Submitted:
02 October 2024
Posted:
03 October 2024
You are already at the latest version
Abstract
Keywords:
| GLOSSARY OF TERMS |
| Sensitivity (True Positive Rate): the proportion of subjects with disease, who have a positive testDetection rate: the proportion of subjects enrolled who have a true positive testOdds ratio (OR): Odds ratio was calculated according to: (TP*TN)/(FP*FN), where TP= True Positives; TN=True Negatives; FN=False Negatives; FP=False PositivesSpecificity (True Negative Rate): the proportion of subjects without disease, who have a negative testPositive Predictive Value (PPV): the proportion of subjects with disease among those with a positive testNegative Predictive Value (NPV): the proportion of subjects without disease among those with a negative testPositivity Rate: the proportion of subjects enrolled who have a positive testNon-Inferiority Margin (NIM): The largest clinically acceptable difference between a new treatment and an active comparator.Triage: The process of prioritizing patients for treatment based on the urgency of their needs.False Positive Rate (1-Specificity): Proportion of subjects without disease, who have a positive testFalse Negative Rate (1-Sensitivity): Proportion of subjects with disease, who have a negative testCN: colorectal neoplasia. (Abnormal growth or mass in the colon or rectum. It includes advanced adenomas, high -risk sessile serrated lesions and colorectal cancers)AA: advanced adenoma.NAA: non-advanced adenoma.CRC: colorectal cancer.SOC: standard of care , or immediate colonoscopy following a FIT+ test.AAP: alternative approach., or colonoscopy iff both FIT+ and QuantiDNA+. |
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Study Population
2.3. Technical Workflow
2.4. Statistical Analysis
3. Results
3.1. Study Population
3.2. Clinical Performance
4. Discussion
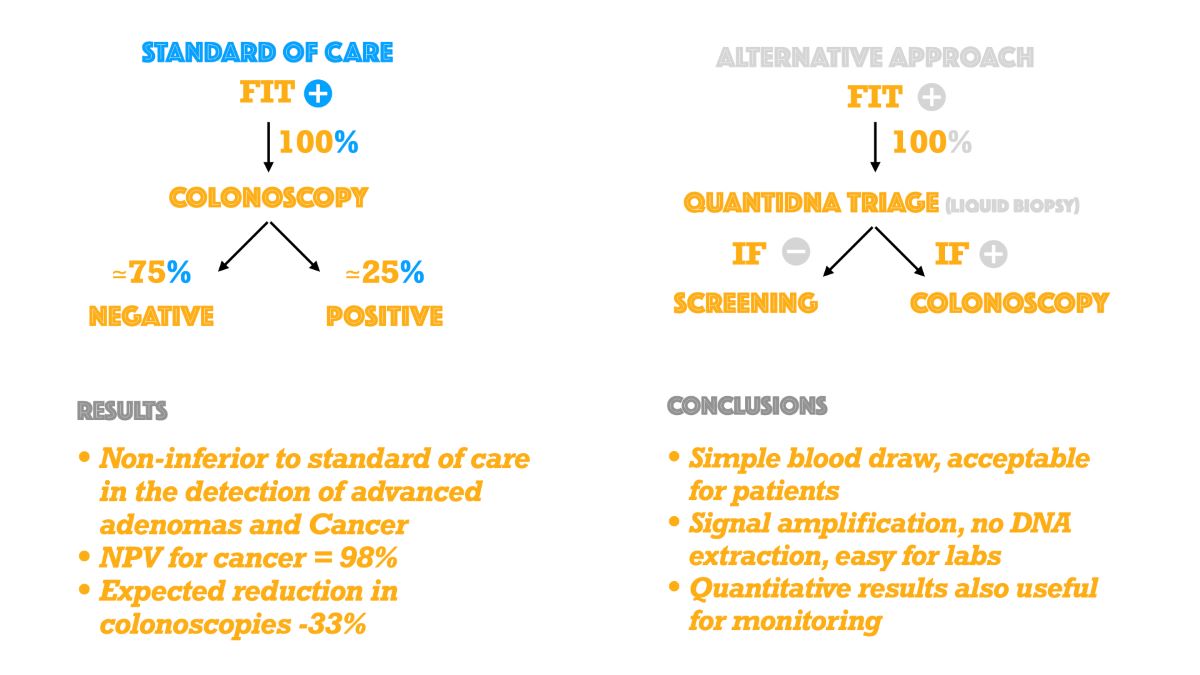
5. Conclusions
Ethics approval statement
Supplementary Materials
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xin Wang, Xia-Qing Shi, Peng-Wei Zeng, Fong-Ming Mo, Zi-Hua Chen. Circulating cell free DNA as the diagnostic marker for colorectal cancer: a systematic review and meta-analysis. Oncotarget, 2018, Vol.9, No.36, pp: 24514-24524.
- Paul Okunieff, Natalie A. Lockney, Randal H. Henderson, Steven G. Swarts, Zhenhuan Zhang, Bingrong Zhang, Jennifer Li, Robert A. Zlotecki, Cristopher G. Morris, Katherine A. Casey-Sawicki. Measuring Radiation Toxicity Using Circulating Cell-free DNA in Prostate Cancer Patients. IJPT, 2021.
- Scimia M, Du J, Pepe F, Bianco MA, Russo Spena S, Patell-Socha F, Sun Q, Powell MJ, Malapelle U, Troncone G. Evaluation of a novel liquid biopsy-based ColoScape assay for mutational analysis of colorectal neoplasia and triage of FIT+ patients: a pilot study. J Chin Pathol, 2018; 0: 1-4.
- Carlo Senore, Partha Basu, Ahti Anttila, Antonio Ponti, Mariano Tomatis, Diama Bhadra Vale, Guglielmo Ronco, Isabelle Soerjomataram, Maja Primic-Zakelj, Emilia Riggi, Joakim Dillner, Miriam Klara Elfstrom, Stefan Lonnberg, Rengaswamy Sankaranarayanan, Nereo Segnan. Performance of colorectal cancer screening in the European Union Member States: data from the second European screening report. Gut 2018; 0: 1-13.
- Chen Qian, Shaoqing Ju, Jing Qi, Jianmei Zhao, Xianjuan Shen, Rongrong Jing, Juan Yu, Li Li, Yingjuan Shi, Lurong Zhang, Zhiwei Wang, Hui Kong. Alu–based cell-free DNA: a novel biomarker for screening of gastric cancer. Oncotarget, advance publications 2016.
- Maria Antonia Bianco, Gianluca Rotondano, Maria Lucia Garofano, Livio Cipolletta. Discovering a magnified world. Superficial neoplastic lesions… and beyond. Area Qualità, first published September 2005.
- EMA Guideline on the choice of the non-inferiority margin. Doc. Ref. EMEA/CPMP/EWP/2158/99.
- FDA Non-inferiority clinical trials to establish effectiveness. Guidance for Industry.
- Jack Cuzick, Peter Sasieni. Interpreting the results of non-inferiority trials — a review. British Journal of Cancer, 2022; 127:1755-1759.
- Thomas F Imperiale, David F Ransohoff, Steven H. Itzkowitz, Theodore R Levin, Philip Lavin, Graham P Midgard, David A. Ahlquist, Barry M. Berger. Multitarget Stool DNA Testing for Colorectal-Cancer Screening. The New England Journal of Medicine, 2014.
- Liang Huang, Yue Hu, Shan Liu, Bo Jin, Bin Lu. The analysis of multilevel factors affecting adenoma detection rates for colonoscopies: a large-scale retrospective study. BMC Gastroenterology, 2021; 21; 403.
- Hayato Yamaguchi, Masakatsu Fukuzawa, Hirohito Minami, Tadashi Ichimiya, Hiroshi Takahashi, Yubu Matsue, Mitsuyoshi Honjo, Yasutake Hirayama, Daisuke Nutahara, Junichi Taira, Hironori Nakamura, Takashi Kawai, Takao Itoi. The relationship between Post-colonoscopy Colorectal Cancer and Quality Indicators of Colonoscopy: The Latest Single-centre Cohort Study with a Review of the Literature. Intern. Med., 2020; 59: 141-1488.
- Manuel Zorzi, Giulio Antonelli, Claudio Barbiellini Amidei, Jessica Battagello, Bastianello Germanà, Flavio Valiante, Stefano Benvenuti, Alberto Tringali, Francesco Bortoluzzi, Erica Cervellin, Davide Giacomin, Tamara Meggiato, Erik Rosa-Rizzotto, Diego Fregonese, Manuela Dinca, Gianluca Baldassarre, Paola Scalon, Maurizio Pantalena, Luisa Milan, Gianmarco Bulighin, Daniele Di Piramo, Maurizio Azzurro, Armando Gabrielli, Alessandro Repici, Douglas K Rex, Massimo Rugge, Cesare Hassan, Veneto Screening Endoscopists Working Group, Anna Giacomin, Andrea Buda, Deborah Costa, Davide Checchin, Renato Marin, Elisabetta Patarnello, Aldo Ceriani, Ennio Guido, Perla Bertomoro, Nicoletta Merlini, Francesca Murer, Ephrem Ntakirutimana, Luca Benamato, Maria Cristina Conti Bellocchi. Adenoma detection rate and colorectal cancer risk in fecal immunochemical test screening programs: an observational cohort study. Ann Intern Med. 2023; 176:303-310.
- Michael Greenspan, Kumar Bharat Rajan, Adil Baig, Todd Beck, Sohrab Mobarhan, Joshua Melson. Advanced adenoma detection rate is independent of nonadvanced adenoma detection rate. Am J Gastroenterol. 2013;108:1286-92.
- Aasma Shaukat, Amy A. Gravely, Adam S Kim, Jeffery Rank, Timothy R Church, John I Allen. Rates of Detection of Adenoma, Sessile Serrated Adenoma, and Advanced Adenoma Are Stable Over Time and Modifiable. Gastroenterology AGA, Brief communication. Volume 156, Issue 3, P816-817, 2019.
- Aasma Shaukat, Jennifer Holub, Irving M Pike, Mark Pochapin, David Greenwald, Colleen Schmitt, Glenn Eisen. Benchmarking Adenoma Detection Rates for Colonoscopy: Results from a US-Based Registry. Am J Gastroenterol. 2021 1; 116: 1946-1949.
- Manuel Zorzi. Giscor Survey of Italian Colorectal cancer screening programs 2022-2023. www.giscor.it.
- Mark Schiffman, Diane Solomon. Findings to date from the ASCUS-LSIL Triage Study (ALTS). Arch Pathol Lab Med. 2003;127:946-9.
- Efrat L Amitay, Katarina Cuk, Tobias Niedermaier, Korbinian Weigl, Hermann Brenner. Factors associated with false-positive fecal immunochemical tests in a large German colorectal cancer screening study. Int J Cancer. 2019 15;144:2419-2427.
- Gregory J Tsongalis. Branched DNA technology in molecular diagnostics. Am J Clin Pathol. 2006; 126:448-53.
- T B Hao, W Shi, X J Shen, J Qi, X H Wu, Y Wu, Y Y Tang, S Q Ju. Circulating cell-free DNA in serum as a biomarker for the diagnosis and prognostic prediction of CRC. BJC, 2014, 111, 1482–1489;
- <i>22. </i>V. Heinemann, J.Y. V. Heinemann, J.Y. Douillard, M. Ducreux, M. Peeters. Targeted therapy in Metastatic Colorectal Cancer - An example of personalised medicine in action. Cancer Treatment Reviews, 2013, vol.39: 592-601.
- Matthew, G. Krebs, Umberto Malapelle, Fabrice Andrè, Luis Paz-Ares, Martin Schuler, David M. Thomas, Gilad Vainer, Takayuki Yoshino, Christian Rolfo. Practical Considerations for the Use of Circulating tumor DNA in the Treatment of Patients with Cancer: a Narrative Review. JAMA Oncology, 2022.
- Umberto Malapelle, Pasquale Pisapia, Francesco Pepe, Gianluca Russo, Mauro Buono, Alessandro Russo, Jorge Gomez, Ola Khorshid, Philip C. Mack, Christian Rolfo, Giancarlo Troncone. The Evolving role of Liquid Biopsy in Lung Cancer. Lung Cancer, 2022, 172: 53-64.
| Demographic Characteristics | Number * | % | |
|---|---|---|---|
| Gender | Female | 348 | 51.9 |
| Male | 323 | 48.1 | |
| Total | 671 | 100.0 | |
| Race | Caucasian | 668 | 99.6 |
| Black or African | 0 | 0.0 | |
| Middle Eastern or North African | 0 | 0.0 | |
| South Asian | 0 | 0.0 | |
| East Asian | 0 | 0.0 | |
| Eastern European | 2 | 0.3 | |
| Western European | 1 | 0.1 | |
| Southeast Asian or Pacific Islander | 0 | 0.0 | |
| Other | 0 | 0.0 | |
| Total | 671 | 100.0 |
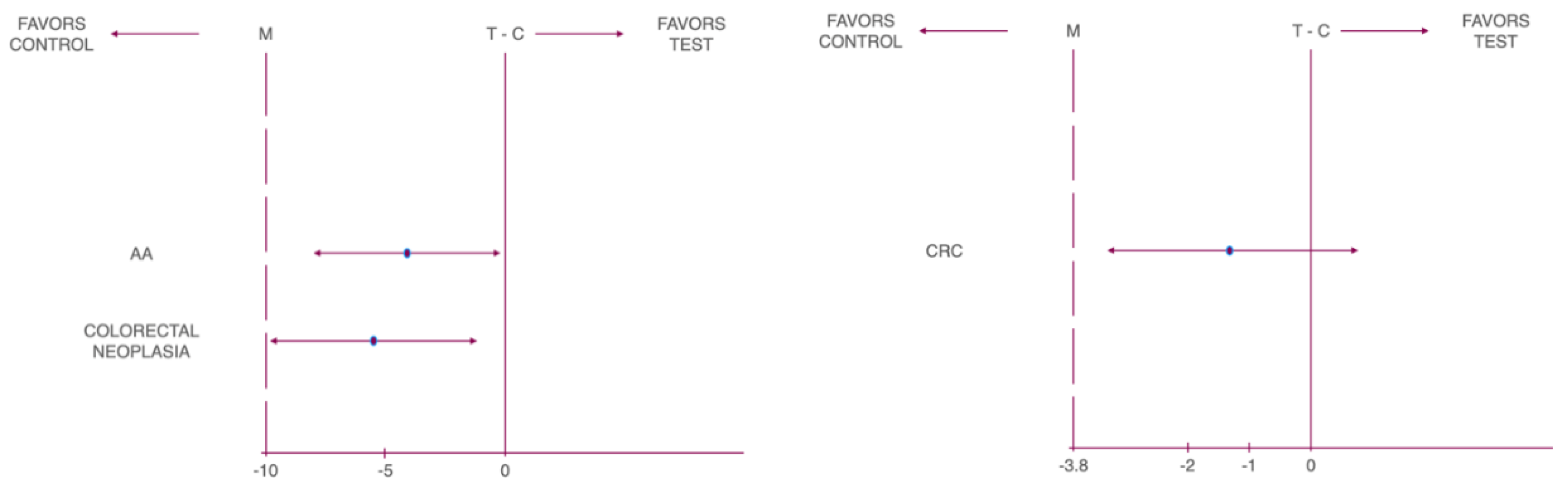
| Cancer Type | Non-inferiority Margin (%) | Risk Difference (%) | Lower 95% CL | Upper 95% CL | P-Value |
|---|---|---|---|---|---|
| CN | -10 | -5.07 | -9.23 | -0.90 | 0.010 |
| AA | -10 | -4.02 | -7.89 | -0.16 | 0.001 |
| CRC | -3.8 | -1.04 | -3.16 | 1.07 | 0.005 |
| CN | Parameter | Total N | True Outcome | Estimate (%) | Lower 95% CL | Upper 95% CL | |
| Sensitivity | 141 | 107 | 75.9 | 68.0 | 82.7 | ||
| Specificity | 530 | 190 | 35.8 | 31.8 | 40.1 | ||
| PPV | 447 | 107 | 23.9 | 21.9 | 26.0 | ||
| NPV | 224 | 190 | 84.8 | 80.3 | 88.4 | ||
| DR | 671 | 107 | 15.9 | 13.3 | 18.9 | ||
| PR | 671 | 447 | 66.6 | 62.9 | 70.2 | ||
| NLR | 0.67 | 0.49 | 0.92 | ||||
| PLR | 1.18 | 1.06 | 1.32 | ||||
| OR | 1.76 | 1.15 | 2.69 | ||||
| Youden's J statistic | 0.117 | ||||||
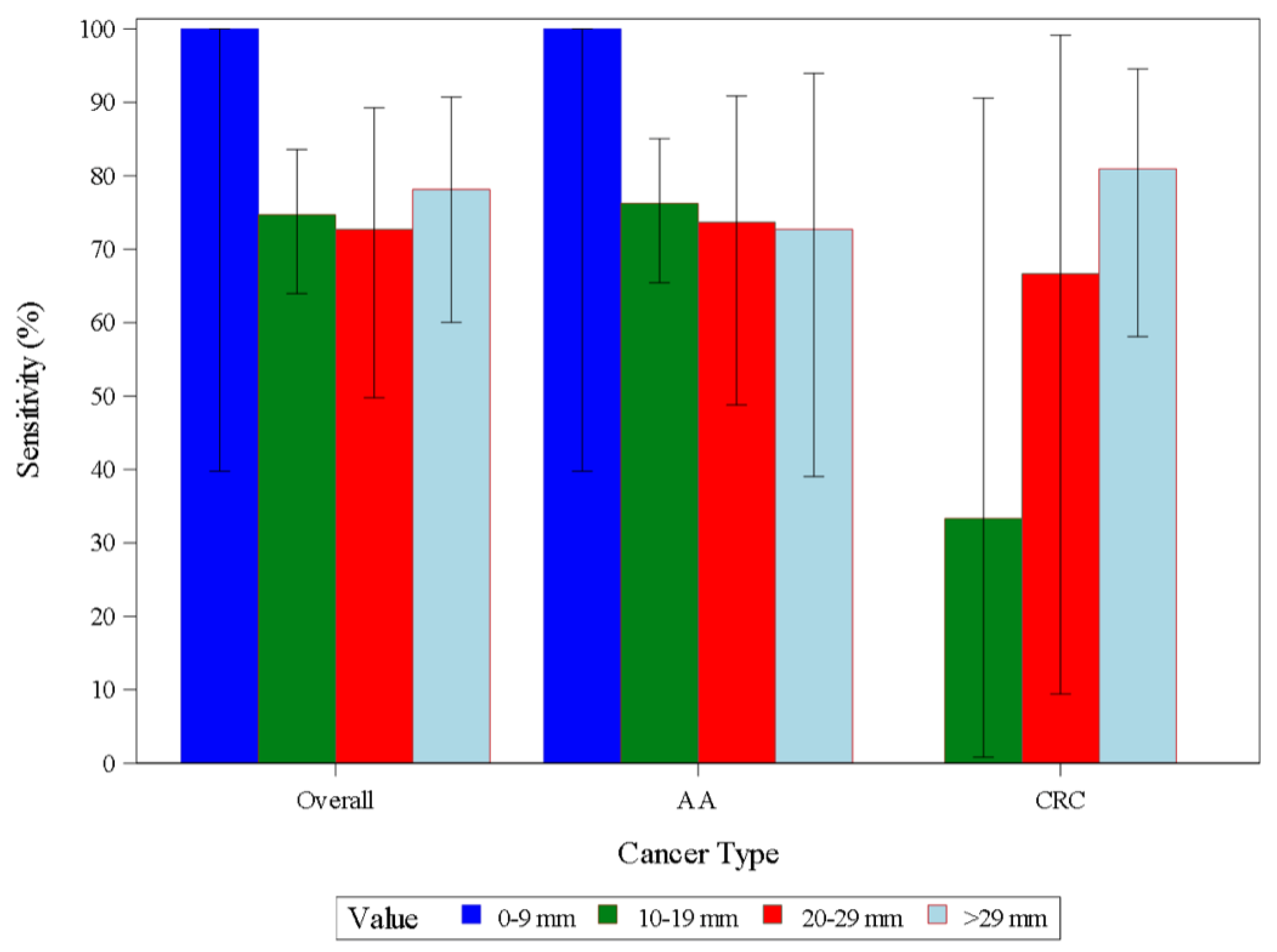
| AA | Sensitivity | 114 | 87 | 76.3 | 67.4 | 83.8 | |
| Specificity | 557 | 197 | 35.4 | 31.4 | 39.5 | ||
| PPV | 447 | 87 | 19.5 | 17.7 | 21.4 | ||
| NPV | 224 | 197 | 87.9 | 83.7 | 91.2 | ||
| DR | 671 | 87 | 13.0 | 10.5 | 15.7 | ||
| PR | 671 | 447 | 66.6 | 62.9 | 70.2 | ||
| NLR | 0.67 | 0.47 | 0.95 | ||||
| PLR | 1.18 | 1.05 | 1.33 | ||||
| OR | 1.76 | 1.11 | 2.81 | 0.017 | |||
| Youden's J statistic | 0.117 | ||||||
| CRC | Sensitivity | 27 | 20 | 74.1 | 53.7 | 88.9 | |
| Specificity | 644 | 217 | 33.7 | 30.0 | 37.5 | ||
| PPV | 447 | 20 | 4.5 | 3.6 | 5.6 | ||
| NPV | 224 | 217 | 96.9 | 94.2 | 98.3 | ||
| DR | 671 | 20 | 3.0 | 1.8 | 4.6 | ||
| PR | 671 | 447 | 66.6 | 62.9 | 70.2 | ||
| NLR | 0.77 | 0.40 | 1.47 | ||||
| PLR | 1.12 | 0.89 | 1.41 | ||||
| OR | 1.45 | 0.60 | 3.49 | 0.404 | |||
| Youden's J statistic | 0.078 |
| Parameter | Total N | True Outcome | Estimate (%) | Lower 95% CL | Upper 95% CL |
| Sensitivity | 27 | 20 | 74.1 | 53.7 | 88.9 |
| PPV | 447 | 20 | 4.5 | 3.6 | 5.6 |
| NPV | 224 | 217 | 96.9 | 94.2 | 98.3 |
| DR | 671 | 20 | 3.0 | 1.8 | 4.6 |
| PR | 671 | 447 | 66.6 | 62.9 | 70.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





