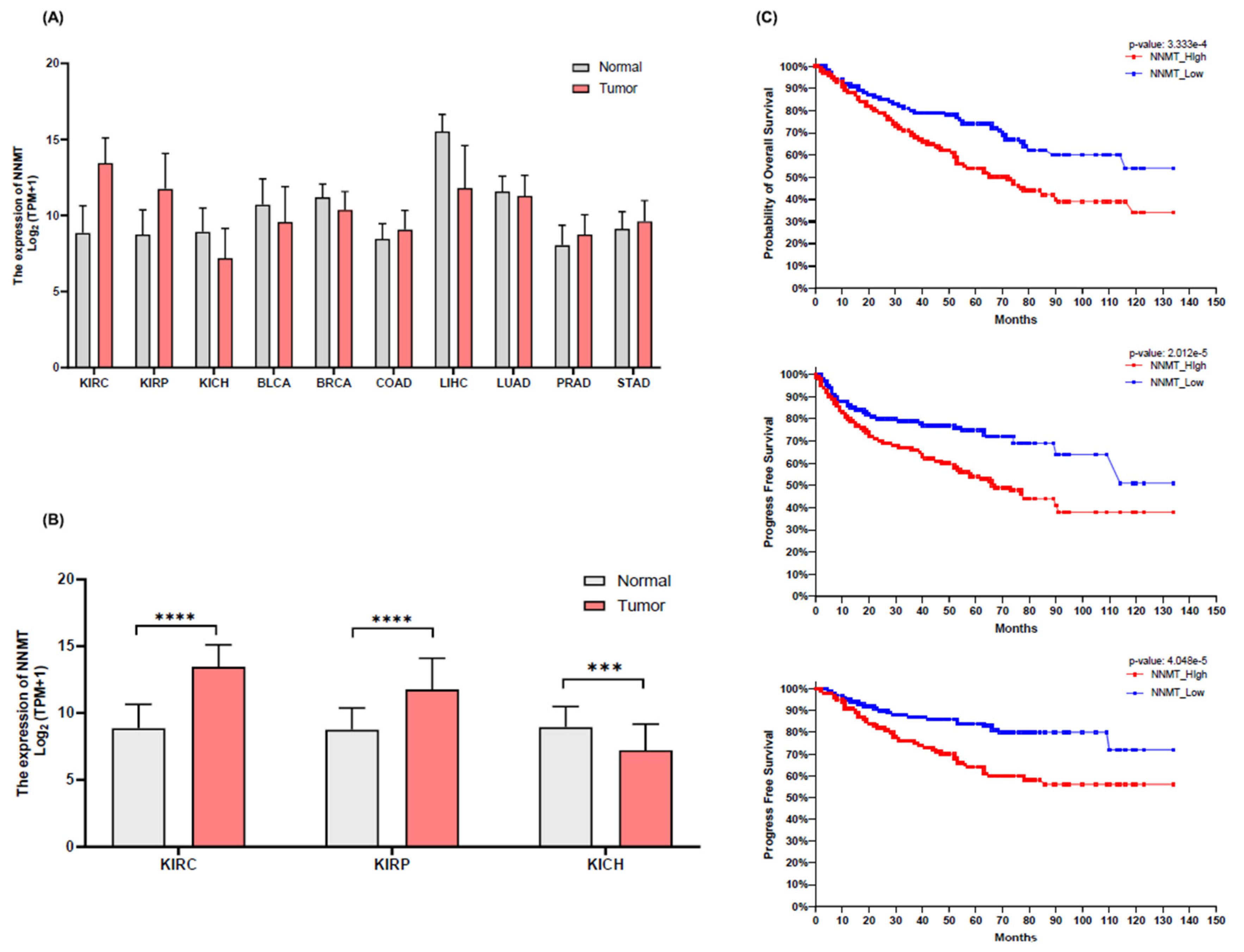
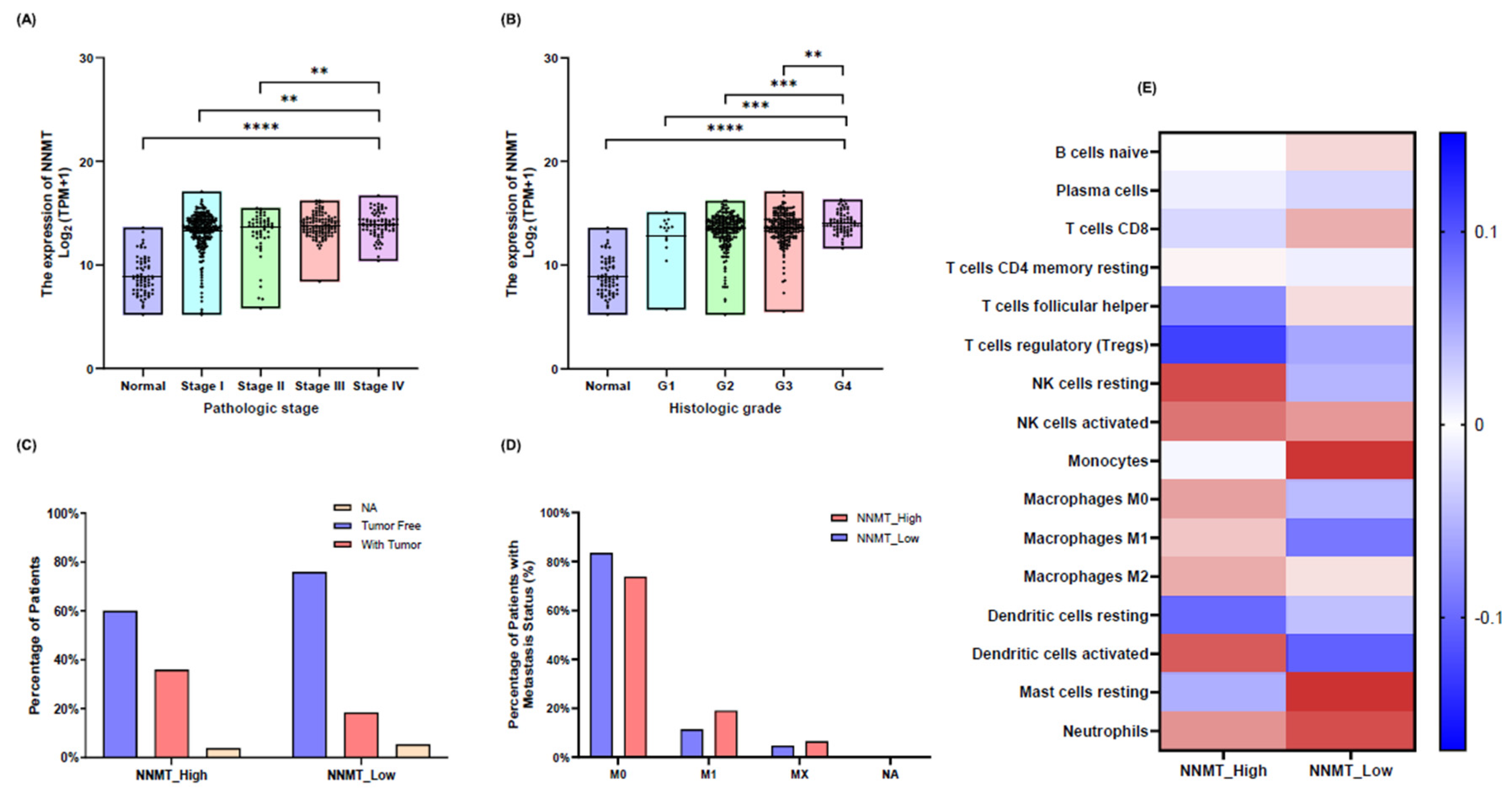
NNMT Expression and Its Correlation with Patient Survival in Renal Cell Carcinoma
NNMT Expression and Its Impact on Clinical Pathological Factors in KIRC
Immune Cell Infiltration and NNMT Expression in KIRC
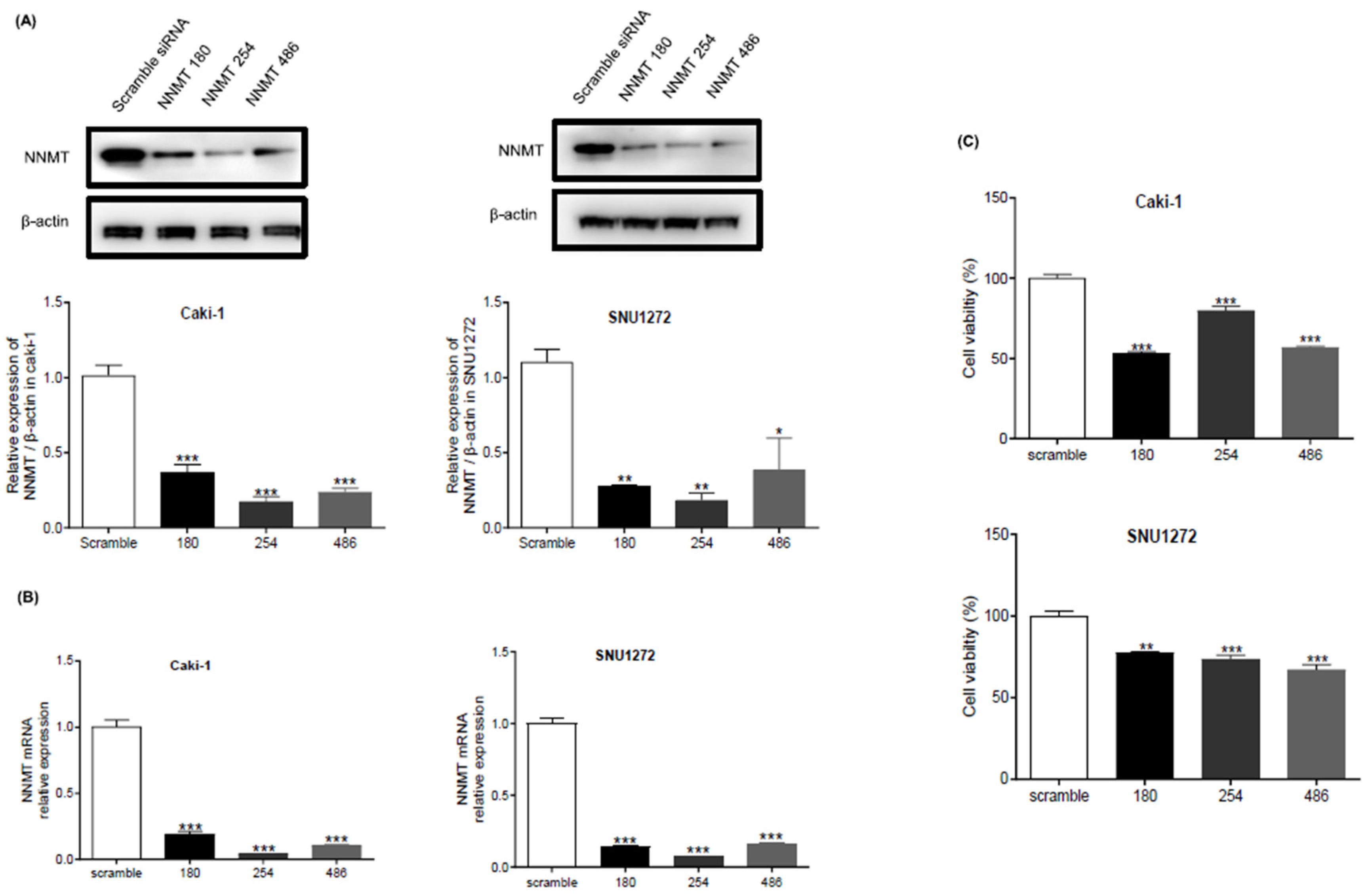
NNMT Knockdown Suppresses ccRCC Cell Proliferation
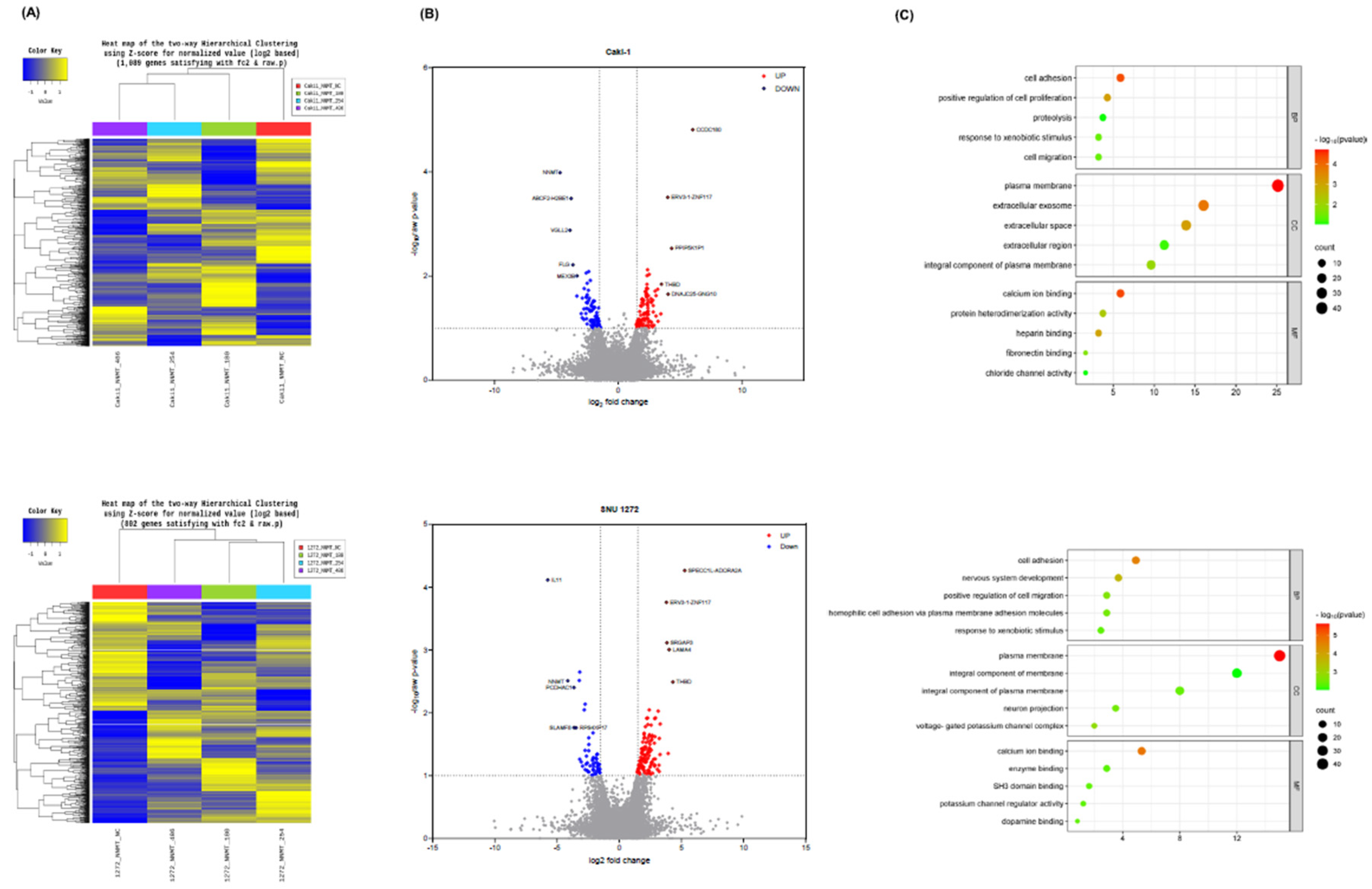
NNMT Knockdown in ccRCC Cells Alters Gene Expression Profiles
Discussion
Conclusion
Author Contributions
Funding Information
Data availability
Acknowledgments
Competing interests
References
- Padala, S.A., et al. Epidemiology of renal cell carcinoma. World J. Oncol. 2020, 11, 79. [CrossRef] [PubMed]
- Hsieh, J.J., et al. Renal cell carcinoma. Nat. Rev. Dis. Primers 2017, 3, 1–19.
- Bahadoram, S., et al. Renal cell carcinoma: an overview of the epidemiology, diagnosis, and treatment. G Ital. Nefrol. 2022, 39, 1.
- Cairns, P. Renal cell carcinoma. Cancer Biomark. 2011, 9, 461–473. [Google Scholar] [CrossRef]
- Cohen, H.T.; McGovern, F.J. Renal-cell carcinoma. New Engl. J. Med. 2005, 353, 2477–2490. [Google Scholar] [CrossRef]
- Porta, C., et al. The adjuvant treatment of kidney cancer: a multidisciplinary outlook. Nat. Rev. Nephrol. 2019, 15, 423–433. [CrossRef]
- Krabbe, L.-M., et al. Surgical management of renal cell carcinoma. In Seminars in Interventional Radiology 2014.
- Antonelli, A., et al. Surgical treatment of atypical metastasis from renal cell carcinoma (RCC). BJU Int. 2012, 110, E559–E563.
- Linehan, W.M. Genetic basis of kidney cancer: role of genomics for the development of disease-based therapeutics. Genome Res. 2012, 22, 2089–2100. [Google Scholar] [CrossRef]
- Frew, I.J.; Moch, H. A clearer view of the molecular complexity of clear cell renal cell carcinoma. Annu. Rev. Pathol. : Mech. Dis. 2015, 10, 263–289. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. et al. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y., et al., Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 2013, 45, 860–867. [CrossRef] [PubMed]
- Grignon, D.J.; Che, M. Clear cell renal cell carcinoma. Clin. Lab. Med. 2005, 25, 305–316. [Google Scholar] [CrossRef]
- Schiliro, C.; Firestein, B.L. Mechanisms of metabolic reprogramming in cancer cells supporting enhanced growth and proliferation. Cells 2021, 10, 1056. [Google Scholar] [CrossRef]
- Wei, Q., et al. Metabolic rewiring in the promotion of cancer metastasis: mechanisms and therapeutic implications. Oncogene 2020, 39, 6139–6156. [CrossRef]
- Bergers, G.; Fendt, S.-M. The metabolism of cancer cells during metastasis. Nat. Rev. Cancer 2021, 21, 162–180. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.; Kim, S.S.; Lee, J. Cancer cell metabolism: implications for therapeutic targets. Exp. Mol. Med. 2013, 45, e45–e45. [Google Scholar] [CrossRef]
- Martínez-Reyes, I.; Chandel, N.S. Cancer metabolism: looking forward. Nat. Rev. Cancer 2021, 21, 669–680. [Google Scholar] [CrossRef]
- Furuta, E., et al. Metabolic genes in cancer: their roles in tumor progression and clinical implications. Biochim. Et Biophys. Acta (BBA)-Rev. Cancer 2010, 1805, 141–152. [CrossRef]
- Kim, D.S., et al. Scale-up evaluation of a composite tumor marker assay for the early detection of renal cell carcinoma. Diagnostics 2020, 10, 750. [CrossRef]
- Kim, D.S., et al. Panel of candidate biomarkers for renal cell carcinoma. J. Proteome Res. 2010, 9, 3710–3719. [CrossRef] [PubMed]
- Shin, J.H., et al. NNMT depletion contributes to liver cancer cell survival by enhancing autophagy under nutrient starvation. Oncogenesis 2018, 7, 58. [CrossRef]
- Eckert, M.A., et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature 2019, 569, 723–728. [CrossRef] [PubMed]
- Ramsden, D.B., et al. Nicotinamide N-methyltransferase in health and cancer. Int. J. Tryptophan Res. 2017, 10, 1178646917691739. [CrossRef] [PubMed]
- Roberti, A.; Fernández, A.F.; Fraga, M.F. Nicotinamide N-methyltransferase: At the crossroads between cellular metabolism and epigenetic regulation. Mol. Metab. 2021, 45, 101165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J., et al. Nicotinamide N-methyltransferase protein expression in renal cell cancer. J. Zhejiang Univ. Sci. B 2010, 11, 136–143. [CrossRef]
- Holstein, S., et al. Nicotinamide N-methyltransferase and its precursor substrate methionine directly and indirectly control malignant metabolism during progression of renal cell carcinoma. Anticancer. Res. 2019, 39, 5427–5436. [CrossRef] [PubMed]
- Campagna, R., et al. The Utility of Nicotinamide N-methyltransferase as a potential biomarker to predict the oncological outcomes for urological cancers: an update. Biomolecules 2021, 11, 1214.
- Gupta, S.; Kanwar, S.S. Biomarkers in renal cell carcinoma and their targeted therapies: a review. Explor. Target. Anti-Tumor Ther. 2023, 4, 941. [Google Scholar] [CrossRef]
- Ricketts, C.J., et al. The cancer genome atlas comprehensive molecular characterization of renal cell carcinoma. Cell Rep. 2018, 23, 313–326. [CrossRef]
- Linehan, W.M.; Ricketts, C.J. The Cancer Genome Atlas of renal cell carcinoma: findings and clinical implications. Nat. Rev. Urol. 2019, 16, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. Review The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol./Współczesna Onkol. 2015, 2015, 68–77. [Google Scholar] [CrossRef]
- Dennis, G., et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003, 4, 1–11. [CrossRef]
- Steen, C.B., et al. Profiling cell type abundance and expression in bulk tissues with CIBERSORTx. Stem Cell Transcr. Netw. Methods Protoc. 2020, 135–157.
- Pontén, F., et al. The Human Protein Atlas as a proteomic resource for biomarker discovery. J. Intern. Med. 2011, 270, 428–446. [CrossRef]
- Pontén, F.; Jirström, K.; Uhlen, M. The Human Protein Atlas—a tool for pathology. J. Pathol. A J. Pathol. Soc. Great Br. Irel. 2008, 216, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Thul, P.J.; Lindskog, C. The human protein atlas: a spatial map of the human proteome. Protein Sci. 2018, 27, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Olshan, A.F., et al. Racial difference in histologic subtype of renal cell carcinoma. Cancer Med. 2013, 2, 744–749. [CrossRef]
- Muglia, V.F.; Prando, A. , Renal cell carcinoma: histological classification and correlation with imaging findings. Radiol. Bras. 2015, 48, 166–174. [Google Scholar] [CrossRef]
- Low, G., et al. Review of renal cell carcinoma and its common subtypes in radiology. World J. Radiol. 2016, 8, 484. [CrossRef]
| NO. | Patient ID | Histologic Subtype | Tumor Size |
Nucleolar grade (WHO/ISUP) | Tumor Necrosis | Pathologic stage (AJCC 2017): |
|---|---|---|---|---|---|---|
| #1 | KC-90 | clear cell type | 3.5x3.0x3.0cm | 3/4 | 0% | pT1aNx |
| #2 | KC-91 | clear cell type | 3.2x2.4x2.2cm | 2/4 | 10% | pT1aN0 |
| #3 | KC-92 | clear cell type | 1.8x1.6x1.5cm | 3/4 | 0% | pT1aNx |
| #4 | KC-93 | clear cell type | 1.6x1.5x1.2cm | 2/4 | 10% | pT1aN0 |
| #5 | KC-96 | clear cell type | 3.5x2.5x2.4cm | 3/4 | 20% | pT1aNx |
| #6 | KC-97 | clear cell type | 2.5x1.7x1.5cm | 2/4 | 40% | pT1aNx |
| #7 | KC-99 | clear cell type | 3.6x2.8x2.5cm | 3/4 | 30% | pT1aNx |
| #8 | KC-100 | clear cell type | 1.7x1.7x0.8cm | 2/4 | 0% | pT1aNx |
| #9 | KC-102 | clear cell type | 1.8x1.6x1.6cm | 2/4 | 0% | pT1aNx |
| #10 | KC-104 | clear cell type | 3.2x2.7x2.4cm | 3/4 | 5% | pT1aNx |
| #11 | KC-105 | clear cell type | 2.0x1.8x1.5cm | 3/4 | 20% | pT1aNx |
| #12 | KC-106 | clear cell type | 1.6x1.5x1.0cm | 2/4 | 0% | pT1aNx |
| #13 | KC-109 | clear cell type | 4.2x3.3x2.0cm | 3/4 | 10% | pT1bNx |
| #14 | KC-110 | clear cell type | 2.2x1.2x1.2cm | 2/4 | 20% | pT1aNx |
| Genes | Sequence (5′-3′) | Annealing Temperature (°C) | |
|---|---|---|---|
| 18 S | ForwardReverse | 5′-CTGCCCTATCAACTTTCGATGGTA-3′ 5′- CCGTTTCTCAGGCTCCCTCTC-3′ |
54 °C |
| NNMT | ForwardReverse | 5′-CCATCTGTTCTAAAAGAAGGGC -3′ 5′-GGAGGTGAAGCCTGATTCCA -3′ |
58 °C |
| HILPDA | ForwardReverse | 5′-ACCGACTTTCCTCCGGACTC -3′ 5′-TGCAGAGAAACAGAGCTGCC -3′ |
58 °C |
| ENPP6 | ForwardReverse | 5′-GTGGGTCACTCTGACCAAGG -3′ 5′-GAGCATCGCTGACTGCATTG -3′ |
57 °C |
| SLC13A3 | ForwardReverse | 5′-CTGCTGGTGCTGCTGTTCAC -3′ 5′-CTGGGGTCCTTTCGAACCTC -3′ |
56 °C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





