Submitted:
30 September 2024
Posted:
01 October 2024
You are already at the latest version
Abstract
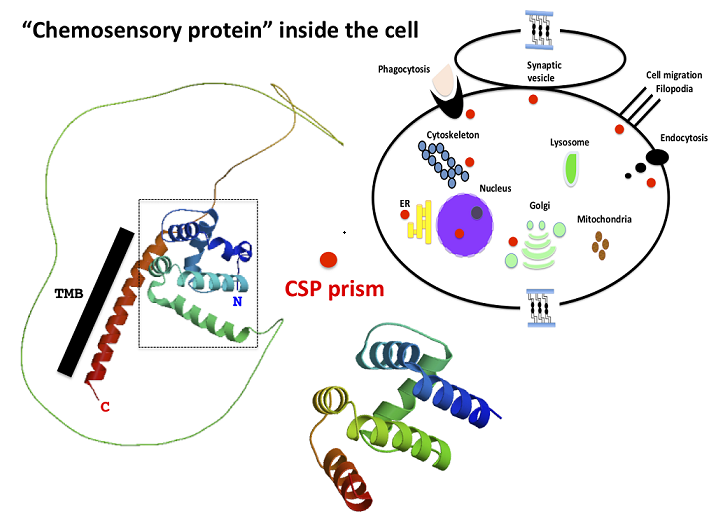
Keywords:
1. Introduction
2. History of the Discovery of “CSPs” and Their Uncertain or Pleiotropic Roles in Insects
3. Broad Survey and Analysis of CSP Sequences in Insects and Hexapods
4. The Evolutionary Trajectory of CSPs
5. Existence of “CSPs” Outside Insects
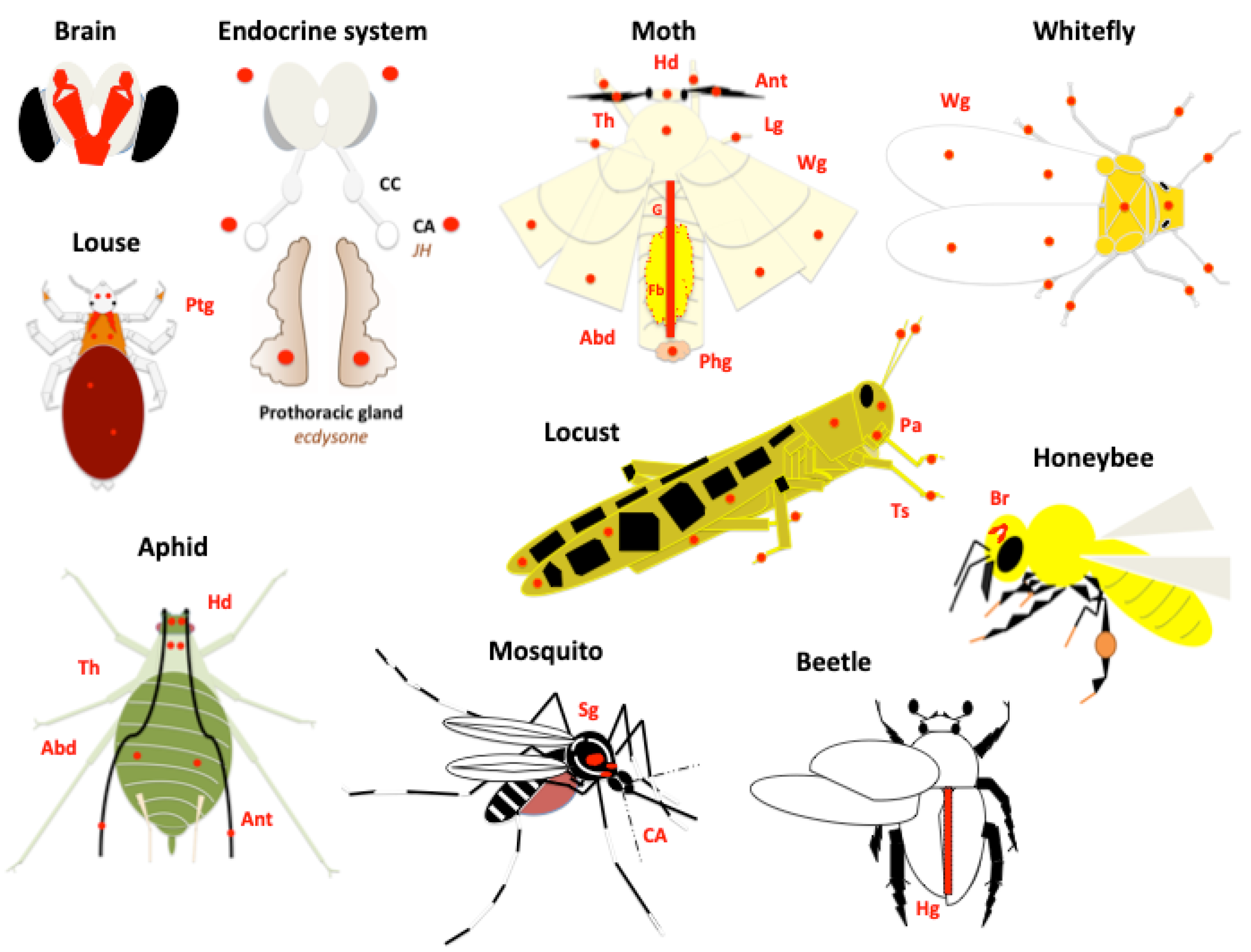
6. Expression Profiling in Organisms, Development, and Tissues
7. Putative Function Outside the Chemosensory/Binding Aspect
8. The Myzus Persicae Mp10 Story Along with Additional Immune-Related Roles
9. CSPs’ Intracellular Functions (Gene Regulation to Stress Response)
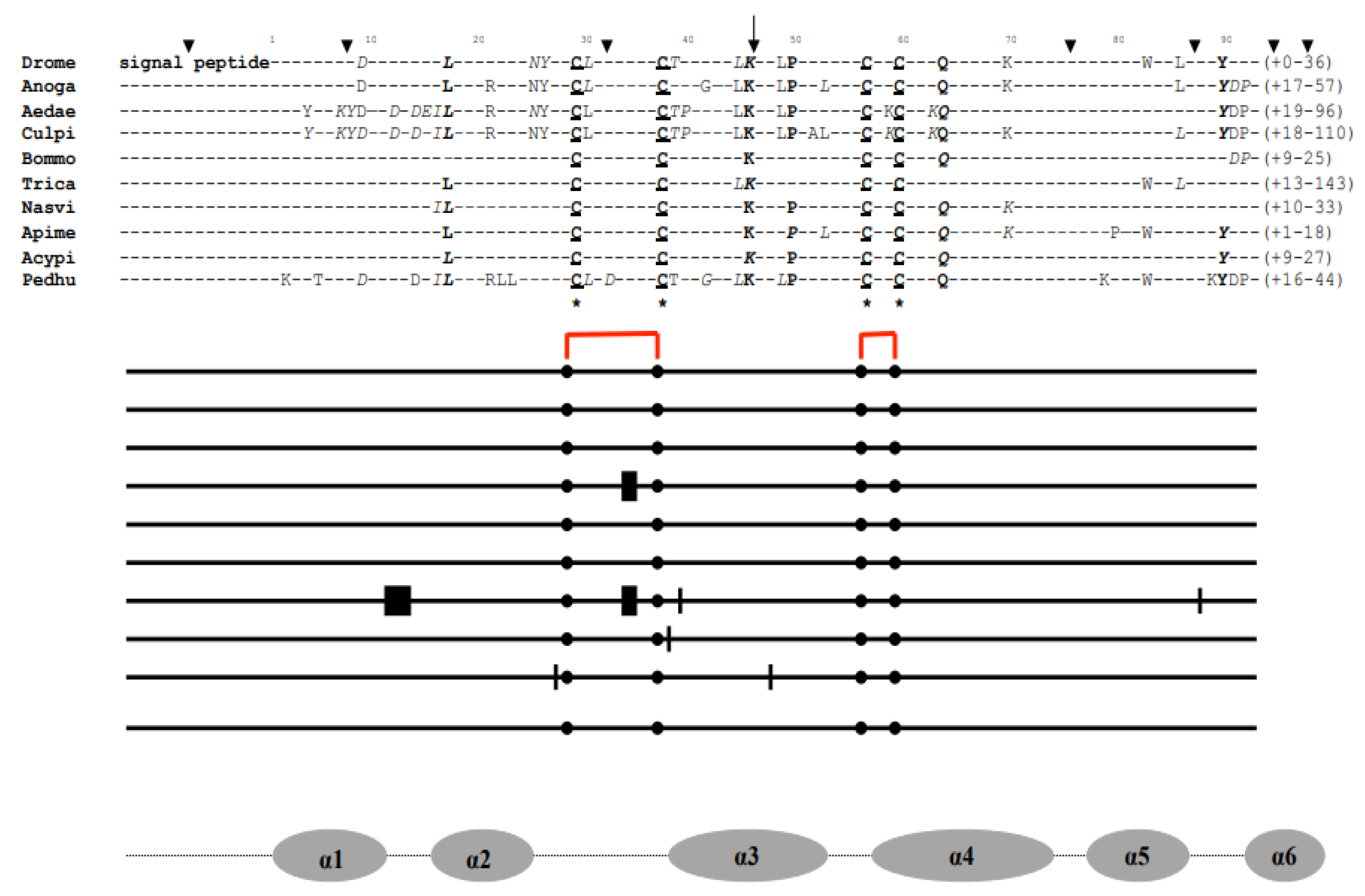
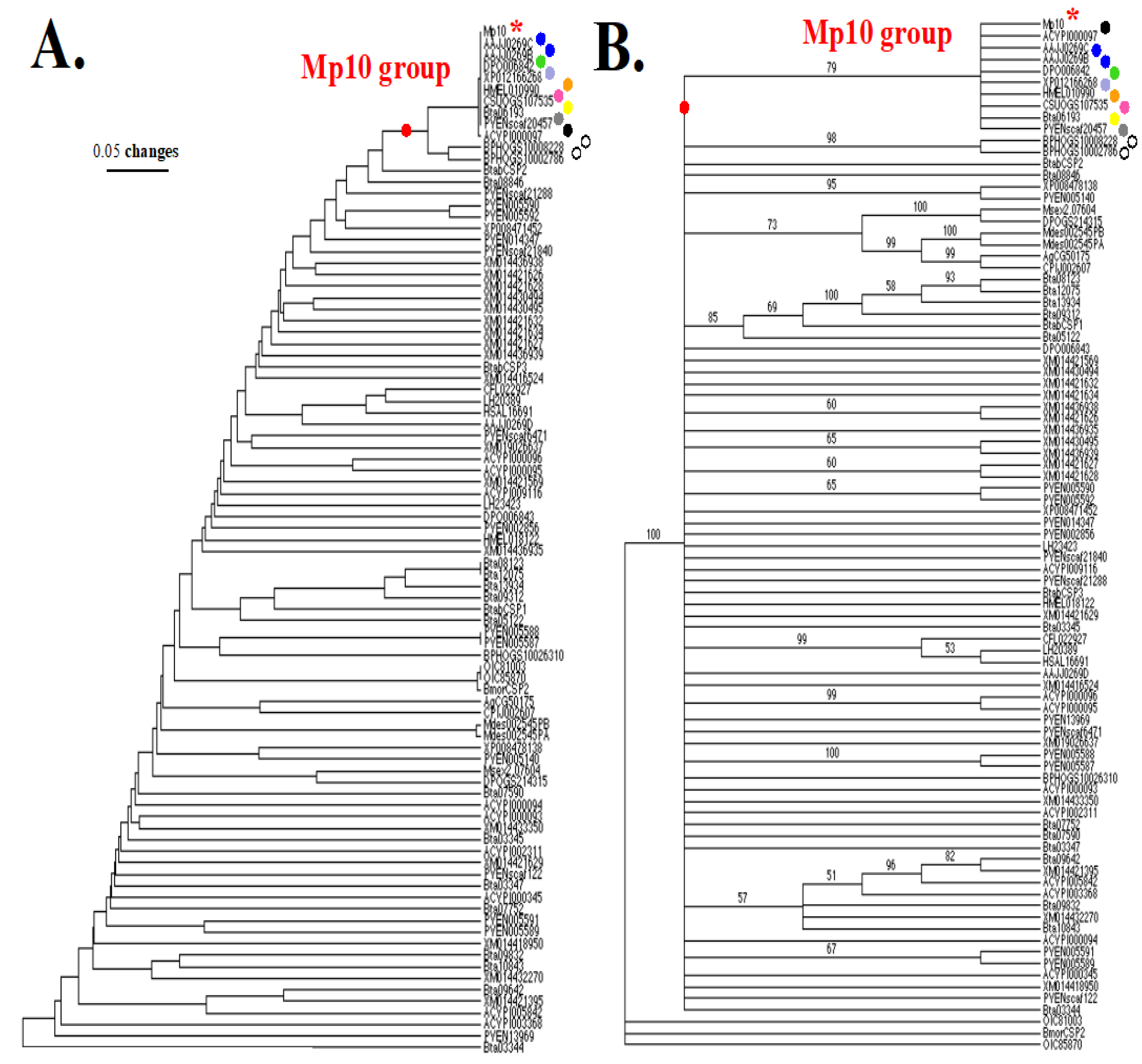
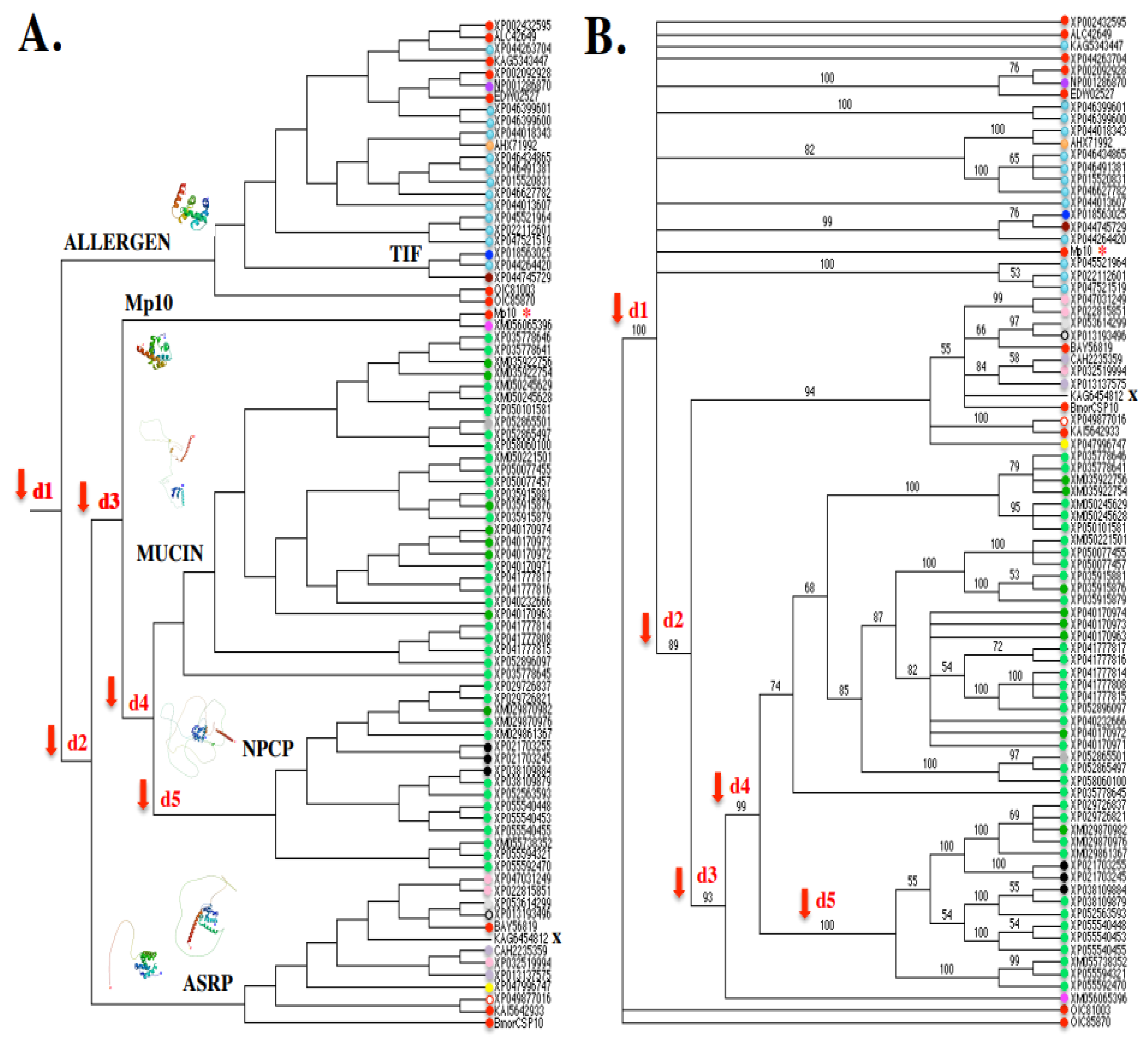
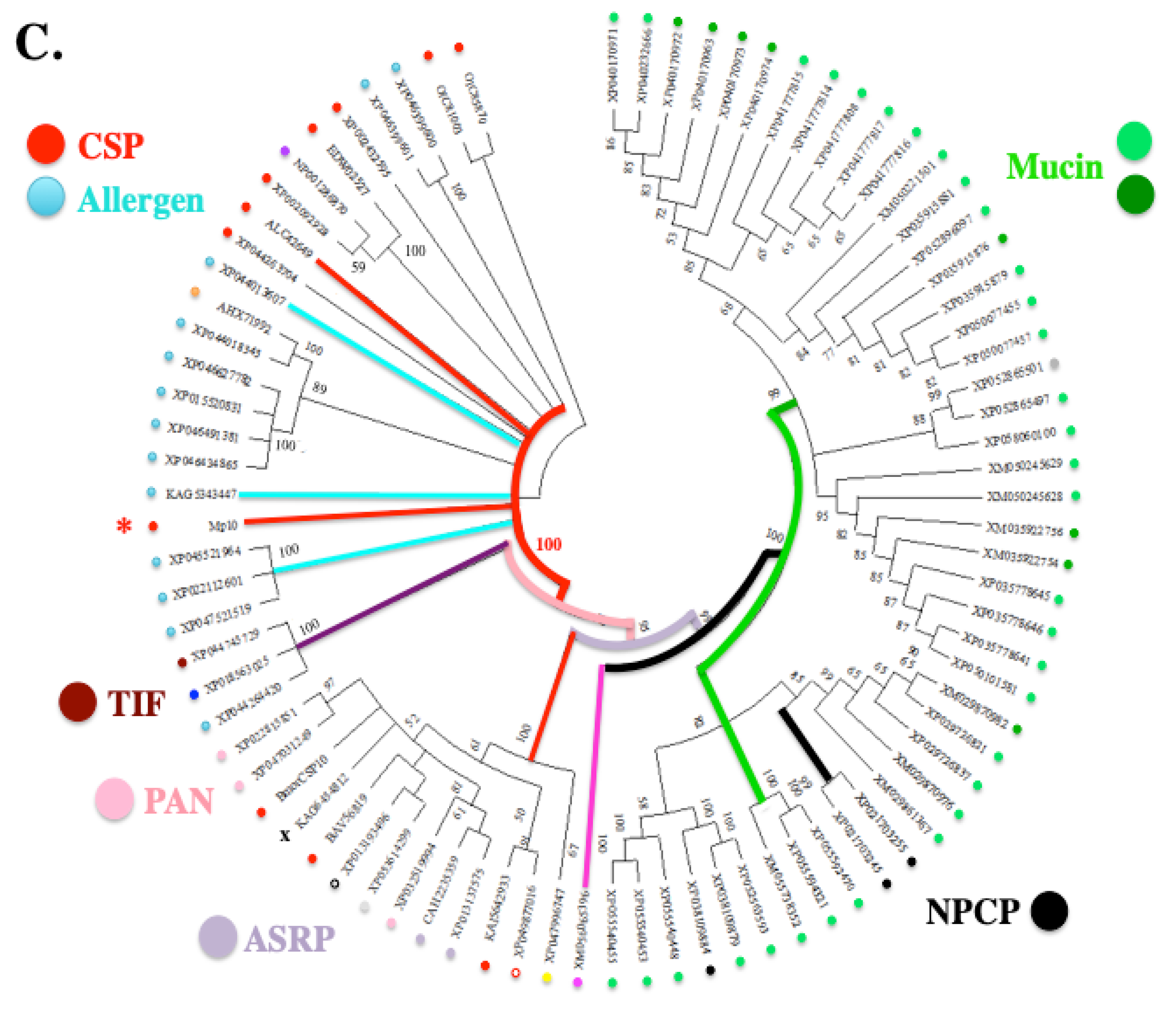
9.1. Evolutionary Evidence Derived from Amino Acid Sequence Phylogenetic Analysis
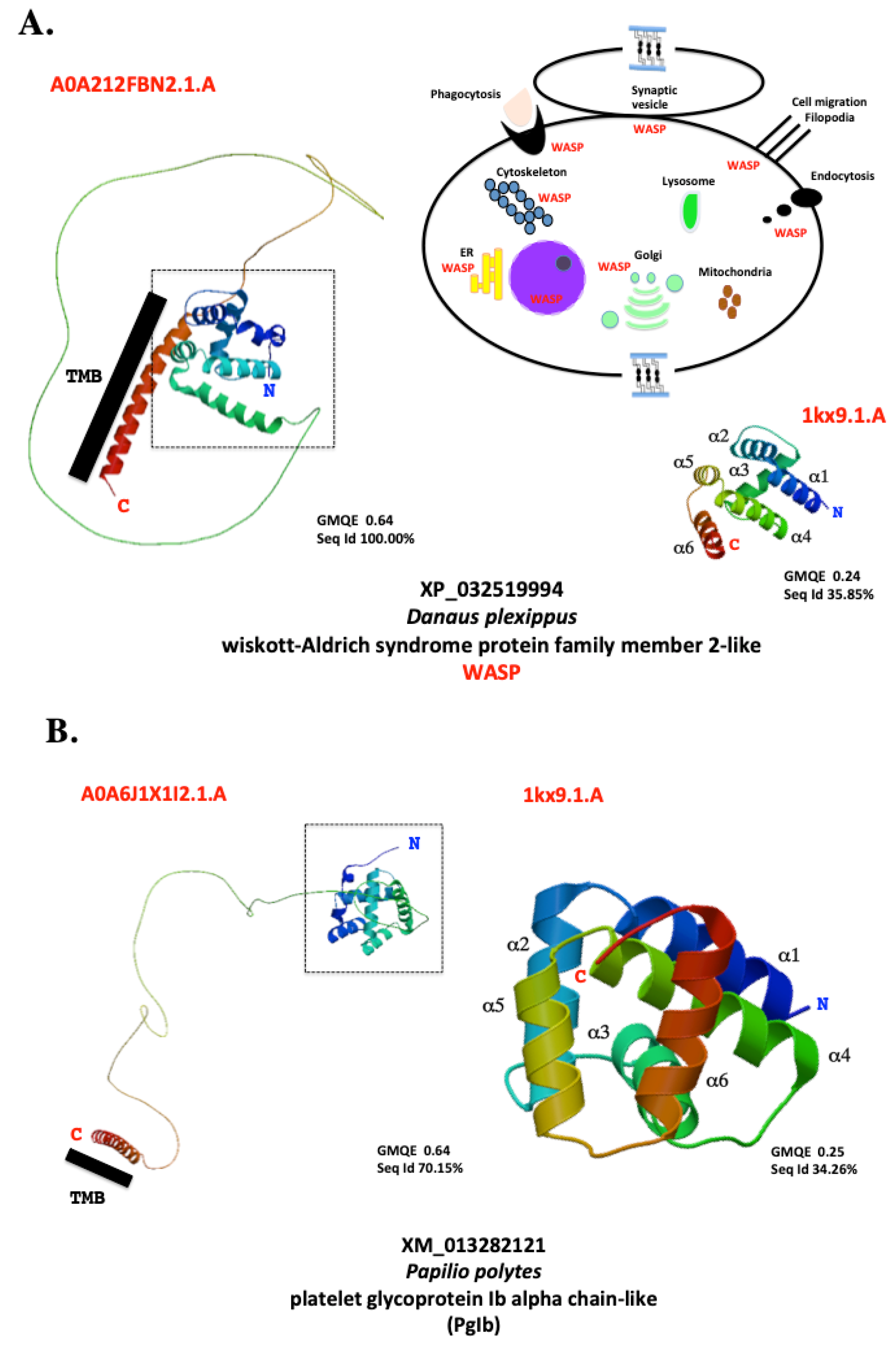
9.2. Molecular Evidence Derived from Amino Acid Sequence Modeling Analysis
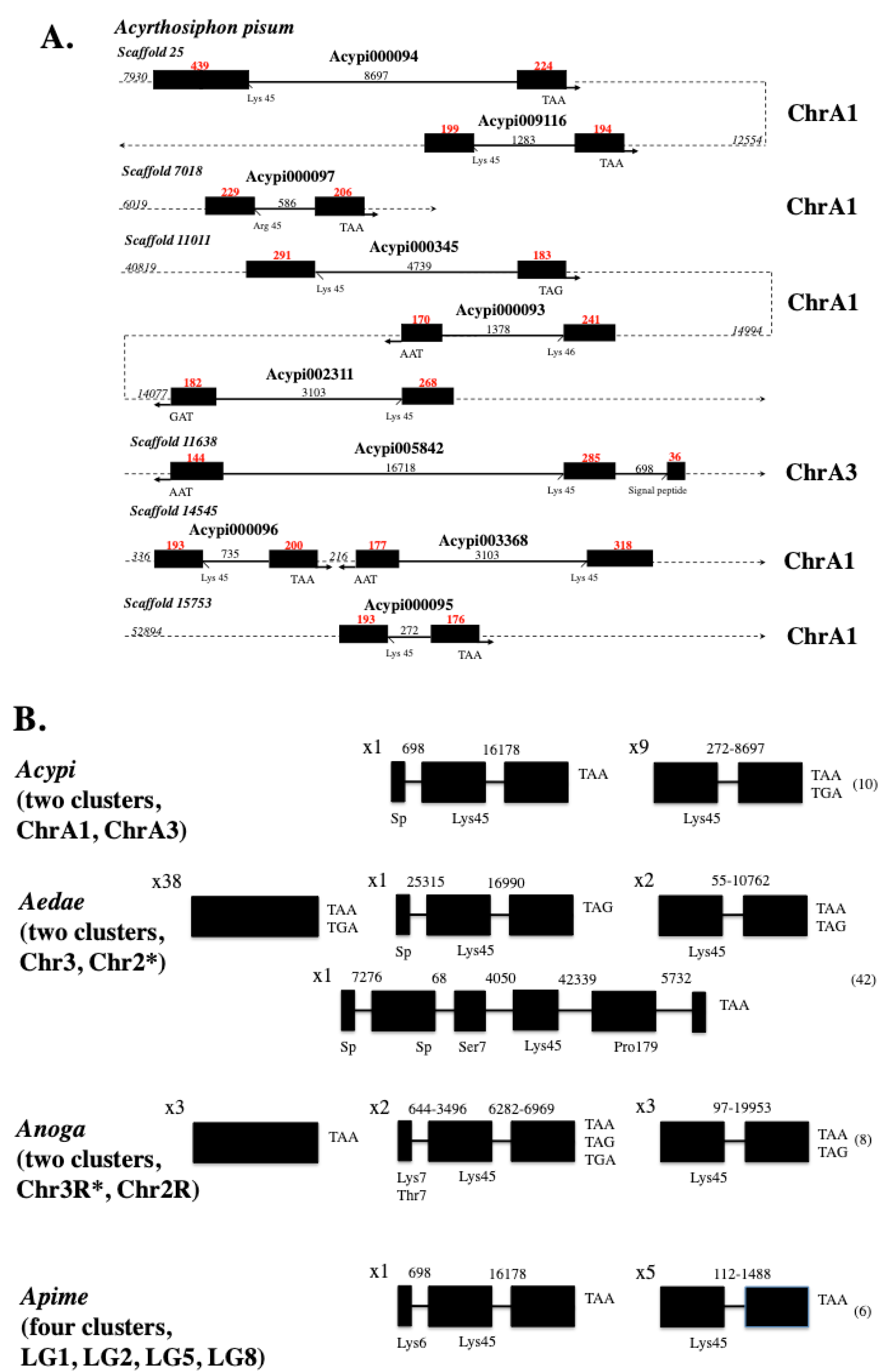
9.3. Cellular Evidence Derived from Location, Size, Structure, and Possible Expression in Viruses and Microbes
10. To design “CSP”, New Terminology Is Needed: Structure or Function?
11. Concluding Remark
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 4CSP | 4 Cysteines Soluble Proteins |
| AA | Arachidonic acid |
| AcrR | Regulator of adjacent acrAB efflux genes - functional protein of the transcriptional regulation system that confers bacterial resistance to the antibiotic tetracycline (see TetR and TFTR) - Regulated by FA lipids |
| Acypi |
Acyrthosiphum pisum (pea aphid) |
| Aedae |
Aedes aegypti (dengue yellow fever mosquito) |
| Anoga |
Anopheles gambiae (malaria mosquito) |
| Allergen Tha p 1 | IgE-binding protein (15 kDa) and major allergen of pine processionary caterpillar (Thaumetopoea pityocampa, Thapi) - variant 1.0101 |
| Apime |
Apis mellifera (honey bee) |
| Arp2/3 | Actin related protein 2/3 complex |
| ASRC | Actin skeleton regulatory complex |
| ASRP | Actin skeleton regulatory protein |
| Avd | Accessory variability determinant |
| Bacil | Bacillus |
| B-CSP | Bacterial “Chemosensory protein” |
| BioNJ | Bio (improved version) of the Neighbor Joining algorithm based on simple model of sequence data |
| BLASTn | Nucleotide BLAST |
| BLLF1 | Epstein-Barr virus envelope glycoprotein encoded by BLLF1 gene |
| Bommo |
Bombyx mori (silkworm moth) |
| BemtaCSP1 |
Bemisia tabaci “Chemosensory Protein”-1 (LA-binding protein) |
| CA | Corpora allata |
| CBP | Chemical Binding Proteins |
| Che | Chemotaxis proteins (histidine kinases) |
| CheA, CheB | Chemosensory proteins-A, Chemosensory proteins-B (Drosophila) |
| Cocse |
Coccinella septempunctata (seven-spot ladybird) |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| CRLPB | Chemical sense-related lipophilic ligand binding proteins (Drosophila) |
| CSP | Chemosensory protein |
| Culpi |
Culex pipiens (common house mosquito) |
| CWA | Cell wall anchored |
| CWP | Cell wall protein |
| CYP | Cytochrome P450 |
| Cys | Cysteine |
| DAN4 | Cell wall mannoprotein expressed under delayed anaerobic conditions (Saccharomyces) |
| Danpl |
Danaus plexippus (monarch butterfly) |
| Dd-Rp | DNA-dependent RNA polymerase |
| DE | Degradative enzyme |
| DGR | Diversity generating retroelement |
| DNA-BP | Deoxyribonucleic acid-binding protein |
| DNA-RP | Deoxyribonucleic acid-regulatory protein |
| DP | Diphosphate |
| Drome | Drosophila melanogaster (fruit fly) |
| Droso |
Drosophila species |
| EA | Elaidic acid |
| Ebsp | Ejaculatory bulb-specific protein |
| Epiba |
Episyrphus balteatus (marmalade hoverfly) |
| ER | Endoplasmic reticulum |
| EST | Expressed Sequence Tag |
| Eupco |
Eupeodes corollae (migrant overfly) |
| FA | Fatty acid |
| GC | Guanine-cytosine content |
| GM2-AP | Ganglioside GM2 -activator protein |
| GMQE | Global Model Quality Estimate |
| GR | Gustatory receptor |
| GTP | Guanosine triphosphate |
| Halha |
Halyomorpha halys (brown marmorated stink bug) |
| HexA | Hexosaminidase A |
| HSV | Herpesvirus |
| IgE-BP | Immunoglobulin E-binding protein |
| Iphpo |
Iphiclides podalirius (scarce swallowtail) |
| Jg5928 | major outer envelope protein |
| JH | Juvenile hormone |
| JHRP | Juvenile hormone-related protein |
| HSV | Herpesvirus |
| Locmi |
Locusta migratoria (migratory locust) |
| LRR | Leucine-rich repeat protein complex |
| M | Mucin |
| Manse |
Manduca sexta (tobacco hornworm, sphinx moth) |
| MCP | Methyl-accepting chemotaxis protein |
| ML | MD-2-related lipid-recognition domain |
| MP | Maximum parsimony |
| Mp10 | Myzpe p10-like protein |
| Myzpe |
Myzus persicae (green peach aphid) |
| NIESMuc |
Nilaparvata lugens eggshell-related mucin |
| Nillu |
Nilaparvata lugens (brown planthopper, BPH) |
| NNK | Nuclear nucleoside kinase |
| NDPK | Nucleoside diphosphate kinase |
| NPCP | Nuclear pore complex protein |
| Nasvi |
Nasonia vitripennis (parasitoid jewel wasp) |
| OBP | Odorant-binding protein |
| OR | Olfactory receptor |
| ORCO | Conserved Odorant Receptor type (Co-receptor) |
| OS-D | Olfactory sensilla-type D protein |
| OXPHOS | Oxidative phosphorylation |
| P | Phosphate |
| P10 | Peram 10 kDa protein from regenerating legs |
| Pacve |
Pachypsylla venusta (hackberry petiole gall psyllid) |
| PAN | Hexameric ATPase complex |
| PAN-1 | Protein encoded by the pan-1 gene in the nematode Caenorhabditis elegans
|
| Pappo |
Papilio polytes (common Mormon) |
| PAUP | Phylogenetic analysis using parsimony |
| Pedhu |
Pediculus humanus humanus (human body louse) |
| Peram |
Periplaneta americana (American cockroach) |
| PBP | Pheromone-binding protein |
| PgIb | Platelet glycoprotein Ib alpha chain-like |
| Phk | Pherokine (hemolymph protein) |
| PKC | Protein kinase C |
| PLC | Phospholipase C |
| Pol | Polymerase |
| Ras | Family of GTPases derived from rat sarcoma virus |
| Rho | Family of GTPases, family of small (~21 kDa) signaling G proteins, subfamily of the Ras superfamily |
| RhoGAP | Rho GTPase-activating protein, regulator of the Rho-related protein family, crucial in many cellular processes, motility, contractility, growth, differentiation, and development |
| RickA |
Rickettsia (conorii) surface protein A (activator of Arp2/3) |
| RNA-BP | Ribonucleic acid-binding protein |
| RNAi | RNA interference |
| SamkC | Serine/Threonine-protein kinase (from Social amoeba) |
| SA | Stearic acid |
| SAP | Sensory appendage protein |
| Sec31 | Protein transport protein encoded by the SEC31A gene (human) |
| SF | Splicing factor |
| SRP | Signal recognition particle |
| TERT | Telomerase reverse transcriptase |
| TetR | Tetracycline repressor |
| TFTR | TetR-family transcriptional regulator |
| TIF | Transcription initiation factor |
| TP | Triphosphate |
| Trica |
Tribolium castaneum (red flour beetle) |
| TSSC1 | Tumor suppressing subtransferable candidate 1 |
| UL36 | Large tegument protein deneddylase encoded by UL36 gene (herpesviridae) |
| UPGMA | Unweighted pair group method with arithmetic mean |
| Uralo |
Uranotaenia lowii (pale-footed Uranotaenia) |
| WAS/WASL | Wiskott-Aldrich Syndrome/ Wiskott-Aldrich Syndrome-like protein |
| WASP | Wiskott-Aldrich Syndrome protein |
| XRE (or Xre) | Xenobiotic response element family of DNA-binding transcriptional regulators |
| YhbY | RNA-binding protein (folded like TIF) encoded by YhbY gene (Escherichia coli) |
| YLPM1 | YLP motif containing 1 encoded by YLPM1 gene (human) |
References
- Hoffmann, K.H.; Meyerinng-Vos, M.; Lorenz, M.W. Allatostatins and allatotropins: Is the regulation of corpora allata activity their primary function? Eur. J. Entomol. 1999, 96, 255–266. [Google Scholar]
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nature, 1981, 293, 161–163. [Google Scholar] [CrossRef] [PubMed]
- Picimbon, J.F.; Leal, W.S. Olfactory soluble proteins of cockroaches. Insect Biochem. Mol. Biol. 1999, 30, 973–978. [Google Scholar]
- Angeli, S.; Ceron, F.; Scaloni, A.; Monti, M.; Monteforti, G.; Minnocci, A.; Petacchi, R.; Pelosi, P. Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria. Eur. J. Biochem. 1999, 262, 745–754. [Google Scholar] [CrossRef]
- Picimbon, J.F. Biochemistry and evolution of CSP and OBP proteins. In Insect Pheromone Biochemistry and Molecular Biology, The Biosynthesis and Detection of Pheromones and Plant Volatiles; Blomquist, G.J., Vogt, R.G., Eds; Elsevier Academic Press, London & San Diego, UK & USA, 2003; pp. 539-566.
- Lartigue, A.; Campanacci, V.; Roussel, A.; Larsson, A.M.; Jones, T.A.; Tegoni, M.; Cambillau, C. X-ray structure and ligand binding study of a moth chemosensory protein. J. Biol. Chem. 2002, 277, 32094–32098. [Google Scholar] [CrossRef]
- Jansen, S.; Zídek, L.; Löfstedt, C.; Picimbon, J.F.; Sklenar, V. 1H, 13C, and 15N resonance assignment of Bombyx mori chemosensory protein 1 (BmorCSP1). J. Biomol. NMR 2006, 36, 47. [Google Scholar] [CrossRef]
- Jansen, S.; Chmelik, J.; Zídek, L.; Padrta, P.; Novak, P.; Zdrahal, Z.; Picimbon, J.F.; Löfstedt, C.; Sklenar, V. Structure of Bombyx mori Chemosensory Protein 1 in solution. Arch. Insect Biochem. Physiol. 2007, 66, 135–145. [Google Scholar] [CrossRef]
- Tomaselli, S.; Crescenzi, O.; Sanfelice, D.; Ab, E.; Wechsel- berger, R.; Angeli, S.; Scaloni, A.; Boelens, R.; Tancredi, T.; Pelosi, P.; Picone, D. Solution structure of a chemosensory protein from the desert locust Schistocerca gregaria. Biochemistry 2006, 45, 1606–1613. [Google Scholar] [CrossRef]
- Jia, Q.; Zeng, H.; Zhang, J.; Gao, S.; Xiao, N.; Tang, J.; Dong, X.; Xie, W. The crystal structure of the Spodoptera litura Chemosensory Protein CSP8. Insects 2021, 12, 602. [Google Scholar] [CrossRef] [PubMed]
- Campanacci, V.; Lartigue, A.; Hällberg, B.M.; Jones, T.A.; Giuici-Orticoni, M.T.; Tegoni, M.; Cambillau, C. Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding. Proc. Natl. Acad. Sci. USA 2003, 100, 5069–5074. [Google Scholar] [CrossRef]
- Mosbah, A.; Campanacci, V.; Lartigue, A.; Tegoni, M.; Cambillau, C.; Darbon, H. Solution structure of a chemosensory protein from the moth Mamestra brassicae. Biochem. J. 2003, 369, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Picimbon, J.F.; Dietrich, K.; Angeli, S.; Scaloni, A.; Krieger, J.; Breer, H.; Pelosi, P. Purification and molecular cloning of chemosensory proteins from Bombyx mori. Arch. Insect Biochem. Physiol. 2000, 44, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Picimbon, J.F.; Dietrich, K.; Krieger, J.; Breer, H. Identity and expression pattern of chemosensory proteins in Heliothis virescens (Lepidoptera, Noctuidae). Insect Biochem. Mol. Biol. 2001, 31, 1173–1181. [Google Scholar]
- Wanner, K.W.; Isman, M.B.; Feng, Q.; Plettner, E.; Theilmann, D.A. Developmental expression patterns of four chemosensory protein genes from the Eastern spruce budworm, Choristoneura fumiferana. Insect Mol. Biol. 2005, 14, 289–300. [Google Scholar]
- Picimbon, J.F. Chapter three—Bioinformatic, genomic and evolutionary analysis of genes: A case study in Dipteran CSPs. Meth. Enzymol. 2020, 642, 35–79. [Google Scholar]
- Picimbon, J.F.; Dietrich, K.; Breer, H.; Krieger, J. Chemosensory proteins of Locusta migratoria (Orthoptera: Acrididae). Insect Biochem. Mol. Biol. 2000, 30, 233–241. [Google Scholar] [CrossRef]
- Guo, W.; Wang, X.; Ma, Z.; Xue, L.; Han, J.; Yu, D.; Kang, L. CSP and Takeout genes modulate the switch between attraction and repulsion during behavioral phase change in the migratory locust. PLoS Genet. 2011, 7, e1001291. [Google Scholar] [CrossRef]
- Martín-Blázquez, R.; Chen, B.; Kang, L.; Bakkali, M. Evolution, expression and association of the chemosensory protein genes with the outbreak phase of the two main pest locusts. Sci. Rep. 2018, 7, 6653. [Google Scholar] [CrossRef]
- Xuan, N.; Guo, X.; Xie, H.Y.; Lou, Q.N.; Bo, L.X.; Liu, G.X.; Picimbon, J.F. Increased expression of CSP and CYP genes in adult silkworm females exposed to avermectins. Insect Sci. 2015, 22, 203–219. [Google Scholar] [CrossRef]
- Ban, L.; Scaloni, A.; Brandazza, A.; Angeli, S.; Zhang, Y.; Pelosi, P. Chemosensory proteins of Locusta migratoria. Insect Mol. Biol. 2003, 12, 125–134. [Google Scholar] [CrossRef]
- Xuan, N.; Bu, X.; Liu, Y.Y.; Yang, X.; Liu, G.X.; Fan, Z.X.; Bi, Y.P.; Yang, L.Q.; Lu, Q.N.; Rajashekar, B.; Leppik, G.; Kasvandik, S.; Picimbon, J.F. Molecular evidence of RNA editing in the Bombyx chemosensory protein family. PLoS ONE 2014, 9, e86932. [Google Scholar] [CrossRef] [PubMed]
- Picimbon, J.F. Evolution of protein physical structures in insect chemosensory systems. In Olfactory Concepts of Insect Control-Alternative to Insecticides; Picimbon, J.F., Ed; Vol. 2, Springer Nature AG, Cham, Switzerland, 2019; pp. 231-263.
- Khuhro, S.A.; Yan, Q.; Liao, H.; Zhu, G.H.; Sun, J.B.; Dong, S.L. Expression profile and functional characterization suggesting the involvement of three chemosensory proteins in perception of host plant volatiles in Chilo suppressalis (Lepidoptera: Pyralidae). J. Insect Sci. 2018, 18, 6. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Picimbon, J.F.; Ji, S.; Kan, Y.; Chuanling, Q.; Zhou, J.J.; Pelosi, P. Multiple functions of an odorant-binding protein in the mosquito Aedes aegypti. Biochem. Biophys. Res. Commun. 2008, 372, 464–468. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.E.; Delannay, C.; Goindin, D.; Deng, L.; Guan, S.; Lu, X.; Fouque, F.; Vega-Rúa, A.; Picimbon, J.F. Cartography of odor chemicals in the dengue vector mosquito (Aedes aegypti L., Diptera/Culicidae). Sci. Rep. 2019, 9, 8510. [Google Scholar] [CrossRef]
- Sabatier, L.; Jouanguy, E.; Dostert, C.; Zachary, D.; Dimarcq, J.L.; Bulet, P.; Imler, J.C. Pherokine-2 and -3: Two Drosophila molecules related to pheromone/odor-binding proteins induced by viral and bacterial infections. Eur. J. Biol. 2003, 270, 3398–3407. [Google Scholar] [CrossRef]
- Liu, G.X.; et al. Biotype expression and insecticide response of Bemisia tabaci chemosensory protein-1. Arch. Insect Biochem. Physiol. 2014, 85, 137–151. [Google Scholar] [CrossRef]
- Einhorn, E.; Imler, J.L. Insect immunity; from systemic to chemosensory organs protection. In Olfactory Concepts of Insect Control-Alternative to Insecticides; Picimbon, J.F., Ed; Vol. 2 Springer Nature AG, Cham, Switzerland, 2019; pp. 205-229.
- Oduol, F.; Xu, J.; Niare, O.; Natajaran, R.; Vernick, K.D. Genes identified by an expression screen of the vector mosquito Anopheles gambiae display differential molecular immune response to malaria parasites and bacteria. Proc. Natl. Acad. Sci. USA 2000, 97, 11397–11402. [Google Scholar] [CrossRef]
- Sim, S.; Ramirez, J.L.; Dimopoulos, G. Dengue virus infection of the Aedes aegypti salivary gland and chemosensory apparatus induces genes that modulate infection and blood-feeding behavior. PLoS Pathog. 2012, 8, e1002631. [Google Scholar] [CrossRef]
- Hou, C.X.; Qin, G.X.; Liu, T.; Mei, X.L.; Li, B.; Shen, Z.Y.; Guo, X.J. Differentially expressed genes in the cuticle and hemolymph of the silkworm, Bombyx mori, injected with the fungus Beauvaria bassiana. J. Insect Sci. 2013, 13, 138. [Google Scholar] [CrossRef]
- Guo, W.; Song, J.; Yang, P.; Chen, X.; Chen, D.; Ren, D.; Kang, L.; Wang, X. Juvenile hormone suppresses aggregation behavior through influencing antennal gene expression in locusts. PLoS Genet. 2020, 16, e1008762. [Google Scholar] [CrossRef]
- Carr, A.L.; Rinker, D.C.; Dong, Y.; Dimopoulos, G.; Zwiebel, L.J. Transcriptome profiles of Anopheles gambiae harboring natural low-level Plasmodium infection reveal advantages for the mosquito. Sci. Rep. 2021, 11, 22578. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Xiang, Y.; Chen, D.; Hu, J.; Liu, F. Comparative transcriptome analysis of chemoreception organs of Laodelphax striatellus in response to rice stripe virus infection. Int. J. Mol. Sci. 2021, 22, 10299. [Google Scholar] [CrossRef] [PubMed]
- Traverso, L.; Latorre Estivalis, J.M.; da Rocha Fernandes, G.; Fronza, G.; Lobbia, P.; Mougabure Cueto, G.; Ons, S. Transcriptomic modulation in response to an intoxication with deltamethrin in a population of Triatoma infestans with low resistance to pyrethroids. PLoS Negl. Trop. Dis. 2022, 16, e0010060. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, R.; Gao, J.; Xiao, X.; Yin, X.; Hu, S.; Zhang, Y.; Liang, P.; Gu, S. Two cuticle-enriched chemosensory proteins confer multi-insecticide resistance in Spodoptera frugiperda. Int. J. Biol. Macromol. 2024, 266, 130941. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.X.; Ma, H.M.; Xie, Y.N.; Xuan, N.; Guo, X.; Fan, Z.X.; Rajashekar, B.; Arnaud, P.; Offmann, B.; Picimbon, J.F. Biotype characterization, developmental profiling, insecticide response and binding property of Bemisia tabaci chemosensory proteins: Role of CSP in insect defense. PLoS ONE 2016, 11, e0154706. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.X.; Xuan, N.; Rajashekar, B.; Arnaud, P.; Offmann, B.; Picimbon, J.F. Comprehensive history of CSP genes: Evolution, phylogenetic distribution, and functions. Genes 2020, 11, 413. [Google Scholar] [CrossRef]
- Nomura, A.; Kawasaki, K.; Kubo, T.; Natori, S. Purification and localization of p10, a novel protein that increases in nymphal regenerating legs of Periplaneta americana (American cockroach). Int. J. Dev. Biol. 1992, 36, 391–398. [Google Scholar]
- Pikielny, C.W.; Hasan, G.; Rouyer, F.; Rosbach, M. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron 1994, 12, 35–49. [Google Scholar] [CrossRef]
- McKenna, M.P.; Hekmat-Scafe, D.S.; Gaines, P.; Carlson, J.R. Putative Drosophila pheromone-binding-proteins expressed in a subregion of the olfactory system. J. Biol. Chem. 1994, 269, 16340–16347. [Google Scholar] [CrossRef]
- Robertson, H.M.; Martos, R.; Sears, C.R.; Todres, E.Z.; Walden, K.K.; Nardi, J.B. Diversity of odourant binding proteins revealed by an expressed sequence tag project on male Manduca sexta moth antennae. Insect Mol. Biol. 1999, 8, 501–518. [Google Scholar] [CrossRef]
- Bos, J.I.; Prince, D.; Pitino, M.; Maffei, M.E.; Win, J.; Hogenhout, S.A. A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet. 2010, 6, e1001210. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.X.; Yue, S.; Rajashekar, B.; Picimbon, J.F. Expression of chemosensory protein (CSP) structures in Pediculus humanus corporis and Acinetobacter baumannii. SOJ Microbiol. Infect. Dis. 2019, 7, 1–17. [Google Scholar]
- Picimbon, J.F. Molecular phylogeny of ”Chemosensory Proteins” in bacteria and crustaceans: CSP as an extremely ancient gene. J. Mol. Evol. revised. 2024. [Google Scholar]
- Picimbon, J.F.; Regnault-Roger, C. Composés sémiochimiques volatils, phytoprotection et olfaction: Cibles moléculaires de la lutte intégrée. In Biopesticides d’Origine Végétale; Regnault-Roger, C., Philogène, B., Vincent, C., Eds; Lavoisier Tech & Doc, Paris, France, 2008; pp. 383-415.
- Gissendanner, C.R.; Kelley, T.D. The C. elegans gene pan-1 encodes novel transmembrane and cytoplasmic leucine-rich repeat proteins and promotes molting and the larva to adult transition. BMC Dev. Biol. 2013, 13, 21. [Google Scholar] [CrossRef]
- Gonzáles-Gonzáles, A. , Yañez, O.; Ballesteros, G.I.; Palma-Millanao, R.; Figueroa, C.C.; Niemeyer, H.M.; RamÍrez, C.C.. A mutation increases the specificity to plant compounds in an insect chemosensory protein. J. Mol. Graph. Model. 2022, 114, 108191. [Google Scholar] [CrossRef]
- Yue, S.; He, Q.X.; Picimbon, J.F. Lactobacillus for ribosome peptide cancer. Clin. Transl. Oncol. 2023, 25, 1522–1544. [Google Scholar] [CrossRef]
- Breer, H.; Fleischer, J.; Pregitzer, P.; Krieger, J. Molecular mechanisms of insect olfaction: Olfactory receptors. In Olfactory Concepts of Insect Control-Alternative to Insecticides; Picimbon, J.F., Ed; Vol. 2, Springer Nature AG, Cham, Switzerland, 2019; pp. 93-114.
- Blomquist, G.J.; Tittiger, C.; Jurenka, R. Cuticular hydrocarbons and pheromones of arthropods. In Oils and Lipids: Diversity, Origin, Chemistry and Fate; Wilkes, H., Ed; Hydrocarbons, Handbook of Hydrocarbon and Lipid Microbiology, Springer, Cham, Swizerland, 2018; pp. 1-32.
- Davis, J.S.; Pearcy, M.J.; Yew, J.Y.; Moyle, L.C. A shift to shorter cuticular hydrocarbons accompanies sexual isolation among Drosophila americana group population. Evol. Lett. 2021, 5, 521–540. [Google Scholar] [CrossRef]
- Ha, T.S.; Smith, D.P. A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J. Neurosci. 2006, 26, 8727–8733. [Google Scholar] [CrossRef]
- Lebreton, S.; Borrero-Echeverry, F.; Gonzales, F.; Solum, M.; Wallin, E.A.; Hedenström, E.; Hansson, B.S.; Gustavsson, A.L.; Bengtsson, M.; Birgersson, G.; Walker, W.B., 3rd; Dweck, H.K.M.; Becher, P.G.; Witzgall, P.A. A Drosophila female pheromone elicits species-specific long-range attraction via an olfactory channel with dual specificity for sex and food. BMC Biol. 2017, 15, 88. [Google Scholar] [CrossRef]
- Starostina, E.; Xu, A.; Lin, H.; Pikielny, C.W. A Drosophila protein family implicated in pheromone perception is related to Tay-Sachs GM2-activator protein. J. Biol. Chem. 2009, 284, 585–594. [Google Scholar] [CrossRef]
- Pikielny, C.W. Drosophila CheB proteins involved in gustatory detection of pheromones are related to human neurodegeneration factor. Vitam. Horm. 2010, 83, 273–287. [Google Scholar] [PubMed]
- Shang, J.; Tang, G.; Yang, J.; Lu, M.; Wang, C.Z.; Wang, C. Sensing of a spore surface protein by a Drosophila chemosensory protein induces behavioral defense against fungal parasitic infections. Curr. Biol. 2023, 99, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Bontonou, G.; Saint-Leandre, B.; Kafle, T.; Baticle, T.; Hassan, A.; Sánchez-Alcañiz, J.A.; Arguello, J.R. Evolution of chemosensory tissues and cells across ecologically diverse Drosophilids. Nat. Commun. 2024, 15, 1047. [Google Scholar] [CrossRef] [PubMed]
- Torres-Oliva, M.; Almeida, F.C.; Sánchez-Gracia, A.; Rozas, J. Comparative genomics uncovers unique gene turnover and evolutionary rates in a gene family involved in the detection of insect cuticular pheromones. Genome Biol. Evol. 2016, 8, 1734–1747. [Google Scholar] [CrossRef]
- Wendeler, M.; Werth, N.; Maier, T.; Schwarzmann, G.; Kolter, T.; Schoeniger, M.; Hoffmann, D.; Lemm, T.; Saenger, W.; Sandhoff, K. The enzyme-binding region of human GM2-activator protein. FEBS J. 2006, 273, 982–991. [Google Scholar] [CrossRef]
- Spealman, P.; Burrelli, J.; Gresham, D. Inverted duplicate DNA sequences increase translocation rates through sequencing nanopores resulting in reduced base calling accuracy. Nucl. Acids Res. 2020, 48, 4940–4945. [Google Scholar] [CrossRef]
- Chau, L.M.; Goodisman, M.A.D. Gene duplication and the evolution of phenotypic diversity in insect societies. Evolution 2017, 71, 2871–2884. [Google Scholar] [CrossRef]
- Wellenreuther, M.; Bernatchez, L. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol. Evol. 2018, 33, 427–440. [Google Scholar] [CrossRef]
- Gloss, A.D.; Abbot, P.; Whiteman, N.K. How interactions with plant chemicals shape insect genomes. Curr. Opin. Insect Sci. 2019, 36, 149–156. [Google Scholar] [CrossRef]
- Nosil, P.; Soria-Carrasco, V.; Villoutreix, R.; De-la-Mora, M.; de Carvaho, C.F.; Parchman, T.; Feder, J.L.; Gompert, Z. Complex evolutionary processes maintain an ancient chromosomal inversion. Proc. Natl. Acad. Sci. USA 2023, 120, e2300673120. [Google Scholar] [CrossRef]
- Ding, *!!! REPLACE !!!*; et al. Adaptive functions of structural variants in human brain development. Science 2024, 10. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Zhu, B.; Jia, Y.; Zhang, Y.; Wang, W.; Shen, Y.; Zhong, Y.; Zheng, Y.; Wang, Y.; Tong, Y.; Zhang, W.; Li, S. Single-cell transcriptomics reveals the brain evolution of web-building spiders. Nat. Ecol. Evol. 2023, 7, 2125–2142. [Google Scholar] [CrossRef] [PubMed]
- Maleszka, J. , Forêt, S., Saint, R., Maleszka, R. RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev. Genes Evol. 2007, 217, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Blobel, G. Protein targeting (Nobel lecture). Chembiochem. 2000, 1, 86–102. [Google Scholar] [CrossRef]
- Matlin, K.S. The strange case of the signal recognition particle. Nat. Rev. Mol. Cell Biol. 2002, 3, 538–542. [Google Scholar] [CrossRef]
- Doudna, J.A.; Batey, R.T. Structural insights into the signal recognition particle. Annu. Rev. Biochem. 2004, 73, 539–557. [Google Scholar] [CrossRef]
- Jin, X.; Brandazza, A.; Navarrini, A.; Ban, L.; Zhang, S.; Steinbrecht, R.A.; Zhang, L.; Pelosi, P. Expression and immunolocalization of odorant-binding and chemosensory proteins in locusts. Cell. Mol. Life Sci. 2005, 62, 1156–1166. [Google Scholar] [CrossRef]
- Steinbrecht, R.A.; Ozaki, M.; Ziegelberger, G. Immunocytochemical localization of pheromone-binding protein in moth antennae. Cell Tissue Res. 1992, 270, 287–302. [Google Scholar] [CrossRef]
- Liu, G.X.; Ma, H.M.; Xie, H.Y.; Xuan, N.; Picimbon, J.F. Sequence variation of Bemisia tabaci Chemosensory protein 2 in cryptic species B and Q: New DNA markers for whitefly recognition. Gene 2016, 576, 284–291. [Google Scholar] [CrossRef]
- Zhu, J.; Iovinella, I.; Dani, F.R.; Pelosi, P.; Wang, G. Chemosensory proteins: A versatile binding family. In Olfactory Concepts of Insect Control-Alternative to Insecticides; Picimbon, J.F., Ed; Vol. 2, Springer Nature AG, Cham, Switzerland, 2019; pp. 147-169.
- Pino, J.; Godoy, R.; Venthur, H.; Larama, G.; Quiroz, A.; Mutis, A. Identification and ligand binding of a chemosensory protein from sea louse Caligus rogercresseyi (Crustacea: Copepoda). Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 2023, 265, 110830. [Google Scholar] [CrossRef]
- Ghai, R.; Mizuno, C.M.; Picazo, A.; Camacho, A.; Rodriguez-Valera, F. Metagenomics uncovers a new group of low GC and ultra-small marine Actinobacteria. Sci. Rep. 2013, 3, 2471. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Gonsior, M.; Schmitt-Kopplin, P.; Zhan, Y.; Zhang, R.; Jiao, N.; Chen, F. Microbial transformation of virus-induced dissolved organic matter from picocyanobacteria: Coupling of bacterial diversity and DOM chemodiversity. ISME J. 2019, 13, 2551–2565. [Google Scholar] [CrossRef] [PubMed]
- Doré, H.; Guyet, U.; Leconte, J.; Farrant, G.K.; Alric, B.; Ratin, M.; Ostrowski, M.; Ferrieux, M.; Brillet-Guéguen, L.; Hoebeke, M.; Siltanen, J.; Le Corguillé, G.; Corre, E.; Wincker, P.; Scanlan, D.J.; Eveillard, D.; Partensky, F.; Garczarek, L. Differential global distribution of marine picocyanobacteria gene clusters reveals distinct niche-related adaptive strategies. ISME J. 2023, 17, 720–732. [Google Scholar] [CrossRef]
- Bos, L.D.J.; Sterk, P.J.; Schultz, M.J. Volatile metabolites of pathogens: A systemic review. PLoS Pathog. 2013, 9, e1003311. [Google Scholar] [CrossRef] [PubMed]
- Taga, M.E.; Bassler, B.L. Chemical communication among bacteria. Proc. Natl. Acad. Sci. USA 2003, 100, 14549–14554. [Google Scholar] [CrossRef]
- Silva-Junior, E.A.; Ruzzini, A.C.; Paludo, C.R.; Nascimento, F.S.; Currie, C.R.; Clardy, J.; Pupo, M.T. Pyrazines from bacteria and ants: Convergent chemistry within an ecological niche. Sci. Rep. 2018, 8, 2595. [Google Scholar] [CrossRef]
- Salah Ud-Din, A.I.M.; Roujeinikova, A. Methyl-accepting chemotaxis proteins: A core sensing element in prokaryotes and archaea. Cell Mol. Life Sci. 2017, 74, 3293–3303. [Google Scholar] [CrossRef]
- Baker, M.D.; Wolanin, P.W.; Stock, J.B. Systems biology of bacterial chemotaxis. Curr. Opin. Microbiol. 2006, 9, 187–192. [Google Scholar] [CrossRef]
- Galperin, M.Y. Sensory transduction in bacteria. Encyclopedia of Microbiology (Third Edition), Academic Press, 2009, 447-463.
- Stock, A.; Chen, T.; Welsh, D.; Stock, J. CheA protein, a central regulator of bacterial chemotaxis, belongs to a family of proteins that control gene expression in response to changing environmental conditions. Proc. Natl. Acad. Sci. USA 1988, 85, 1403–1407. [Google Scholar] [CrossRef]
- Huang, Z.; Pan, X.; Xu, N.; Guo, M. Bacterial chemotaxis coupling protein: Structure, function and diversity. Microbiol. Res. 2019, 219, 40–48. [Google Scholar] [CrossRef]
- Perkin, L.C.; Friesen, K.S.; Flinn, P.W.; Oppert, B. Venom gland components of the ectoparasitoid wasp, Anisopteromalus calandrae. J. Venom Res. 2015, 6, 19–37. [Google Scholar] [PubMed]
- Celorio-Mancera, M.L.; Sundmalm, S.M.; Vogel, H., Rutishauser; Ytterberg, A.J.; Zubarv, R.A.; Janz, N. Chemosensory proteins, major salivary factors in caterpillar mandibular glands. Insect Biochem. Mol. Biol. 2012, 42, 796–805. [Google Scholar]
- González-Caballero, N.; Valenzuela, J.G.; Ribeiro, J.M.C.; Cuervo, P.; Brazil, R.P. Transcriptome exploration of the sex pheromone gland of Lutzomyia longipalpis (Diptera: Psychodidae: Phlebotominae). Parasit. Vectors 2013, 6, 56. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.L.; Guo, H.; Huang, L.Q.; Pelosi, P.; Wang, C.Z. Unique function of a chemosensory protein in the proboscis of two Helicoverpa species. J. Exp. Biol. 2014, 217, 1821–1826. [Google Scholar] [CrossRef]
- Zhu, J.; Iovinella, I.; Dani, F.R.; Liu, Y.L.; Huang, L.Q.; Liu, Y.; Wang, C.Z.; Pelosi, P.; Wang, G. Conserved chemosensory proteins in the proboscis and eyes of Lepidoptera. Int. J. Biol. Sci. 2016, 12, 1394–1404. [Google Scholar] [CrossRef]
- Ma, C.; Cui, S.; Tian, Z.; Zhang, Y.; Chen, G.; Gao, X.; Tian, Z.; Chen, H.; Guo, J.; Zhou, Z. OcomCSP12, a chemosensory protein expressed specifically by ovary, mediates reproduction in Ophraella communa (Coleoptera: Chrysomelidae). Front. Physiol. 2019, 10, 1290. [Google Scholar] [CrossRef]
- Xu, A.; Park, S.K.; D’Mello, S.; Kim, E.; Wang, Q.; Pikielny, C.W. Novel genes expressed in subsets of chemosensory sensilla on the front legs of male Drosophila melanogaster. Cell Tissue Res. 2002, 307, 381–392. [Google Scholar]
- Park, S.K.; Mann, K.J.; Lin, H.; Starostina, E.; Kolski-Andreaco, A.; Pikielny, C.W. A Drosophila protein specific to pheromone-sensing gustatory hairs delays males’ copulation attempts. Curr. Biol. 2006, 16, 1154–1159. [Google Scholar] [CrossRef]
- Singh, S.S.; Mittal, A.M.; Chepurwar, S.; Gupta, N. Olfactory systems in insects : Similarities and differences between species In Olfactory Concepts of Insect Control-Alternative to Insecticides; Picimbon, J.F., Ed; Vol. 2, Springer Nature AG, Cham, Switzerland, 2019; pp.
- 29-48.
- Ozaki, M. Odorant-binding proteins in taste systems: Putative roles in taste sensation and behavior. In Olfactory Concepts of Insect Control-Alternative to Insecticides; Picimbon, J.F., Ed; Vol. 2, Springer Nature AG, Cham, Switzerland, 2019; pp. 93-114.
- Pedra, J.H.; Brandt, A.; Li, H.M.; Westerman, R.; Romero-Serverson, J.; Pollack, R.J.; Murdock, L.L.; Pittendrigh, B.R. Transcriptome identification of putative genes involved in protein catabolism and innate immune response in human body louse (Pediculicidae: Pediculus humanus). Insect Biochem. Mol. Biol. 2003, 33, 1135–1143. [Google Scholar] [CrossRef]
- Kirkness, E.F.; et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic life. Proc. Natl Acad. Sci. USA 2010, 107, 12168–12173. [Google Scholar] [CrossRef]
- Pittendrigh, B.R.; Clark, J.M.; Lee, S.H.; Yoon, K.S.; Sun, W.; Steele, L.D.; Seong, K.M. Body lice: From the genome project to functional genomics and reverse genetics. In Short views on insect genomics and proteomics, Entomology in focus; Raman, C., Goldsmith, M., Agunbiade, T., Eds; Springer, Cham, Switzerland, 2015 ; pp. 1-18.
- Shigenobu, S.; Richards, S.; Cree, A.G.; Morioka, M.; Fukatsu, T.; Kudo, T.; Miyagishima, S.; Gibbs, R.A.; Stern, D.L.; Nakabashi, A. A full-length cDNA resource for the pea aphid, Acyrtosiphon pisum. Insect Mol. Biol. 2010, 19, 23–31. [Google Scholar]
- Ollivier, M.; Legeai, F.; Rispe, C. Comparative analysis of the Acyrthosiphon pisum genome and expressed sequence tag-based gene sets from other aphid species. Insect Mol. Biol. 2019, 19, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.X.; Arnaud, P.; Offmann, B.; Picimbon, J.F. Genotyping and bio-sensing chemosensory proteins in insects. Sensors 2017, 17, 1801. [Google Scholar] [CrossRef] [PubMed]
- Loftus, B.J.; Utterback, T.; Pertea, G.; Koo, H.; Mori, A.; Schneider, J.; Lovin, D.; de Bruyn, B.; Song, Z.; Raikhel, A.; de Fatima, B.M.; Casavant, T.; Soares, B.; Severson, D. Aedes aegypti cDNA sequencing. NCBI 2005, #DV263125, DV263127, DV289920, DV289921, DV297938, DV314711, DV314712, DV316589, DV316619, DV334734, DV334735, DV335057, DV335058, DV344048, DV347318, DV347319, DV365747, DV365766, DV357763, DV368339, DV368340, DV393559, DV400785, DV400787, “…”.
- Verjovski-Almeida, S.; Eiglmeier, K.; El-Dorry, H.; Gomes, S.L.; Menck, S.F.M.; Nascimento, A.L.; Roth, C.W. FAPESP and Institut Pasteur/AMSUD Network Aedes aegypti cDNA sequencing project. NCBI 2005, #EG001037.
- Noriega, F.G.; Ribeiro, J.M.C.; Koener, J.F.; Valenzuela, J.G.; Hernandez-Martinez, S.; Pham, V.M.; Feyereisen, R. Comparative genomics of insect juvenile hormone biosynthesis. Insect Biochem. Mol. Biol. 2006, 36, 366–374. [Google Scholar] [CrossRef]
- Nene, V.; Worthman, J.R.; Lawson, D.; et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science 2007, 22, 1718–1723. [Google Scholar] [CrossRef]
- Wei, Z.; Ortiz-Urquiza, A.; Keyhani, N.O. Altered expression of chemosensory and odorant binding proteins in response to fungal infection in the red imported fire ant, Solenopsis invicta. Front. Physiol. 2021, 12, 596571. [Google Scholar] [CrossRef]
- Xu, H.; Yan, K.; Ding, Y.; Lv, Y.; Li, J.; Yang, F.; Chen, X.; Gao, X.; Pan, Y.; Shang, Q. Chemosensory proteins are associated with thiametoxam and Spirotetramat tolerance in Aphis gossypii Glover. Int. J. Mol. Sci. 2022, 23, 2356. [Google Scholar] [CrossRef]
- Gao, P.; Zhang, S.; Tan, J.J.; Li, X.; Chen, M. Chemosensory proteins are associated with thiametoxam tolerance in bird cherry-oat aphid Rhopalosiphum padi. Pestic. Biochem. Physiol. 2023, 192, 105393. [Google Scholar] [CrossRef]
- Xu, H.; Pan, Y.; Li, J.; Yang, F.; Chen, X.; Gao, X.; Wen, S.; Shang, Q. Chemosensory proteins confer adaptation to the ryanoid anthranilic diamide insecticide cyantraniliprole in Aphis gossypii glover. Pestic. Biochem. Physiol. 2022, 184, 105076. [Google Scholar] [CrossRef]
- Yi, X.; Qi, J.; Zhou, X.; Hu, M.Y.; Zhong, G.H. Differential expression of chemosensory-protein genes in midguts in response to diet of Spodoptera litura. Sci. Rep. 2017, 7, 296. [Google Scholar] [CrossRef]
- Guo, X.; Xuan, N.; Liu, G.X.; Xie, H.; Lou, Q.; Arnaud, P.; Offmann, B.; Picimbon, J.F. An expanded survey of the moth PBP/GOBP clade in Bombyx mori: New insight into expression and functional roles. Front. Physiol. 2021, 12, 712593. [Google Scholar] [CrossRef] [PubMed]
- Celorio-Mancera, M.L.; Ytterberg, A.; Rutishauser, D.; Janz, N.; Zubarev, R.A. Effect of host plant and immune challenge on the levels of chemosensory proteins in catterpillar salivary glands. Insect Biochem. Mol. Biol. 2015, 61, 34–45. [Google Scholar]
- Bian, H.X.; Chen, B.D.; Zheng, X.X. , Ma, H.F.; Li, Y.P.; Li, Q.; Xia, R.X.; Wang, H.; Jiang, Y.R.; Liu, Y.Q.; Qin, L. Transcriptomic analysis of the prothoracic gland from two lepidopteran insects, domesticated silkmoth Bombyx mori and wild silkmoth Antheraea pernyi. Sci. Rep. 2019, 9, 5313. [Google Scholar] [CrossRef] [PubMed]
- Prajapati, V.K. .; Varma, M.; Vadassery, J. In silico identification of effector proteins from generalist herbivore Spodoptera litura. BMC genomics 2020, 21, 816. [Google Scholar] [CrossRef]
- Zeng, Y.; Merchant, A.; Wu, Q.; Wang, S.; Kong, L.; Zhou, X.; Xie, W.; Zhang, Y. A chemosensory protein BtabCSP11 mediates reproduction in Bemisia tabaci. Front. Physiol. 2020, 11, 709. [Google Scholar] [CrossRef]
- Wang, Q.; Wen, X.; Lu, Y.; Wen, J. Comparison and functional analysis of chemosensory protein genes from Eucryptorrhynchus scrobiculatus Motschulsky and Eucryptorrhynchus brandti Harold. Front. Physiol. 2021, 12, 661310. [Google Scholar] [CrossRef]
- Kim, I.H.; Pham, V.; Jablonka, W.; Goodman, W.G.; Ribeiro, J.M.C.; Andersen, J.F. A mosquito hemolymph odorant-binding protein family member specifically binds juvenile hormone. J. Biol. Chem. 2017, 292, 15329–15339. [Google Scholar] [CrossRef]
- Laufer, H.; Borst, D.; Baker, F.C.; Reuter, C.C.; Tsai, L.W.; Schooley, D.A.; Carrasco, C.; Sinkus, M. Identification of a juvenile-hormone-like compound in a Crustacean. Science 1987, 235, 202–205. [Google Scholar] [CrossRef]
- Jindra, M.; Palli, S.R.; Riddiford, L.M. The juvenile hormone signaling pathway in insect development. Annu. Rev. Entomol. 2013, 58, 181–204. [Google Scholar] [CrossRef]
- Cayre, M.; Strambi, C.; Strambi, A. Neurogenesis in an adult insect brain and its hormonal control. Nature 1994, 368, 57–59. [Google Scholar] [CrossRef]
- Fahrbach, S.E.; Moore, D.; Capaldi, E.A.; Farris, S.M.; Robinson, G.E. Experience-expectant plasticity in the mushroom bodies of the honeybee. Learn. Mem. 1998, 5, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Anton, S.; Rossler, W. Plasticity and modulation of olfactory circuits in insects. Cell Tiss. Res. 2021, 383, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Hrithik, M.T.H.; Roy, M.C.; Bode, H.; Kim, Y. Phurealipids, produced by the entomopathogenic bacteria, Photorhabdus, mimic juvenile hormone to suppress insect immunity and immature development. J. Invertebr. Pathol. 2022, 193, 107799. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, M.; Wada-Katsumata, A.; Fujikawa, K.; Iwasaki, M.; Yokohari, F.; Satoji, Y.; Nisimura, T.; Yamaoka, R. Ant nestmate and non-nestmate discrimination by a chemosensory sensillum. Science 2005, 309, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.A.; Ahmed, S.; Kim, Y. Biosynthetic pathway of arachidonic acid in Spodoptera exigua in response to bacterial challenge. Insect Biochem. Mol. Biol. 2019, 111, 103179. [Google Scholar] [CrossRef]
- Vatanparast, M.; Ahmed, S.; Lee, D.H.; Hwang, S.H.; Hammock, B.; Kim, Y. EpOMEs act as immune suppressors in a lepidoptera insect, Spodoptera exigua. Sci. Rep. 2020, 10, 20183. [Google Scholar] [CrossRef]
- Liaci, A.M.; Förster, F. Take me home, protein roads: Structural insights into signal peptide interactions during ER translocation. Int. J. Mol. Sci. 2021, 22, 11871. [Google Scholar] [CrossRef]
- Malcicka, M.; Visser, B.; Ellers, J. An evolutionary perspective on linoleic acid synthesis in animals. Evol. Biol. 2018, 45, 15–26. [Google Scholar] [CrossRef]
- Herrera, H.; Barros-Parada, W.; Bergmann, J. Linoleic acid and stearic acid are biosynthetic precursors of (7Z,10Z)-7,10-hexadecadienal, the major component of the sex pheromone of Chilecomadia valdiviana (Lepidoptera: Cossidae). PLoS ONE 2019, 14, e0215769. [Google Scholar] [CrossRef]
- Shibao, H.; Kutsukake, M.; Matsuyama, S.; Fukatsu, T. Linoleic acid as corpse recognition signal in a social aphid. Zoological Lett. 2022, 8, 2. [Google Scholar] [CrossRef]
- Buehlmann, C.; Graham, P.; Hansson, B.S.; Knaden, M. Desert ants locate food by combining high sensitivity to food odors with extensive crosswind runs. Curr. Biol. 2014, 24, 960–964. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.A.; Stam, R.; Warbroek, T.; Bos, J.I. Mp10 and Mp42 from the aphid species Myzus persicae trigger plant defenses in Nicotiana benthamiana through different activities. Mol. Plant. Microbe. Interact. 2014, 27, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Shan, G.; Guo, D.; Wang, S.; Ma, X.; Xiao, H.; Liu, J.; Zhang, Z.; Liu, Y.; Zhang, Y.; Yu, K.; Huang, S.; Li, F. InsectBase: A resource for insect genomes and transcriptomes. Nucl. Acids Res. 2016, 44, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Sáez, A.; Morales, C.L.; Ramos, L.Y.; Aizen, M.A. Extremely frequent bee visits increase pollen decomposition but reduce drupelet set in raspberry. J. Appl. Ecol. 2014, 51, 1603–1612. [Google Scholar] [CrossRef]
- Jacobsen, D.J.; Raguso, R.A. Lingering effects of herbivory and plant defenses on pollinators. Curr. Biol. 2018, 28, R1164–R11. [Google Scholar] [CrossRef]
- Barre, A.; Pichereaux, C.; Simplicien, M.; Burlet-Schiltz, O.; Benoist, H.; Rougé, P. A proteomic- and bioinformatic-based identification of specific allergens from edible insects: Probes for future detection as food ingredients. Foods 2021, 10, 280. [Google Scholar] [CrossRef]
- Moon, S.Y.; Zheng, Y. (2003) Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003, 13, P13–22. [Google Scholar] [CrossRef]
- Gouin, E.; Egile, C.; Dehoux, P.; Villiers, V.; Adams, J.; Gertler, F.; Li, R.; Cossart, P. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature 2004, 427, 457–461. [Google Scholar] [CrossRef]
- Cronmiller, E.; Toor, D.; Shao, N.C.; Kariyawasam, T.; Wang, M.H.; Lee, J.H. Cell wall integrity signaling regulates cell wall-related gene expression in Chlamydomonas reinhardtii. Sci. Rep. 2019, 9, 12204. [Google Scholar] [CrossRef]
- Ho, S. The molecular clock and estimating species divergence. Nat. Educ. 2008, 1, 168. [Google Scholar]
- Wang, P.; Granados, R.R. An intestinal mucin is the target substrate for a baculovirus enhancin. Proc. Natl. Acad. Sci. USA 1997, 94, 6977–6982. [Google Scholar] [CrossRef] [PubMed]
- Dias, R.O.; Cardoso, C.; Pimentel, A.C.; Damasceno, T.F.; Ferreira, C.; Terra, W.R. The roles of mucus-forming mucins, peritrophins and peritrophins with mucin domains in the insect midgut. Insect Mol. Biol. 2018, 27, 46–60. [Google Scholar]
- Schachat, S.R.; Goldstein, P.Z.; Desalle, R.; Bobo, D.M.; Boyce, K.; Payne, J.L.; Labandeira, C.C. Illusion of flight? Absence, evidence and the age of winged insects. Biol. J. Linn. Soc. 2023, 138, 143–168. [Google Scholar] [CrossRef]
- Armstrong, L.; Lako, M.; van Herpe, I.; Evans, J.; Saretzki, G.; Hole, N. A role for nucleotide Zap3 in the reduction of telomerase activity during embryonic stem cell differentiation. Mech. Dev. 2004, 121, 1509–1522. [Google Scholar] [CrossRef] [PubMed]
- Teparic, R.; Lozancic, M.; Mrsa, V. Evolutionary overview of molecular interactions and enzymatic activities in the yeast cell walls. Int. J. Mol. Sci. 2020, 21, 8996. [Google Scholar] [CrossRef] [PubMed]
- Adachi, N.; Senda, T.; Horikoshi, M. Uncovering ancient transcription systems with a novel evolutionary indicator. Sci. Rep. 2016, 6, 217922. [Google Scholar] [CrossRef]
- Akil, C.; Kitaoku, Y.; Tran, L.T.; Liebl, D.; Choe, H.; Muengsaen, D.; Suginta, W.; Schulte, A.; Robinson, R.C. Mythical origins of the actin cytoskeleton. Curr. Opin. Cell Biol. 2021, 68, 55–63. [Google Scholar] [CrossRef]
- Rodriguez-Oliveira, T.; Wollweber, F.; Ponce-Toledo, R.I.; Xu, J.; Rittmann, S.K.-M.R.; Klingl, A.; Pilhofer, M.; Schleper, C. Actin cytoskeleton and complex cell architecture in an Asgard archaeon. Nature 2023, 613, 332–339. [Google Scholar] [CrossRef]
- Charles-Orszag, A.; Petek-Seoane, N.A.; Mullins, R.D. Archaeal actins and the origin of a multifunctional cytoskeleton. J. Bacteriol. 2024, 206, e00348–23. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; Lepore, R.; Schwede, T. SWISS-MODEL: Homology modelling of protein structures and complexes. Nuc. Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Ho, H.-Y.H.; Rohatgi, R.; Ma, L.; Kirschner, M.W. CR16 forms a complex with N-WASP in brain and is a novel member of a conserved proline-rich actin-binding protein family. Proc. Natl. Acad. Sci. USA 2001, 98, 11306–11311. [Google Scholar] [CrossRef] [PubMed]
- Chereau, D.; Kerff, F.; Graceffa, P.; Grabarek, Z.; Langsetmo, K.; Dominguez, R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. USA 2005, 102, 16644–16649. [Google Scholar] [CrossRef] [PubMed]
- [156] Rogerio, A.P.; Anibal, F.F. Role of leukotrienes on protozoan and helminth infections. Mediators Inflamm. 2012, 2012, 595694. [157] Stanley, D.; Kim, Y. Chapter Eight – Insect prostaglandins and other eicosanoids: From molecular to physiological actions. Adv. Insect Physiol. 2019, 56, 283-343. [CrossRef]
- Roy, M.C.; Nam, K.; Kim, J.; Stanley, D.; Kim, Y. Thromboxane mobilizes insect blood cells to infection foci. Front. Immunol. 2021, 12, 791319. [Google Scholar] [CrossRef]
- El-Yassimi, A.; Hichami, A.; Besnard, P.; Khan, N.A. Linoleic acid induces calcium signaling, Src kinase phosphorylation, and neurotransmitter release in mouse CD36-positive gustatory cells. J. Biol. Chem. 2008, 283, 12949–12959. [Google Scholar] [CrossRef]
- Feng, D.D.; Zhao, Y.F.; Luo, Z.Q.; Keating, D.J.; Chen, C. Linoleic acid induces Ca2+-induced inactivation of voltage-dependent Ca2+- currents in rat pancreatic -cells. J. Endocrinol. 2008, 196, 377–384. [Google Scholar] [CrossRef]
- Yang, B.; Zhou, Y.; Wu, M.; Li, X.; Mai, K.; Ai, Q. -6 Polyunsaturated fatty acids (linoleic acid) activate both autophagy ands antioxidation in a synergistic feedback loop via TOR-dependent and TOR-independent signaling pathways. Cell Death Dis. 2020, 11, 607. [Google Scholar] [CrossRef]
- Hidalgo, M.A.; Carretta, M.D.; Burgos, R.A. Long chain fatty acids as modulators of immune cells function: Contribution of FFA1 and FFA4 receptors. Front. Physiol. 2021, 12, 668330. [Google Scholar] [CrossRef]
- Blomquist, G.J.; Borgeson, C.E.; Vundla, M. Polyunsaturated fatty acids and eicosanoids in insects. Insect Biochem. 1991, 21, 99–106. [Google Scholar] [CrossRef]
- Marmunti, M.; Catalá, A. Incorporation of 1-(14)C linoleic acid in rat liver nuclei and chromatin fractions. Int. J. Biochem. Cell Biol. 2001, 33, 261–267. [Google Scholar] [CrossRef]
- Ando, H.; Wen, Z.M.; Kim, H.Y.; Valencia, J.C.; Costin, G.E.; Watabe, H.; Yasumoto, K.; Niki, Y.; Kondoh, H.; Ichihashi, M.; Hearing, V.J. Intracellular composition of fatty acid affects the processing and function of tyrosinase through the ubiquitin-proteasome pathway. Biochem. J. 2006, 394, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Masner, M.; Lujea, N.; Bisbal, M.; Acosta, C.; Kunda, P. Linoleic and oleic acids enhance cell migration by altering the dynamics of microtubules and the remodeling of the actin cytoskeleton at the leading edge. Sci. Rep. 2021, 11, 14984. [Google Scholar] [CrossRef] [PubMed]
- Lauson, C.B.N.; et al. Linoleic acid potentiates CD8+ T cell metabolic fitness and antitumor activity. Cell Metabol. 2023, 35, 633–650. [Google Scholar] [CrossRef] [PubMed]
- Vesala, L.; Basikhina, Y.; Tuomela, T.; Nurminen, A.; Siukola, E.; Vale, P.F.; Salminen, T.S. Mitochondrial perturbation in immune cells enhances cell-mediated innate immunity in Drosophila. BMC Biol. 2024, 22, 60. [Google Scholar] [CrossRef]
- Wrońska, A.K.; Kaczmarek, A.; Boguś, M.I.; Kuna, A. Lipids as a key element of insect defense systems. Front. Genet. 2023, 14, 1183659. [Google Scholar] [CrossRef]
- Blank, H.M.; Maitra, N.; Polymenis, M. Lipid biosynthesis: When the cell cycle meets protein synthesis? Cell Cycle 2017, 16, 905–906. [Google Scholar] [CrossRef]
- Blank, H.M.; Perez, R.; He, C.; Maitra, N.; Metz, R.; Hill, J.; Lin, Y.; Johnson, C.D.; Bankaitis, V.A.; Kennedy, B.K.; Aramayo, R.; Polymenis, M. Translational control of lipogenic enzymes in the cell cycle of synchronous, growing yeast cells. EMBO Journal 2017, 36, 487–502. [Google Scholar] [CrossRef]
- Feyereisen, R. Insect P450 enzymes. Annu. Rev. Entomol. 1999, 44, 507–533. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Wada, K.; Kabuta, T. Lysosomal degradation of intracellular nucleic acids—Multiple autophagic pathways. J. Biochem. 2017, 161, 145–154. [Google Scholar] [CrossRef]
- Yokoyama, N.; Fónagy, A.; Tatsuki, S.; Arie, T.; Yamashita, S.; Matsumoto, S. Ultrastructural studies on the pheromone-producing cells in the silkmoth, Bombyx mori: Formation of cytoplasmic lipid droplets before adult eclosion. Acta Biol. Hung. 2003, 54, 299–311. [Google Scholar] [CrossRef]
- Hull, J.; Fonagy, A. Molecular basis of pheromonogenesis regulation in moths. In Olfactory Concepts of Insect Control-Alternative to Insecticides; Picimbon, J.F., Ed; Vol. 1, Springer Nature AG, Cham, Switzerland, 2019; pp. 115-202.
- Ibarra, A.; Hetzer, M.W. Nuclear pore proteins and the control of genome functions. Genes Dev. 2015, 29, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.H.; Hoelz, A. The structure of the Nuclear Pore Complex (an update). Annu. Rev. Biochem. 2019, 88, 725–783. [Google Scholar] [CrossRef] [PubMed]
- Kenis, S.; Istiban, M.N.; Van Damme, S.; Vandewyer, E.; Watteyne, J.; Schoofs, L.; Beets, I. Ancestral glycoprotein hormone-receptor pathway controls growth in C. elegans. Front. Endocrinol. 2023, 14, 1200407. [Google Scholar] [CrossRef] [PubMed]
- Lang, T.; Hansson, G.C.; Samuelsson, T. Gel-forming mucins appeared early in metazoan evolution. Proc. Natl. Acad. Sci. USA 2007, 104, 16209–16214. [Google Scholar] [CrossRef] [PubMed]
- Lang, T.; Klasson, S.; Larsson, E.; Johansson, M.E.; Hansson, G.C.; Samuelsson, T. Searching the evolutionary origin of epithelial mucus protein components-Mucin and FCGBP. Mol. Biol. Evol. 2016, 33, 1921–1936. [Google Scholar] [CrossRef]
- McShane, A.; Bath, J.; Jaramillo, A.M.; Ridley, C.; Walsh, A.A.; Evans, C.M.; Thornton, D.J.; Ribbeck, K. Mucus. Curr. Biol. 2021, 31, R931–R947. [Google Scholar] [CrossRef]
- Syed, Z.A.; Hard, T.; Uv, A.; van Dijk-Hard, I.F. A potential role for Drosophila mucins in development and physiology. PLoS ONE 2008, 3, e3041. [Google Scholar] [CrossRef]
- Huang, Y.; Li, L.; Rong, Y.S. JiangShi (僵尸): A widely distributed Mucin-like protein essential for Drosophila development. G3 (Bethesda) 2022, 12, jkac126. [Google Scholar] [CrossRef]
- Ibrahim, S.P.; Dias, R.O.; Ferreira, C.; Silva, C.P.; Terra, W.R. Histochemistry and transcriptomics of mucins and peritrophic membrane (PM) proteins along the midgut of a beetle with incomplete PM and their complementary function. Insect Biochem. Mol. Biol. 2023, 162, 104027. [Google Scholar] [CrossRef]
- Moriyama, M.; Hayashi, T.; Fukatsu, T. A mucin protein predominantly expressed in the female-specific symbiotic organ of the stinkbug Plautia stali. Sci. Rep. 2022, 12, 7782. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, J.; Yang, J.; Niu, N.; Zhang, J.; Yang, Q. Mucin family genes are essential for the growth and development of the migratory locust, Locusta migratoria. Insect Biochem. Mol. Biol. 2020, 123, 103404. [Google Scholar]
- Shangguan, X.; Zhang, J.; Liu, B.; Zhao, Y.; Wang, H.; Wang, Z.; Guo, J.; Rao, W.; Jing, S.; Guan, W.; Ma, Y.; Wu, Y.; Hu, L.; Chen, R.; Du, B.; Zhu, L.; Yu, D.; He, G. A mucin-like protein of planthopper is required for feeding and induces immunity responses in plants. Plant Physiol. 2018, 176, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.H.; Shen, Y.; Li, D.T.; Huang, H.J.; Lu, J.B.; Zhang, C.X. A mucin-like protein is essential for oviposition in Nilaparvata lugens. Front. Physiol. 2019, 10, 551. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Wu, S.; Wu, Y.; Liu, X.; Wu, P.; Zhai, Z. Identification of mucins and their expression in the vector mosquito Aedes albopictus. J. Vector Ecol. 2020, 45, 297–305. [Google Scholar] [CrossRef]
- Linden, S.K.; Sutton, P.; Karlsson, N.G.; Korolik, V.; McGuckin, M.A. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008, 1, 183–197. [Google Scholar] [CrossRef]
- Sheng, Y.H.; Hasnain, S.Z. Mucus and mucins: The underappreciated host defence system. Front. Cell. Infect. Microbiol. 2022, 12, 856962. [Google Scholar] [CrossRef]
- Muthusamy, N.; Sommerville, L.J.; Moeser, A.J.; Stumpo, D.J.; Sannes, P.; Adler, K.; Blackshear, P.J.; Weimer, J.M.; Ghashgaei, H.T. MARCKS-dependent mucin clearance and lipid metabolism in ependymal cells are required for maintenance of forebrain homeostasis duing aging. Aging Cell. 2015, 14, 764–773. [Google Scholar] [CrossRef]
- Campanacci, V.; Krieger, J.; Bette, S.; Sturgis, J.N.; Lartigue, A.; Cambillau, C.; Breer, H.; Tegoni, M. Revisiting the specificity of Mamestra brassicae and Antheraea polyphemus pheromone-binding proteins with a fluorescence binding assay. J. Biol. Chem. 2001, 276, 20078–20084. [Google Scholar] [CrossRef]
- Picimbon, J.F. Insect chemosensory proteins against COVID-19. Med Discov. 2024, 3, 1152. [Google Scholar] [CrossRef]
- Gong, D.H.; Turner, B.; Bhaskar, K.R.; Lamont, J.T. Lipid binding to gastric mucin: Protective effect against oxygen radicals. Am. J. Physiol. 1990, 259, G681–686. [Google Scholar] [CrossRef]
- Chang, Y.; McClements, D.J. Characterization of mucin-lipid droplet interactions: Influence on potential fate of fish oil-in-water emulsions under simulated gastrointestinal conditions. Food Hydrocoll. 2016, 56, 425–433. [Google Scholar] [CrossRef]
- Hansson, G.C. Mucins and the microbiome. Annu. Rev. Biochem. 2020, 89, 769–793. [Google Scholar] [CrossRef] [PubMed]
- Ingham, V.A.; Anthousi, A.; Douris, V.; Harding, N.J.; Lycett, G.; Morris, M.; Vontas, J.; Ranson, H. A sensory appendage protein protects malaria vectors from pyrethroids. Nature 2020, 577, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Diez-Hermano, S.; Ganfornina, M.D.; Skerra, A.; Guttiérez, G.; Sanchez, D. An evolutionary perspective of the lipocalin protein family. Front. Physiol. 2021, 12, 718983. [Google Scholar] [CrossRef] [PubMed]
- Kevorkian, Y.; Shen, A. Revisiting the role of Csp family proteins in regulating Clostridium difficile spore germination. J. Bacteriol. 2017, 199, e00266–17. [Google Scholar] [CrossRef]
- Keto-Timonen, R.; Hietala, N.; Palonen, E.; Hakakorpi, A.; Lindström, M.; Korkeala, H. ; Cold Shock Proteins: A mini-review with special emphasis on Csp-family of enteropathogenic Yersinia. Front. Microbiol. 2016, 7, 1151. [Google Scholar] [CrossRef]
- Steinbrecht, R.A. What can we learn from localizing lipoclistins? European Symposium For Insect Taste and Olfaction (ESITO VIII) 2003, July 2-7th, Harstad, Norway.
- Steinbrecht, R.A. Fine structure immunocytochemistry—An important tool for research on odorant-binding proteins. Meth. Enzymol. 2020, 642, 259–278. [Google Scholar]
- Novak, A.; Goyal, N.; Gronostajski, RM. Four conserved cysteine residues are required for the DNA binding activity of nuclear factor I. J. Biol. Chem. 1992, 267, 12986–12990. [Google Scholar] [CrossRef]
- Shimasaki, T.; Ohtsuka, H.; Naito, C.; Azuma, K.; Tenno, T.; Hiroaki, H.; Murakami, H.; Aiba, H. Ecl1 is a zinc-binding protein involved in the zinc-limitation-dependent extension of chronological life span in fission yeast. Mol. Genet. Genomics 2017, 292, 475–481. [Google Scholar] [CrossRef]
- Miller, M.C.; Mayo, K.H. Chemokines from a structural perspective. Int. J. Mol. Sci. 2017, 18, 2088. [Google Scholar] [CrossRef]
- DiRusso, C.C.; Black, P.N. Bacterial long chain fatty acid transport: Gateway to a fatty acid-responsive signaling system. J. Biol. Chem. 2004, 279, 49563–49566. [Google Scholar] [CrossRef] [PubMed]

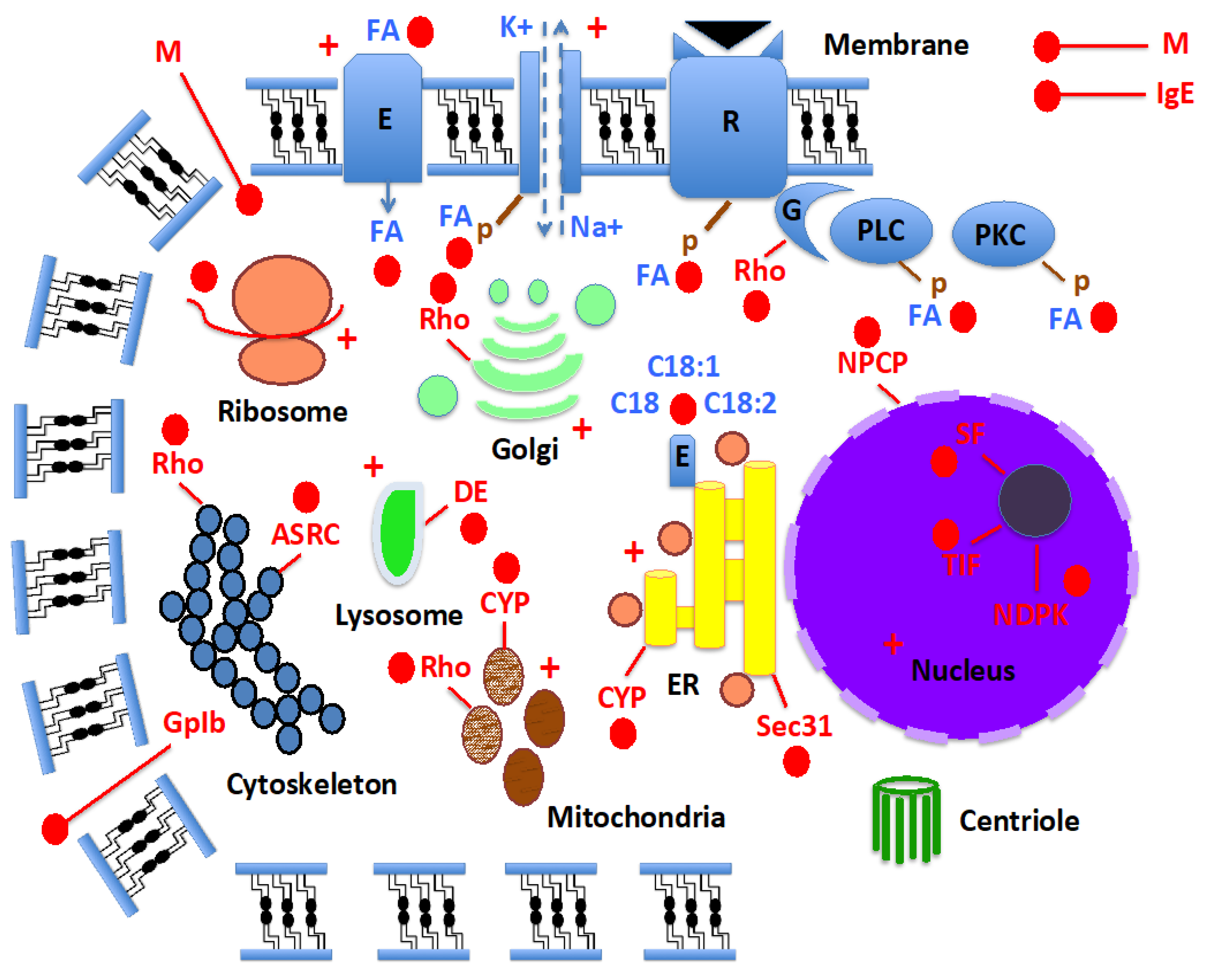
| Name | Organism | Authors | Year | Reference |
|---|---|---|---|---|
| p10 | Periplaneta americana | Nomura et al. | 1992 | |
| Ebsp-3/PebIII | Drosophila melanogaster | Dyanov et al. | 1994 | AAA87058* |
| A10 | Drosophila melanogaster | Pikielny et al. | 1994 | [41] |
| OS-D | Drosophila melanogaster | McKenna et al. | 1994 | [42] |
| Pam | Periplaneta americana | Picimbon & Leal | 1999 | [3] |
| CSP | Schistocerca gregaria | Angeli et al. | 1999 | [4] |
| SAP | Manduca sexta | Robertson et al. | 1999 | [43] |
| Pherokine | Drosophila melanogaster | Sabatier et al. | 2003 | [27] |
| Mp10 | Myzus persicae | Bos et al. | 2010 | [44] |
| Name | Organism | Authors | Year | Reference |
| Lipoclistin | Insects | Steinbrecht | 2003 | [202] |
| LA-BP | Bemisia tabaci | Liu et al. | 2016 | [38] |
| Toxin-BP | Bemisia tabaci | Liu et al. | 2016 | [38] |
| B-CSP | Acinetobacter baumannii | Liu et al. | 2019 | [45] |
| Lipid-BP | Bemisia tabaci | Liu et al. | 2020 | [39] |
| JHRP | Aedes aegypti | Picimbon | 2020 | [16] |
| Mucin module | Aedes aegypti | Liu et al. | 2024 | This paper |
| TIF module | Aedes aegypti | Liu et al. | 2024 | This paper |
| ASRP module | Aedes aegypti | Liu et al. | 2024 | This paper |
| NPCP module | Aedes aegypti | Liu et al. | 2024 | This paper |
| 4CSP* | Insects, crustaceans, and microbes | Liu et al. | 2024 | This paper |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




