Submitted:
30 September 2024
Posted:
01 October 2024
You are already at the latest version
Abstract
Keywords:
Introduction
- The disease
- Burden of the disease
- Pathogenesis of DFU
- Callus Formation: The initial stage involves the formation of a callus, a thickening of the skin under the foot that appears as a yellowish tinge. This condition is often caused by peripheral neuropathy, resulting from nerve damage in the foot, which can occasionally affect leg nerves as well. Symptoms of peripheral neuropathy include numbness, cramps, and muscle fatigue.
- Motor Neuropathy: In this stage, motor neurons in the foot are damaged, leading to weakness and deformation of the feet.
- Sensory Neuropathy: The final stage involves damage to sensory neurons, resulting in loss of sensation, which can lead to trauma, skin drying, and autonomic neuropathy. Frequent damage to the callus can result in subcutaneous hemorrhage, ultimately forming an ulcer [13].
- Grade 0: No visible ulcer
- Grade 1: Superficial ulcer
- Grade 2: Deep tissue ulceration
- Grade 3: Abscess formation involving bone
- Grade 4: Gangrene formation at the toe
- Grade 5: Extensive gangrene and necrosis throughout the foot [27]
- Detection of biofilms in DFU
- Methods to Study Biofilms in Diabetic Foot Ulcers
- In Vitro Methods
- 2-Dimensional cell culture model
- 3-D DFU model
- In Vivo methods
- The Ischemic animal ulcer model
- The Neuropathic animal ulcer model
- Diabetic Ulcer Model
- Data Analysis Methodology
- Biofilm formation
- Biofilm formation of gram-positive organisms in Diabetic Foot Ulcer
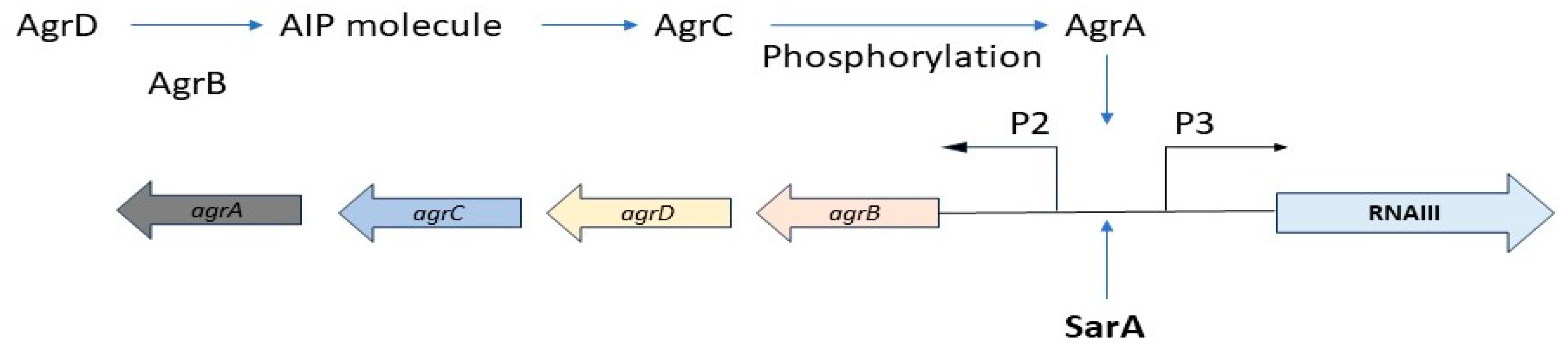
- Mechanism of Biofilm Formation in Staphylococcus aureus
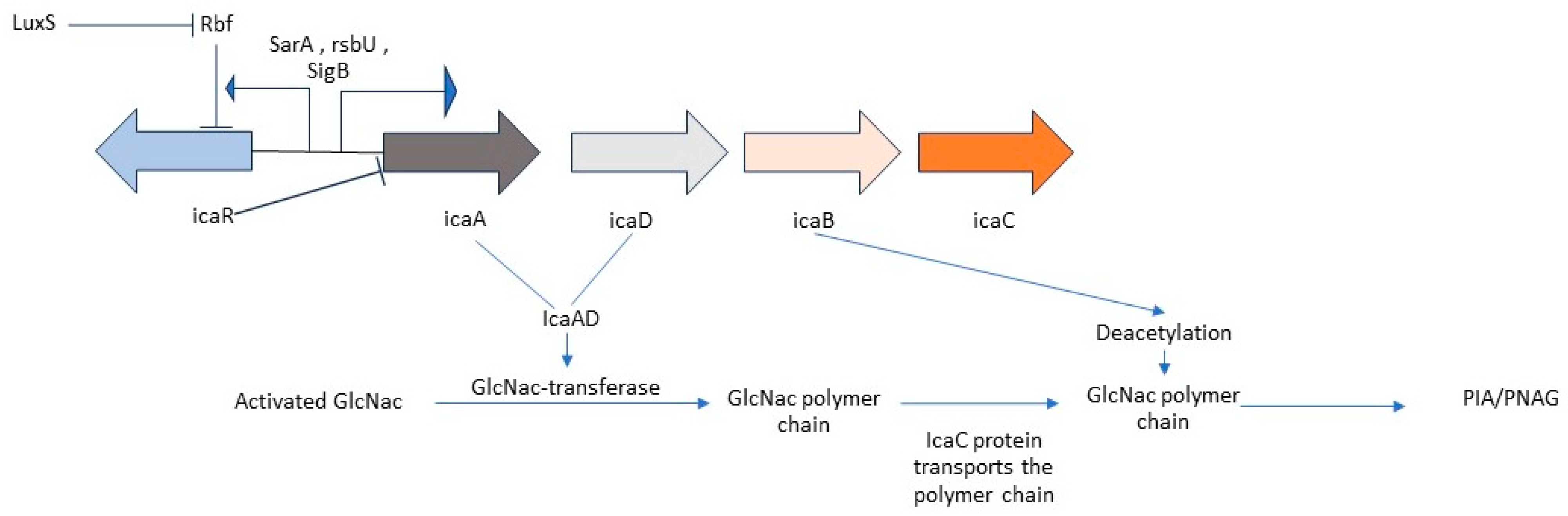
- Ica mediated biofilm formation
- Ica independent Biofilm formation in MRSA
- Biofilm formation of Gram-negative organisms in Diabetic Foot Ulcer
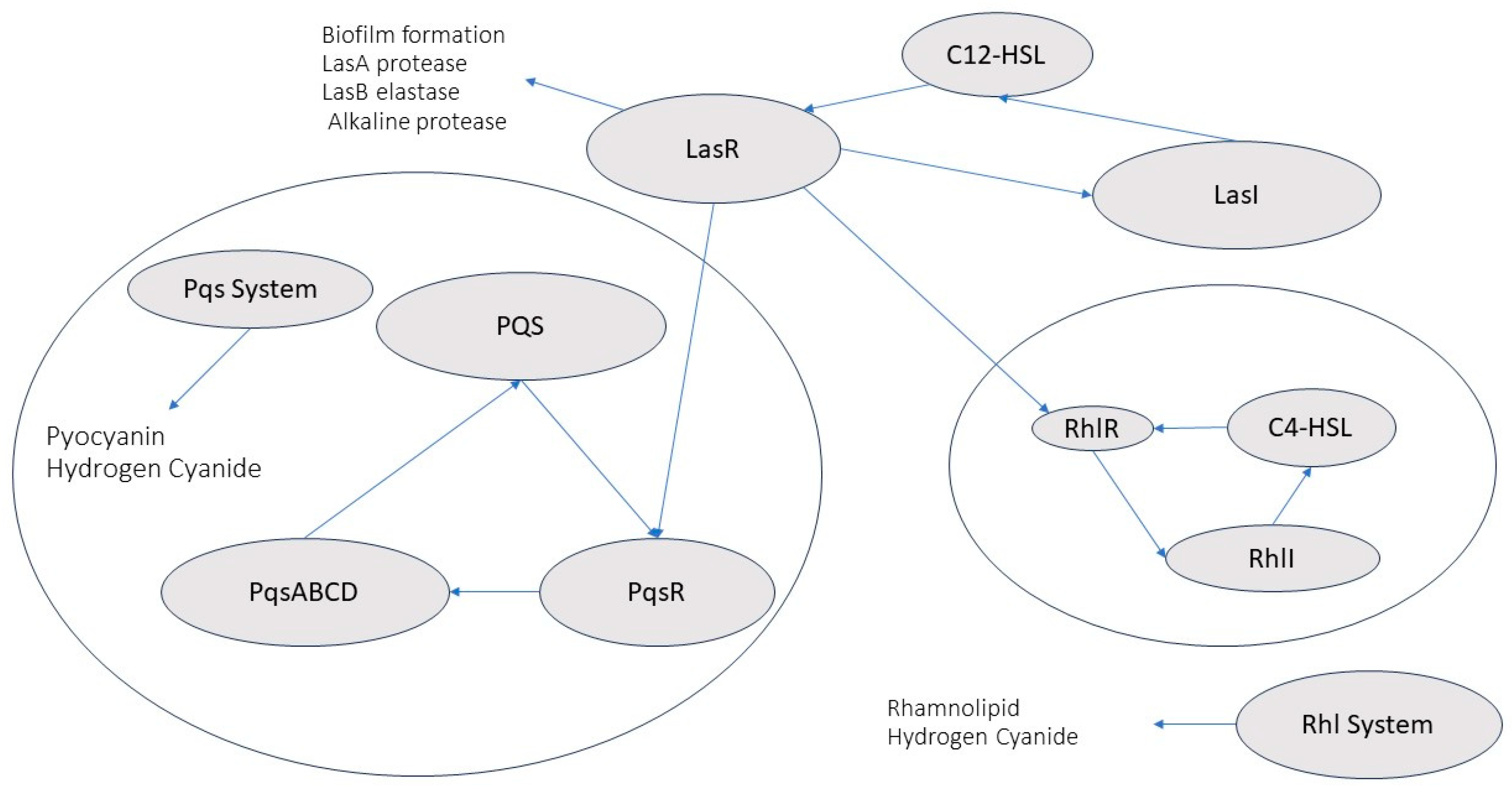
- Mechanism of Biofilm Formation in Pseudomonas aeruginosa
- Mechanism of Biofilm Formation in E.coli_
- Mechanism of Biofilm Formation in Klebsiella pneumoniae
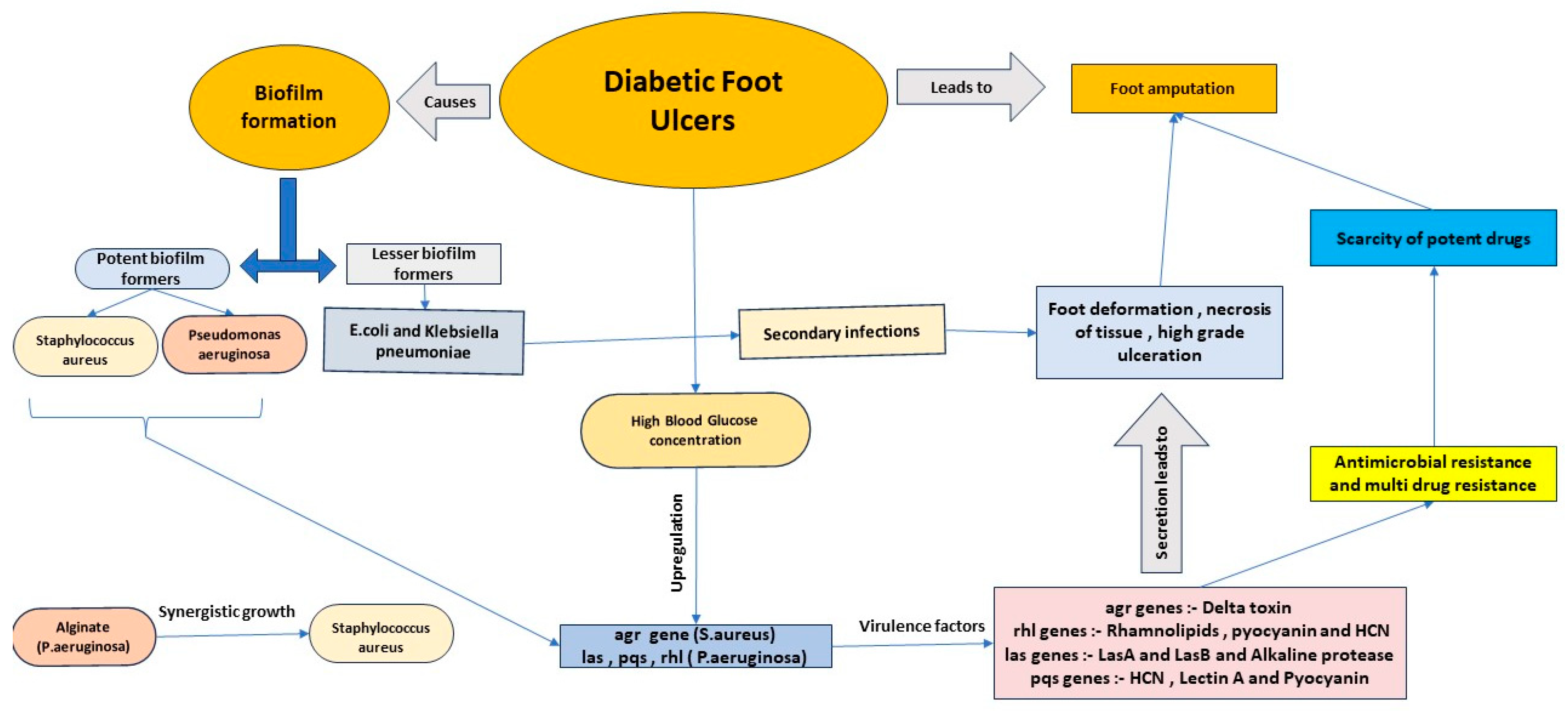
- Pathogenesis of Biofilm-Infected Diabetic Foot Ulcers (DFUs)
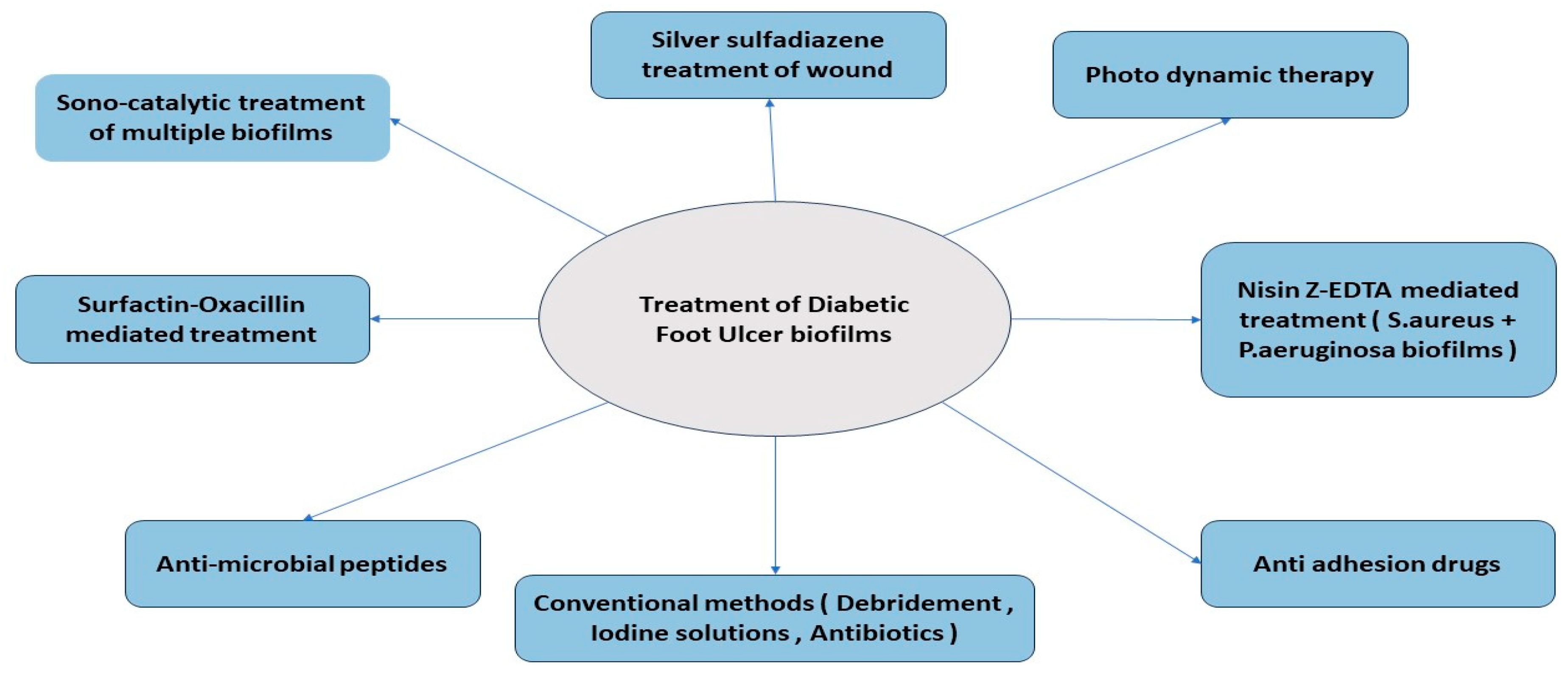
- Treatment of Biofilm in DFU
- Conventional Treatment
- Alternative Treatment Methods
- Other Anti-Biofilm Agents
| Treatment | Description | References |
|---|---|---|
|
Vancomycin Powder Bolus |
Large initial antibiotic concentration Concentration of Antibiotics decreases steadily and rapidly dropping below detectable levels (Rapid washout) No Zone of inhibition Potential side effect:
|
[219,221] |
| Calcium sulphate beads with PMMA loaded space (Vancomycin) |
Greater area under the concentration-time curve (AUC) compared to antibiotics-loaded PMMA space alone. Excessive wound drainage Potentially cytotoxic |
[221] |
| Tobramycin powder bolus | Large initial antibiotic concentration | [221] |
| Calcium sulphate beads with PMMA loaded space (Tobramycin) |
Greater area under the concentration-time curve (AUC) compared to antibiotics loaded PMMA space alone Largest concentration of antibiotics Potentially cytotoxic |
[221] |
| 26% (26 percent degree of substitution of the quaternary ammonium) HACC- loaded PMMA | Cytotoxic and interferes with proliferation and osteogenic differentiation of human bone marrow-derived mesenchymal stem cells with increasing degree of substitution of the quaternary ammonium. Potential obstacle:
|
[222] |
| Gentamicin-loaded PMMA |
|
[222] |
| Limonene |
|
[223] |
| Silver nanoparticle functionalized silicone elastomer |
|
[224] |
| Cold atmospheric plasma | Break peptidoglycan bond of gram-positive bacteria in biofilm | [225] |
| Phage lysins |
|
[226] |
| Mannosidase and glucanasa | Hydrolyses mannan-glucan in C.auris biofilm | [226] |
| Alginate lyase | Removes exopolysaccharide from the surface promoting biofilm eradication | [226] |
| Magnetic iron oxide nanoparticles with magnet fields | Causes mechanical damage to the matrix of the biofilm leading to eradication. | [226] |
| Magneto-responsive gallium-based liquid metal droplet | Disrupts matrix, results in bacterial lysis | [226] |
| DNase |
|
[226] |
| Carolacton |
|
[226] |
| Rhamnolipid | Disrupts and eradicate S. aureus biofilms | [226] |
| D-amino acids incorporation |
Potential side effect:
|
[226,227] |
| Rhamnolipid coated silver and iron oxide nanoparticles |
|
[226,228] |
| Linezolid | Targets Methicillin resistant S.aureus, Streptococci, Enterococci | [122,123] |
| Piperacillin /Tazobactam, Ticarcillin/ Clavulanic acid, Ampicillin/ Sulbactam |
Targets both gram-negative and gram-positive bacteria | [124,125] |
| Active Debridement | Removes necrotic tissue containing bacterial biofilms | [127,128] |
| Iodine solutions (Povidone Iodine 10%, Cadexomer Iodine) | Targets bacterial biofilms | [129,130,131] |
| Sono-catalytically activated C3N4 | Eradication of all types of bacterial biofilms and planktonic cells |
[132] |
| Photodynamic Therapy by Toluidine blue-chitosan coated Gold-Silver nano particles | Eradication of polymicrobial biofilms of P.aeruginosa and S.aureus |
[133,134] |
| Surfactin mediated Oxacillin treatment | Specifically targets S.aureus biofilms and cells (both multi-drug resistant and sensitive) | [135,136] |
| Nisin-EDTA mediated Treatment | Targets polymicrobial biofilms of S.aureus and P.aeruginosa | [137,138] |
| Silver Sulfadiazine | Effective for both S.aureus and P.aeruginosa (P.aeruginosa is more sensitive) | [139,140,141] |
| AMP 108 | Eradicates biofilms of A.baumanii , P.aeruginosa , K.pneumoniae ,S.aureus | [145,146,147] |
| Gingkgo biloba extract | Inhibits adhesion and curli genes of E.coli | [142,143] |
| Eugenol | Inhibits adhesion of E.coli | [142] |
| Phloretin | Inhibits adhesion of E.coli | [142,144] |
Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- P. She et al., ‘Effects of exogenous glucose on Pseudomonas aeruginosa biofilm formation and antibiotic resistance’, MicrobiologyOpen, vol. 8, no. 12, p. e933, Dec. 2019. [CrossRef]
- E. O’Neill et al., ‘A Novel Staphylococcus aureus Biofilm Phenotype Mediated by the Fibronectin-Binding Proteins, FnBPA and FnBPB’, J. Bacteriol., vol. 190, no. 11, pp. 3835–3850, Jun. 2008. [CrossRef]
- T. Bjarnsholt, ‘The role of bacterial biofilms in chronic infections’, APMIS, vol. 121, no. s136, pp. 1–58, May 2013. [CrossRef]
- L. Hall-Stoodley, J. W. Costerton, and P. Stoodley, ‘Bacterial biofilms: from the Natural environment to infectious diseases’, Nat. Rev. Microbiol., vol. 2, no. 2, pp. 95–108, Feb. 2004. [CrossRef]
- T.-F. C. Mah and G. A. O’Toole, ‘Mechanisms of biofilm resistance to antimicrobial agents’, Trends Microbiol., vol. 9, no. 1, pp. 34–39, Jan. 2001. [CrossRef]
- J. Hirschfeld, ‘Dynamic interactions of neutrophils and biofilms’, J. Oral Microbiol., vol. 6, no. 1, p. 26102, Jan. 2014. [CrossRef]
- K. K. Jefferson, ‘What drives bacteria to produce a biofilm?’, FEMS Microbiol. Lett., vol. 236, no. 2, pp. 163–173, Jul. 2004. [CrossRef]
- C. Mottola, J. J. Mendes, J. M. Cristino, P. Cavaco-Silva, L. Tavares, and M. Oliveira, ‘Polymicrobial biofilms by diabetic foot clinical isolates’, Folia Microbiol. (Praha), vol. 61, no. 1, pp. 35–43, Jan. 2016. [CrossRef]
- C. E. Price, D. G. Brown, D. H. Limoli, V. V. Phelan, and G. A. O’Toole, ‘Exogenous Alginate Protects Staphylococcus aureus from Killing by Pseudomonas aeruginosa’, J. Bacteriol., vol. 202, no. 8, Mar. 2020. [CrossRef]
- D. H. Limoli et al., ‘Pseudomonas aeruginosa Alginate Overproduction Promotes Coexistence with Staphylococcus aureus in a Model of Cystic Fibrosis Respiratory Infection’, mBio, vol. 8, no. 2, pp. e00186-17, May 2017. [CrossRef]
- M. Berlanga and R. Guerrero, ‘Living together in biofilms: the microbial cell factory and its biotechnological implications’, Microb. Cell Factories, vol. 15, no. 1, p. 165, Dec. 2016. [CrossRef]
- A. Banu, M. M. N. Hassan, J. Rajkumar, and S. Srinivasa, ‘Spectrum of bacteria associated with diabetic foot ulcer and biofilm formation: A prospective study’, Australas. Med. J., pp. 280–285, Sep. 2015. [CrossRef]
- D. G. Armstrong, A. J. M. Boulton, and S. A. Bus, ‘Diabetic Foot Ulcers and Their Recurrence’, N. Engl. J. Med., vol. 376, no. 24, pp. 2367–2375, Jun. 2017. [CrossRef]
- A. Hotterbeekx, S. Kumar-Singh, H. Goossens, and S. Malhotra-Kumar, ‘In vivo and In vitro Interactions between Pseudomonas aeruginosa and Staphylococcus spp.’, Front. Cell. Infect. Microbiol., vol. 7, Apr. 2017. [CrossRef]
- A. Atlaw, H. B. Kebede, A. A. Abdela, and Y. Woldeamanuel, ‘Bacterial isolates from diabetic foot ulcers and their antimicrobial resistance profile from selected hospitals in Addis Ababa, Ethiopia’, Front. Endocrinol., vol. 13, p. 987487, Aug. 2022. [CrossRef]
- F. W. Wagner, ‘The Dysvascular Foot: A System for Diagnosis and Treatment’, Foot Ankle, vol. 2, no. 2, pp. 64–122, Sep. 1981. [CrossRef]
- S. Vazeille, L. Hawker, R. Chandrasekar, and U. Srinivas-Shankar, ‘Recalcitrant Foot Ulceration in a Patient With Type 1 Diabetes Mellitus’, Cureus, Jun. 2020. [CrossRef]
- X. Liao, S.-H. Li, M. M. El Akkawi, X. Fu, H. Liu, and Y. Huang, ‘Surgical amputation for patients with diabetic foot ulcers: A Chinese expert panel consensus treatment guide’, Front. Surg., vol. 9, p. 1003339, Nov. 2022. [CrossRef]
- N. M. Protzman et al., ‘Placental-Derived Biomaterials and Their Application to Wound Healing: A Review’, Bioengineering, vol. 10, no. 7, p. 829, Jul. 2023. [CrossRef]
- W. Tettelbach et al., ‘A multicentre prospective randomised controlled comparative parallel study of dehydrated human umbilical cord (EpiCord) allograft for the treatment of diabetic foot ulcers’, Int. Wound J., vol. 16, no. 1, pp. 122–130, Feb. 2019. [CrossRef]
- D. Magliano and E. J. Boyko, IDF diabetes atlas, 10th edition. Brussels: International Diabetes Federation, 2021.
- F. Crawford et al., ‘The risk of foot ulceration in people with diabetes screened in community settings: findings from a cohort study’, QJM Int. J. Med., vol. 104, no. 5, pp. 403–410, May 2011. [CrossRef]
- C. A. Abbott et al., ‘The North-West Diabetes Foot Care Study: incidence of, and risk factors for, new diabetic foot ulceration in a community-based patient cohort’, Diabet. Med., vol. 19, no. 5, pp. 377–384, May 2002. [CrossRef]
- A. Raghav, Z. A. Khan, R. K. Labala, J. Ahmad, S. Noor, and B. K. Mishra, ‘Financial burden of diabetic foot ulcers to world: a progressive topic to discuss always’, Ther. Adv. Endocrinol. Metab., vol. 9, no. 1, pp. 29–31, Jan. 2018. [CrossRef]
- A. Ibrahim, ‘IDF Clinical Practice Recommendation on the Diabetic Foot: A guide for healthcare professionals’, Diabetes Res. Clin. Pract., vol. 127, pp. 285–287, May 2017. [CrossRef]
- Z. Yekta, Pourali, and M. Ghasemi-rad, ‘Comparison of demographic and clinical characteristics influencing health-related quality of life in patients with diabetic foot ulcers and those without foot ulcers’, Diabetes Metab. Syndr. Obes. Targets Ther., p. 393, Dec. 2011. [CrossRef]
- P. Shah et al., ‘Wagner’s Classification as a Tool for Treating Diabetic Foot Ulcers: Our Observations at a Suburban Teaching Hospital’, Cureus, Jan. 2022. [CrossRef]
- O. Niță et al., ‘Evaluating Classification Systems of Diabetic Foot Ulcer Severity: A 12-Year Retrospective Study on Factors Impacting Survival’, Healthcare, vol. 11, no. 14, p. 2077, Jul. 2023. [CrossRef]
- K. Ottolino-Perry et al., ‘Improved detection of clinically relevant wound bacteria using autofluorescence image-guided sampling in diabetic foot ulcers’, Int. Wound J., vol. 14, no. 5, pp. 833–841, Oct. 2017. [CrossRef]
- N. Høiby et al., ‘ESCMID∗ guideline for the diagnosis and treatment of biofilm infections 2014’, Clin. Microbiol. Infect., vol. 21, pp. S1–S25, May 2015. [CrossRef]
- S. Sugimoto et al., ‘Staphylococcus epidermidis Esp Degrades Specific Proteins Associated with Staphylococcus aureus Biofilm Formation and Host-Pathogen Interaction’, J. Bacteriol., vol. 195, no. 8, pp. 1645–1655, Apr. 2013. [CrossRef]
- T. S. Murali et al., ‘Characteristics of microbial drug resistance and its correlates in chronic diabetic foot ulcer infections’, J. Med. Microbiol., vol. 63, no. 10, pp. 1377–1385, Oct. 2014. [CrossRef]
- D. Kumar, T. Banerjee, J. Chakravarty, S. Singh, A. Dwivedi, and R. Tilak, ‘Identification, antifungal resistance profile, in vitro biofilm formation and ultrastructural characteristics of Candida species isolated from diabetic foot patients in Northern India’, Indian J. Med. Microbiol., vol. 34, no. 3, pp. 308–314, Jul. 2016. [CrossRef]
- C.-C. E. Lan, C.-S. Wu, H.-Y. Kuo, S.-M. Huang, and G.-S. Chen, ‘Hyperglycaemic conditions hamper keratinocyte locomotion via sequential inhibition of distinct pathways: new insights on poor wound closure in patients with diabetes’, Br. J. Dermatol., vol. 160, no. 6, pp. 1206–1214, Jun. 2009. [CrossRef]
- A. G. Maione et al., ‘Altered ECM deposition by diabetic foot ulcer-derived fibroblasts implicates fibronectin in chronic wound repair’, Wound Repair Regen., vol. 24, no. 4, pp. 630–643, Jul. 2016. [CrossRef]
- B. Wang, P. Chandrasekera, and J. Pippin, ‘Leptin- and Leptin Receptor-Deficient Rodent Models: Relevance for Human Type 2 Diabetes’, Curr. Diabetes Rev., vol. 10, no. 2, pp. 131–145, May 2014. [CrossRef]
- P. M. Pitale and M. S. Gorbatyuk, ‘Diabetic Retinopathy: From Animal Models to Cellular Signaling’, Int. J. Mol. Sci., vol. 23, no. 3, p. 1487, Jan. 2022. [CrossRef]
- J. Michaels et al., ‘db/db mice exhibit severe wound-healing impairments compared with other murine diabetic strains in a silicone-splinted excisional wound model’, Wound Repair Regen., vol. 15, no. 5, pp. 665–670, Sep. 2007. [CrossRef]
- C.-H. Jung, Y. Y. Cho, D. Choi, B.-Y. Kim, C.-H. Kim, and J.-O. Mok, ‘Relationship of Sarcopenia with Microcirculation Measured by Skin Perfusion Pressure in Patients with Type 2 Diabetes’, Endocrinol. Metab., vol. 35, no. 3, pp. 578–586, Sep. 2020. [CrossRef]
- L. M. Rodrigues, C. Rocha, H. Ferreira, and H. Silva, ‘Different lasers reveal different skin microcirculatory flowmotion - data from the wavelet transform analysis of human hindlimb perfusion’, Sci. Rep., vol. 9, no. 1, p. 16951, Nov. 2019. [CrossRef]
- S. A. Cudlip, F. A. Howe, J. R. Griffiths, and B. A. Bell, ‘Magnetic resonance neurography of peripheral nerve following experimental crush injury, and correlation with functional deficit’, J. Neurosurg., vol. 96, no. 4, pp. 755–759, Apr. 2002. [CrossRef]
- S. Jin, M. Zhang, Y. Gao, X. Zhang, G. Cui, and Y. Zhang, ‘The Efficacy of Jing Wan Hong Ointment for Nerve Injury Diabetic Foot Ulcer and Its Mechanisms’, J. Diabetes Res., vol. 2014, pp. 1–9, 2014. [CrossRef]
- J. Rich and J. C. Lee, ‘The Pathogenesis of Staphylococcus aureus Infection in the Diabetic NOD Mouse’, Diabetes, vol. 54, no. 10, pp. 2904–2910, Oct. 2005. [CrossRef]
- M. Y. El-Naggar, Y. M. Gohar, M. A. Sorour, and M. G. Waheeb, ‘Hydrogel Dressing with a Nano-Formula against Methicillin-Resistant Staphylococcus aureus and Pseudomonas aeruginosa Diabetic Foot Bacteria’, J. Microbiol. Biotechnol., vol. 26, no. 2, pp. 408–420, Feb. 2016. [CrossRef]
- J.-P. Rasigade et al., ‘A Prophage in Diabetic Foot Ulcer–Colonizing Staphylococcus aureus Impairs Invasiveness by Limiting Intracellular Growth’, J. Infect. Dis., vol. 214, no. 10, pp. 1605–1608, Nov. 2016. [CrossRef]
- J.-F. Huon et al., ‘Phages versus Antibiotics To Treat Infected Diabetic Wounds in a Mouse Model: a Microbiological and Microbiotic Evaluation’, mSystems, vol. 5, no. 6, pp. e00542-20, Dec. 2020. [CrossRef]
- D. E. Snow et al., ‘The presence of biofilm structures in atherosclerotic plaques of arteries from legs amputated as a complication of diabetic foot ulcers’, J. Wound Care, vol. 25, no. Sup2, pp. S16–S22, Feb. 2016. [CrossRef]
- F. A. Al-Joufi et al., ‘Microbial spectrum, antibiotic susceptibility profile, and biofilm formation of diabetic foot infections (2014–18): a retrospective multicenter analysis’, 3 Biotech, vol. 10, no. 7, p. 325, Jul. 2020. [CrossRef]
- S. J. Steindel, ‘International classification of diseases, 10th edition, clinical modification and procedure coding system: descriptive overview of the next generation HIPAA code sets’, J. Am. Med. Inform. Assoc., vol. 17, no. 3, pp. 274–282, May 2010. [CrossRef]
- Dr. M. Mehraj, ‘A review of Wagner classification and current concepts in management of diabetic foot’, Int. J. Orthop. Sci., vol. 4, no. 1n, pp. 933–935, Jan. 2018. [CrossRef]
- P. Ince et al., ‘Use of the SINBAD Classification System and Score in Comparing Outcome of Foot Ulcer Management on Three Continents’, Diabetes Care, vol. 31, no. 5, pp. 964–967, May 2008. [CrossRef]
- J. L. Mills et al., ‘The Society for Vascular Surgery Lower Extremity Threatened Limb Classification System: Risk stratification based on Wound, Ischemia, and foot Infection (WIfI)’, J. Vasc. Surg., vol. 59, no. 1, pp. 220-234.e2, Jan. 2014. [CrossRef]
- F. R. Martínez-De Jesús, ‘A Checklist System to Score Healing Progress of Diabetic Foot Ulcers’, Int. J. Low. Extrem. Wounds, vol. 9, no. 2, pp. 74–83, Jun. 2010. [CrossRef]
- K. E. Macdonald, S. Boeckh, H. J. Stacey, and J. D. Jones, ‘The microbiology of diabetic foot infections: a meta-analysis’, BMC Infect. Dis., vol. 21, no. 1, p. 770, Dec. 2021. [CrossRef]
- M. J. Islam, K. Bagale, P. P. John, Z. Kurtz, and R. Kulkarni, ‘Glycosuria Alters Uropathogenic Escherichia coli Global Gene Expression and Virulence’, mSphere, vol. 7, no. 3, pp. e00004-22, Jun. 2022. [CrossRef]
- L. Korbel and J. D. Spencer, ‘Diabetes mellitus and infection: an evaluation of hospital utilization and management costs in the United States’, J. Diabetes Complications, vol. 29, no. 2, pp. 192–195, Mar. 2015. [CrossRef]
- S. Mohanty et al., ‘Diabetes downregulates the antimicrobial peptide psoriasin and increases E. coli burden in the urinary bladder’, Nat. Commun., vol. 13, no. 1, p. 4983, Sep. 2022. [CrossRef]
- C. Pouget, C. Dunyach-Remy, A. Pantel, S. Schuldiner, A. Sotto, and J.-P. Lavigne, ‘Biofilms in Diabetic Foot Ulcers: Significance and Clinical Relevance’, Microorganisms, vol. 8, no. 10, p. 1580, Oct. 2020. [CrossRef]
- C. Khadka et al., ‘Extended-spectrum β-lactamases producing Enterobacteriaceae (ESBL-PE) prevalence in Nepal: A systematic review and meta-analysis’, Sci. Total Environ., vol. 901, p. 166164, Nov. 2023. [CrossRef]
- R. Dziri et al., ‘Characterization of extended-spectrum β-lactamase (ESBL)-producing Klebsiella, Enterobacter , and Citrobacter obtained in environmental samples of a Tunisian hospital’, Diagn. Microbiol. Infect. Dis., vol. 86, no. 2, pp. 190–193, Oct. 2016. [CrossRef]
- I. Kyriakidis, E. Vasileiou, Z. D. Pana, and A. Tragiannidis, ‘Acinetobacter baumannii Antibiotic Resistance Mechanisms’, Pathogens, vol. 10, no. 3, p. 373, Mar. 2021. [CrossRef]
- D. Mack et al., ‘The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis’, J. Bacteriol., vol. 178, no. 1, pp. 175–183, Jan. 1996. [CrossRef]
- C. Gerke, A. Kraft, R. Süßmuth, O. Schweitzer, and F. Götz, ‘Characterization of theN-Acetylglucosaminyltransferase Activity Involved in the Biosynthesis of the Staphylococcus epidermidisPolysaccharide Intercellular Adhesin’, J. Biol. Chem., vol. 273, no. 29, pp. 18586–18593, Jul. 1998. [CrossRef]
- C. Vuong et al., ‘A Crucial Role for Exopolysaccharide Modification in Bacterial Biofilm Formation, Immune Evasion, and Virulence’, J. Biol. Chem., vol. 279, no. 52, pp. 54881–54886, Dec. 2004. [CrossRef]
- K. K. Jefferson, D. B. Pier, D. A. Goldmann, and G. B. Pier, ‘The Teicoplanin-Associated Locus Regulator (TcaR) and the Intercellular Adhesin Locus Regulator (IcaR) Are Transcriptional Inhibitors of the ica Locus in Staphylococcus aureus’, J. Bacteriol., vol. 186, no. 8, pp. 2449–2456, Apr. 2004. [CrossRef]
- L. Xu et al., ‘Role of the luxS Quorum-Sensing System in Biofilm Formation and Virulence of Staphylococcus epidermidis’, Infect. Immun., vol. 74, no. 1, pp. 488–496, Jan. 2006. [CrossRef]
- J. K.-M. Knobloch, K. Bartscht, A. Sabottke, H. Rohde, H.-H. Feucht, and D. Mack, ‘Biofilm Formation by Staphylococcus epidermidis Depends on Functional RsbU, an Activator of the sigB Operon: Differential Activation Mechanisms Due to Ethanol and Salt Stress’, J. Bacteriol., vol. 183, no. 8, pp. 2624–2633, Apr. 2001. [CrossRef]
- J. Valle et al., ‘SarA and not σ B is essential for biofilm development by Staphylococcus aureus’, Mol. Microbiol., vol. 48, no. 4, pp. 1075–1087, May 2003. [CrossRef]
- D. Frees et al., ‘Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus’, Mol. Microbiol., vol. 54, no. 5, pp. 1445–1462, Dec. 2004. [CrossRef]
- A. L. Cheung and S. J. Projan, ‘Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr’, J. Bacteriol., vol. 176, no. 13, pp. 4168–4172, Jul. 1994. [CrossRef]
- K. E. Beenken et al., ‘Global Gene Expression in Staphylococcus aureus Biofilms’, J. Bacteriol., vol. 186, no. 14, pp. 4665–4684, Jul. 2004. [CrossRef]
- R. H. Deurenberg and E. E. Stobberingh, ‘The evolution of Staphylococcus aureus’, Infect. Genet. Evol., vol. 8, no. 6, pp. 747–763, Dec. 2008. [CrossRef]
- C. Vuong, H. L. Saenz, F. Götz, and M. Otto, ‘Impact of the agr Quorum-Sensing System on Adherence to Polystyrene in Staphylococcus aureus’, J. Infect. Dis., vol. 182, no. 6, pp. 1688–1693, Dec. 2000. [CrossRef]
- E. O’Neill et al., ‘Association between Methicillin Susceptibility and Biofilm Regulation in Staphylococcus aureus Isolates from Device-Related Infections’, J. Clin. Microbiol., vol. 45, no. 5, pp. 1379–1388, May 2007. [CrossRef]
- F. Fitzpatrick, H. Humphreys, and J. P. O’Gara, ‘Environmental regulation of biofilm development in methicillin-resistant and methicillin-susceptible Staphylococcus aureus clinical isolates’, J. Hosp. Infect., vol. 62, no. 1, pp. 120–122, Jan. 2006. [CrossRef]
- N. Merino et al., ‘Protein A-Mediated Multicellular Behavior in Staphylococcus aureus’, J. Bacteriol., vol. 191, no. 3, pp. 832–843, Feb. 2009. [CrossRef]
- K. C. Rice et al., ‘The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus’, Proc. Natl. Acad. Sci., vol. 104, no. 19, pp. 8113–8118, May 2007. [CrossRef]
- J. L. Bose, M. K. Lehman, P. D. Fey, and K. W. Bayles, ‘Contribution of the Staphylococcus aureus Atl AM and GL Murein Hydrolase Activities in Cell Division, Autolysis, and Biofilm Formation’, PLoS ONE, vol. 7, no. 7, p. e42244, Jul. 2012. [CrossRef]
- P. Houston, S. E. Rowe, C. Pozzi, E. M. Waters, and J. P. O’Gara, ‘Essential Role for the Major Autolysin in the Fibronectin-Binding Protein-Mediated Staphylococcus aureus Biofilm Phenotype’, Infect. Immun., vol. 79, no. 3, pp. 1153–1165, Mar. 2011. [CrossRef]
- M. Vergara-Irigaray et al., ‘Relevant Role of Fibronectin-Binding Proteins in Staphylococcus aureus Biofilm-Associated Foreign-Body Infections’, Infect. Immun., vol. 77, no. 9, pp. 3978–3991, Sep. 2009. [CrossRef]
- D. Passos Da Silva et al., ‘The Pseudomonas aeruginosa lectin LecB binds to the exopolysaccharide Psl and stabilizes the biofilm matrix’, Nat. Commun., vol. 10, no. 1, p. 2183, May 2019. [CrossRef]
- P. Nadal Jimenez, G. Koch, J. A. Thompson, K. B. Xavier, R. H. Cool, and W. J. Quax, ‘The Multiple Signaling Systems Regulating Virulence in Pseudomonas aeruginosa’, Microbiol. Mol. Biol. Rev., vol. 76, no. 1, pp. 46–65, Mar. 2012. [CrossRef]
- K. B. Gilbert, T. H. Kim, R. Gupta, E. P. Greenberg, and M. Schuster, ‘Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR’, Mol. Microbiol., vol. 73, no. 6, pp. 1072–1085, Sep. 2009. [CrossRef]
- D. H. Dusane, S. S. Zinjarde, V. P. Venugopalan, R. J. Mclean, M. M. Weber, and P. K. S. M. Rahman, ‘Quorum sensing: implications on Rhamnolipid biosurfactant production’, Biotechnol. Genet. Eng. Rev., vol. 27, no. 1, pp. 159–184, Jan. 2010. [CrossRef]
- M. Allesen-Holm et al., ‘A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms’, Mol. Microbiol., vol. 59, no. 4, pp. 1114–1128, Feb. 2006. [CrossRef]
- A. Brencic et al., ‘The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs’, Mol. Microbiol., vol. 73, no. 3, pp. 434–445, Aug. 2009. [CrossRef]
- Y. Irie, M. Starkey, A. N. Edwards, D. J. Wozniak, T. Romeo, and M. R. Parsek, ‘Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA’, Mol. Microbiol., vol. 78, no. 1, pp. 158–172, Oct. 2010. [CrossRef]
- A. L. Goodman, B. Kulasekara, A. Rietsch, D. Boyd, R. S. Smith, and S. Lory, ‘A Signaling Network Reciprocally Regulates Genes Associated with Acute Infection and Chronic Persistence in Pseudomonas aeruginosa’, Dev. Cell, vol. 7, no. 5, pp. 745–754, Nov. 2004. [CrossRef]
- D.-G. Ha and G. A. O’Toole, ‘c-di-GMP and its Effects on Biofilm Formation and Dispersion: a Pseudomonas Aeruginosa Review’, Microbiol. Spectr., vol. 3, no. 2, p. 3.2.27, Apr. 2015. [CrossRef]
- D. Apel and M. G. Surette, ‘Bringing order to a complex molecular machine: The assembly of the bacterial flagella’, Biochim. Biophys. Acta BBA - Biomembr., vol. 1778, no. 9, pp. 1851–1858, Sep. 2008. [CrossRef]
- R. M. Macnab, ‘GENETICS AND BIOGENESIS OF BACTERIAL FLAGELLA’, Annu. Rev. Genet., vol. 26, no. 1, pp. 131–158, Dec. 1992. [CrossRef]
- D. M. Fitzgerald, R. P. Bonocora, and J. T. Wade, ‘Comprehensive Mapping of the Escherichia coli Flagellar Regulatory Network’, PLoS Genet., vol. 10, no. 10, p. e1004649, Oct. 2014. [CrossRef]
- G. Capitani, O. Eidam, R. Glockshuber, and M. G. Grütter, ‘Structural and functional insights into the assembly of type 1 pili from Escherichia coli’, Microbes Infect., vol. 8, no. 8, pp. 2284–2290, Jul. 2006. [CrossRef]
- C. Beloin, A. Roux, and J.-M. Ghigo, ‘Escherichia coli Biofilms’, in Bacterial Biofilms, vol. 322, T. Romeo, Ed., in Current Topics in Microbiology and Immunology, vol. 322. , Berlin, Heidelberg: Springer Berlin Heidelberg, 2008, pp. 249–289. [CrossRef]
- M. L. Valenski, S. L. Harris, P. A. Spears, J. R. Horton, and P. E. Orndorff, ‘The Product of the fimI Gene Is Necessary for Escherichia coli Type 1 Pilus Biosynthesis’, J. Bacteriol., vol. 185, no. 16, pp. 5007–5011, Aug. 2003. [CrossRef]
- M. M. Barnhart and M. R. Chapman, ‘Curli Biogenesis and Function’, Annu. Rev. Microbiol., vol. 60, no. 1, pp. 131–147, Oct. 2006. [CrossRef]
- L. Gualdi, L. Tagliabue, S. Bertagnoli, T. Ieranò, C. De Castro, and P. Landini, ‘Cellulose modulates biofilm formation by counteracting curli-mediated colonization of solid surfaces in Escherichia coli’, Microbiology, vol. 154, no. 7, pp. 2017–2024, Jul. 2008. [CrossRef]
- P. Klemm and M. Schembri, ‘Type 1 Fimbriae, Curli, and Antigen 43: Adhesion, Colonization, and Biofilm Formation’, EcoSal Plus, vol. 1, no. 1, p. 10.1128/ecosalplus.8.3.2.6, Dec. 2004. [CrossRef]
- L. Laganenka, R. Colin, and V. Sourjik, ‘Chemotaxis towards autoinducer 2 mediates autoaggregation in Escherichia coli’, Nat. Commun., vol. 7, no. 1, p. 12984, Sep. 2016. [CrossRef]
- Y. Itoh et al., ‘Roles of pgaABCD Genes in Synthesis, Modification, and Export of the Escherichia coli Biofilm Adhesin Poly-β-1,6- N -Acetyl- d -Glucosamine’, J. Bacteriol., vol. 190, no. 10, pp. 3670–3680, May 2008. [CrossRef]
- X. Wang, J. F. Preston, and T. Romeo, ‘The pgaABCD Locus of Escherichia coli Promotes the Synthesis of a Polysaccharide Adhesin Required for Biofilm Formation’, J. Bacteriol., vol. 186, no. 9, pp. 2724–2734, May 2004. [CrossRef]
- D. O. Serra, A. M. Richter, and R. Hengge, ‘Cellulose as an Architectural Element in Spatially Structured Escherichia coli Biofilms’, J. Bacteriol., vol. 195, no. 24, pp. 5540–5554, Dec. 2013. [CrossRef]
- U. Römling, M. Y. Galperin, and M. Gomelsky, ‘Cyclic di-GMP: the First 25 Years of a Universal Bacterial Second Messenger’, Microbiol. Mol. Biol. Rev., vol. 77, no. 1, pp. 1–52, Mar. 2013. [CrossRef]
- J. A. Markova, E. V. Anganova, A. L. Turskaya, V. A. Bybin, and E. D. Savilov, ‘Regulation of Escherichia coli Biofilm Formation (Review)’, Appl. Biochem. Microbiol., vol. 54, no. 1, pp. 1–11, Jan. 2018. [CrossRef]
- C. Prigent-Combaret et al., ‘Complex Regulatory Network Controls Initial Adhesion and Biofilm Formation in Escherichia coli via Regulation of the csgD Gene’, J. Bacteriol., vol. 183, no. 24, pp. 7213–7223, Dec. 2001. [CrossRef]
- B. M. Prüß, C. Besemann, A. Denton, and A. J. Wolfe, ‘A Complex Transcription Network Controls the Early Stages of Biofilm Development by Escherichia coli’, J. Bacteriol., vol. 188, no. 11, pp. 3731–3739, Jun. 2006. [CrossRef]
- N. Majdalani and S. Gottesman, ‘THE RCS PHOSPHORELAY: A Complex Signal Transduction System’, Annu. Rev. Microbiol., vol. 59, no. 1, pp. 379–405, Oct. 2005. [CrossRef]
- C. S. Pereira, J. A. Thompson, and K. B. Xavier, ‘AI-2-mediated signalling in bacteria’, FEMS Microbiol. Rev., vol. 37, no. 2, pp. 156–181, Mar. 2013. [CrossRef]
- T. Pacheco et al., ‘SdiA, a Quorum-Sensing Regulator, Suppresses Fimbriae Expression, Biofilm Formation, and Quorum-Sensing Signaling Molecules Production in Klebsiella pneumoniae’, Front. Microbiol., vol. 12, p. 597735, Jun. 2021. [CrossRef]
- C. De Araujo, D. Balestrino, L. Roth, N. Charbonnel, and C. Forestier, ‘Quorum sensing affects biofilm formation through lipopolysaccharide synthesis in Klebsiella pneumoniae’, Res. Microbiol., vol. 161, no. 7, pp. 595–603, Sep. 2010. [CrossRef]
- J. G. Johnson and S. Clegg, ‘Role of MrkJ, a Phosphodiesterase, in Type 3 Fimbrial Expression and Biofilm Formation in Klebsiella pneumoniae’, J. Bacteriol., vol. 192, no. 15, pp. 3944–3950, Aug. 2010. [CrossRef]
- D. A. Rosen, J. S. Pinkner, J. M. Jones, J. N. Walker, S. Clegg, and S. J. Hultgren, ‘Utilization of an Intracellular Bacterial Community Pathway in Klebsiella pneumoniae Urinary Tract Infection and the Effects of FimK on Type 1 Pilus Expression’, Infect. Immun., vol. 76, no. 7, pp. 3337–3345, Jul. 2008. [CrossRef]
- J. G. Johnson, C. N. Murphy, J. Sippy, T. J. Johnson, and S. Clegg, ‘Type 3 Fimbriae and Biofilm Formation Are Regulated by the Transcriptional Regulators MrkHI in Klebsiella pneumoniae’, J. Bacteriol., vol. 193, no. 14, pp. 3453–3460, Jul. 2011. [CrossRef]
- J. Zheng et al., ‘Biofilm Formation in Klebsiella pneumoniae Bacteremia Strains Was Found to be Associated with CC23 and the Presence of wcaG’, Front. Cell. Infect. Microbiol., vol. 8, p. 21, Feb. 2018. [CrossRef]
- C. Vuotto et al., ‘Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains’, J. Appl. Microbiol., vol. 123, no. 4, pp. 1003–1018, Oct. 2017. [CrossRef]
- J. J. Wilksch et al., ‘MrkH, a Novel c-di-GMP-Dependent Transcriptional Activator, Controls Klebsiella pneumoniae Biofilm Formation by Regulating Type 3 Fimbriae Expression’, PLoS Pathog., vol. 7, no. 8, p. e1002204, Aug. 2011. [CrossRef]
- T.-H. Lin, C.-Y. Tseng, Y.-C. Lai, C.-C. Wu, C.-F. Huang, and C.-T. Lin, ‘IscR Regulation of Type 3 Fimbriae Expression in Klebsiella pneumoniae CG43’, Front. Microbiol., vol. 8, p. 1984, Oct. 2017. [CrossRef]
- J. F. González, M. M. Hahn, and J. S. Gunn, ‘Chronic biofilm-based infections: skewing of the immune response’, Pathog. Dis., vol. 76, no. 3, Apr. 2018. [CrossRef]
- I. Alav, J. M. Sutton, and K. M. Rahman, ‘Role of bacterial efflux pumps in biofilm formation’, J. Antimicrob. Chemother., vol. 73, no. 8, pp. 2003–2020, Aug. 2018. [CrossRef]
- Z. Yekta, Pourali, M. Ghasemi-rad, Ravanyar, and Nezhadrahim, ‘Clinical and behavioral factors associated with management outcome in hospitalized patients with diabetic foot ulcer’, Diabetes Metab. Syndr. Obes. Targets Ther., p. 371, Oct. 2011. [CrossRef]
- J. V. Eggert, E. R. Worth, and C. C. Van Gils, ‘Cost and mortality data of a regional limb salvage and hyperbaric medicine program for Wagner Grade 3 or 4 diabetic foot ulcers’, Undersea Hyperb. Med. J. Undersea Hyperb. Med. Soc. Inc, vol. 43, no. 1, pp. 1–8, 2016.
- A. Shariati, M. Dadashi, M. T. Moghadam, A. Van Belkum, S. Yaslianifard, and D. Darban-Sarokhalil, ‘Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis’, Sci. Rep., vol. 10, no. 1, p. 12689, Jul. 2020. [CrossRef]
- B. P. Howden, J. K. Davies, P. D. R. Johnson, T. P. Stinear, and M. L. Grayson, ‘Reduced Vancomycin Susceptibility in Staphylococcus aureus , Including Vancomycin-Intermediate and Heterogeneous Vancomycin-Intermediate Strains: Resistance Mechanisms, Laboratory Detection, and Clinical Implications’, Clin. Microbiol. Rev., vol. 23, no. 1, pp. 99–139, Jan. 2010. [CrossRef]
- K. Bush and P. A. Bradford, ‘β-Lactams and β-Lactamase Inhibitors: An Overview’, Cold Spring Harb. Perspect. Med., vol. 6, no. 8, p. a025247, Aug. 2016. [CrossRef]
- M. Karmaker, S. K. Sanyal, M. Sultana, and M. A. Hossain, ‘Association of bacteria in diabetic and non-diabetic foot infection – An investigation in patients from Bangladesh’, J. Infect. Public Health, vol. 9, no. 3, pp. 267–277, May 2016. [CrossRef]
- W. D. Aumiller and H. A. Dollahite, ‘Pathogenesis and management of diabetic foot ulcers’, J. Am. Acad. Physician Assist., vol. 28, no. 5, pp. 28–34, May 2015. [CrossRef]
- D. G. Armstrong, L. A. Lavery, B. P. Nixon, and A. J. M. Boulton, ‘It’s Not What You Put On, but What You Take Off: Techniques for Debriding and Off-Loading the Diabetic Foot Wound’, Clin. Infect. Dis., vol. 39, no. Supplement_2, pp. S92–S99, Aug. 2004. [CrossRef]
- K. A. Tayeb, S. D. Bateman, S. Hampton, M. Malone, M. Malone, and J. Fletcher, ‘Managing infection: a holistic approach’, J. Wound Care, vol. 24, no. Sup5b, pp. 20–30, May 2015. [CrossRef]
- L. Gryson, S. Meaume, I. Feldkaemper, and F. Favalli, ‘Anti-biofilm Activity of Povidone-Iodine and Polyhexamethylene Biguanide: Evidence from In Vitro Tests’, Curr. Microbiol., vol. 80, no. 5, p. 161, May 2023. [CrossRef]
- J. A. Schwartz, J. C. Lantis, C. Gendics, A. M. Fuller, W. Payne, and D. Ochs, ‘A prospective, non comparative, multicenter study to investigate the effect of cadexomer iodine on bioburden load and other wound characteristics in diabetic foot ulcers’, Int. Wound J., vol. 10, no. 2, pp. 193–199, Apr. 2013. [CrossRef]
- M. Malone et al., ‘Effect of cadexomer iodine on the microbial load and diversity of chronic non-healing diabetic foot ulcers complicated by biofilm in vivo’, J. Antimicrob. Chemother., vol. 72, no. 7, pp. 2093–2101, Jul. 2017. [CrossRef]
- Q. Xu et al., ‘Sonocatalytic hydrogen/hole-combined therapy for anti-biofilm and infected diabetic wound healing’, Natl. Sci. Rev., vol. 10, no. 5, p. nwad063, Apr. 2023. [CrossRef]
- F. Akhtar et al., ‘A nano phototheranostic approach of toluidine blue conjugated gold silver core shells mediated photodynamic therapy to treat diabetic foot ulcer’, Sci. Rep., vol. 11, no. 1, p. 24464, Dec. 2021. [CrossRef]
- S. Khan, S. N. Khan, R. Meena, A. M. Dar, R. Pal, and A. U. Khan, ‘Photoinactivation of multidrug resistant bacteria by monomeric methylene blue conjugated gold nanoparticles’, J. Photochem. Photobiol. B, vol. 174, pp. 150–161, Sep. 2017. [CrossRef]
- J. Liu et al., ‘Surfactin effectively inhibits Staphylococcus aureus adhesion and biofilm formation on surfaces’, Appl. Microbiol. Biotechnol., vol. 103, no. 11, pp. 4565–4574, Jun. 2019. [CrossRef]
- Z. Li et al., ‘Antibacterial Activity of Surfactin and Synergistic Effect with Conventional Antibiotics Against Methicillin-Resistant Staphylococcus aureus Isolated from Patients with Diabetic Foot Ulcers’, Diabetes Metab. Syndr. Obes., vol. Volume 16, pp. 3727–3737, Nov. 2023. [CrossRef]
- I. Serrano, B. Alhinho, E. Cunha, L. Tavares, A. Trindade, and M. Oliveira, ‘Bacteriostatic and Antibiofilm Efficacy of a Nisin Z Solution against Co-Cultures of Staphylococcus aureus and Pseudomonas aeruginosa from Diabetic Foot Infections’, Life, vol. 13, no. 2, p. 504, Feb. 2023. [CrossRef]
- J. L. Webber et al., ‘Incorporation and antimicrobial activity of nisin Z within carrageenan/chitosan multilayers’, Sci. Rep., vol. 11, no. 1, p. 1690, Jan. 2021. [CrossRef]
- J. B. Wright, K. Lam, and R. E. Burrell, ‘Wound management in an era of increasing bacterial antibiotic resistance: A role for topical silver treatment’, Am. J. Infect. Control, vol. 26, no. 6, pp. 572–577, Dec. 1998. [CrossRef]
- K. Chaloupka, Y. Malam, and A. M. Seifalian, ‘Nanosilver as a new generation of nanoproduct in biomedical applications’, Trends Biotechnol., vol. 28, no. 11, pp. 580–588, Nov. 2010. [CrossRef]
- E. G. Di Domenico et al., ‘Silver Sulfadiazine Eradicates Antibiotic-Tolerant Staphylococcus aureus and Pseudomonas aeruginosa Biofilms in Patients with Infected Diabetic Foot Ulcers’, J. Clin. Med., vol. 9, no. 12, p. 3807, Nov. 2020. [CrossRef]
- L. Lu et al., ‘Developing natural products as potential anti-biofilm agents’, Chin. Med., vol. 14, no. 1, p. 11, Dec. 2019. [CrossRef]
- J.-H. Lee, Y.-G. Kim, S. Y. Ryu, M. H. Cho, and J. Lee, ‘Ginkgolic acids and Ginkgo biloba extract inhibit Escherichia coli O157:H7 and Staphylococcus aureus biofilm formation’, Int. J. Food Microbiol., vol. 174, pp. 47–55, Mar. 2014. [CrossRef]
- J.-H. Lee et al., ‘Apple Flavonoid Phloretin Inhibits Escherichia coli O157:H7 Biofilm Formation and Ameliorates Colon Inflammation in Rats’, Infect. Immun., vol. 79, no. 12, pp. 4819–4827, Dec. 2011. [CrossRef]
- R. Srinivasan, S. Santhakumari, P. Poonguzhali, M. Geetha, M. Dyavaiah, and L. Xiangmin, ‘Bacterial Biofilm Inhibition: A Focused Review on Recent Therapeutic Strategies for Combating the Biofilm Mediated Infections’, Front. Microbiol., vol. 12, p. 676458, May 2021. [CrossRef]
- D. Fleming and K. Rumbaugh, ‘Approaches to Dispersing Medical Biofilms’, Microorganisms, vol. 5, no. 2, p. 15, Apr. 2017. [CrossRef]
- R. Roy, M. Tiwari, G. Donelli, and V. Tiwari, ‘Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action’, Virulence, vol. 9, no. 1, pp. 522–554, Dec. 2018. [CrossRef]
- T. J. Koob et al., ‘Angiogenic properties of dehydrated human amnion/chorion allografts: therapeutic potential for soft tissue repair and regeneration’, Vasc. Cell, vol. 6, no. 1, p. 10, 2014. [CrossRef]
- Y. Mao et al., ‘An in vitro comparison of human corneal epithelial cell activity and inflammatory response on differently designed ocular amniotic membranes and a clinical case study’, J. Biomed. Mater. Res. B Appl. Biomater., vol. 111, no. 3, pp. 684–700, Mar. 2023. [CrossRef]
- T. Ž. Ramuta, T. Šket, M. Starčič Erjavec, and M. E. Kreft, ‘Antimicrobial Activity of Human Fetal Membranes: From Biological Function to Clinical Use’, Front. Bioeng. Biotechnol., vol. 9, p. 691522, May 2021. [CrossRef]
- K. M. Mamede and L. B. Sant’Anna, ‘Antifibrotic effects of total or partial application of amniotic membrane in hepatic fibrosis’, An. Acad. Bras. Ciênc., vol. 91, no. 3, p. e20190220, 2019. [CrossRef]
- C.-H. Wassmer and E. Berishvili, ‘Immunomodulatory Properties of Amniotic Membrane Derivatives and Their Potential in Regenerative Medicine’, Curr. Diab. Rep., vol. 20, no. 8, p. 31, Aug. 2020. [CrossRef]
- J. C. Warning, S. A. McCracken, and J. M. Morris, ‘A balancing act: mechanisms by which the fetus avoids rejection by the maternal immune system’, REPRODUCTION, vol. 141, no. 6, pp. 715–724, Jun. 2011. [CrossRef]
- A. Clinton and T. Carter, ‘Chronic Wound Biofilms: Pathogenesis and Potential Therapies’, Lab. Med., vol. 46, no. 4, pp. 277–284, Nov. 2015. [CrossRef]
- A. Roy, M. Mantay, C. Brannan, and S. Griffiths, ‘Placental Tissues as Biomaterials in Regenerative Medicine’, BioMed Res. Int., vol. 2022, pp. 1–26, Apr. 2022. [CrossRef]
- Z. Chen, Y. Wang, and C. Shi, ‘Therapeutic Implications of Newly Identified Stem Cell Populations from the Skin Dermis’, Cell Transplant., vol. 24, no. 8, pp. 1405–1422, Aug. 2015. [CrossRef]
- L. E. Kokai, K. Marra, and J. P. Rubin, ‘Adipose stem cells: biology and clinical applications for tissue repair and regeneration’, Transl. Res., vol. 163, no. 4, pp. 399–408, Apr. 2014. [CrossRef]
- J. D. Bullard, J. Lei, J. J. Lim, M. Massee, A. M. Fallon, and T. J. Koob, ‘Evaluation of dehydrated human umbilical cord biological properties for wound care and soft tissue healing’, J. Biomed. Mater. Res. B Appl. Biomater., vol. 107, no. 4, pp. 1035–1046, May 2019. [CrossRef]
| Occurrence of Biofilms |
No of articles | No of articles: Biofilm makeup |
No of articles: Specific species |
||
|---|---|---|---|---|---|
|
Present |
66/88 |
Provided | 45/66 |
Provided | 42/45 |
| Not provided | 21/66 |
Not provided | 3/45 | ||
|
Absent |
22/88 |
Not applicable |
Not applicable |
||
| Method of Study | Aim of Study | References |
|---|---|---|
| In Vitro method | ||
| 2-Dimensional Cell Culture | Wound healing and wound formation | [34] |
| 3-Dimensional Cell Culture | Mechanism of wound healing (angiogenesis , re-epithelialization) | [35] |
| In Vivo method | [37],[38] |
|
| Ischemic animal ulcer model | primary ischemic therapy | [39],[40] |
| Neuropathic animal Ulcer model | mechanism of neuropathic infections | [41],[42] |
| Infected Diabetic Ulcer Model | wound formation, progression and healing in diabetic mice | [43], [44], [45], [46], [47], [48] |
| Statistical Data Analysis | Statistical significance of Diabetic foot ulcer data | [49],[50],[51],[52],[53] |
| Bacteria | Multi Drug resistance status | Biofilm formation | References |
|---|---|---|---|
| Escherichia coli | Yes | ++ | [12], [8], [58], [59], [60] |
| Pseudomonas aeruginosa | Yes | ++++ | [12], [8], [58] |
| Proteus sp. | Yes | + | [12], [8], [58] |
| Klebsiella pneumoniae | Yes | + | [12], [8], [60], [58] |
| Staphylococcus aureus | Yes | ++++ | [12], [8], [58] |
| Citrobacter sp. | Yes | ++ | [12], [8], [60], [58] |
| Acinetobacter baumanii | Yes | +++ | [12], [8], [61], [58] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





