Submitted:
03 October 2024
Posted:
04 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Genome-Wide Identification and Annotation of the CaMs and CMLs in C. rosea
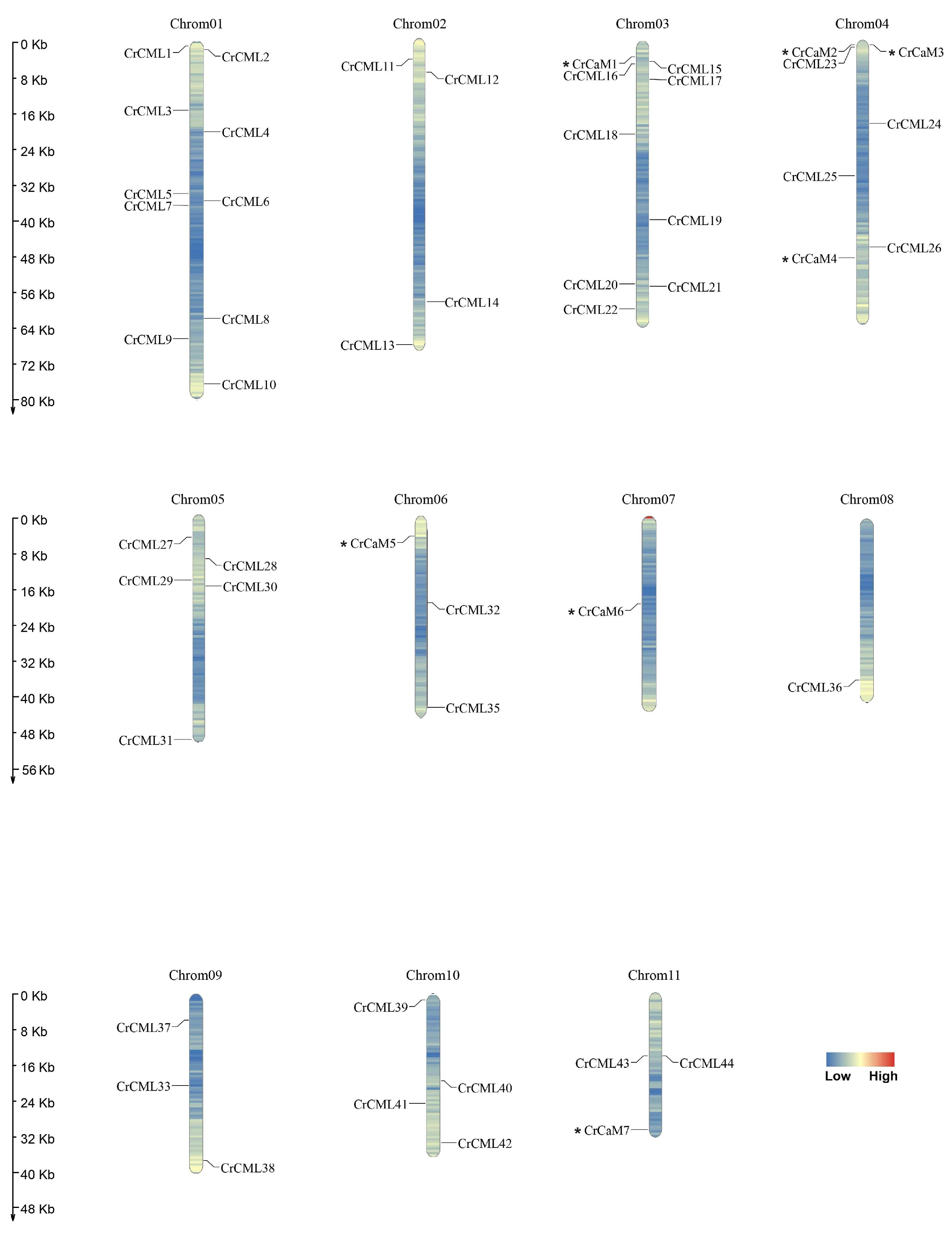
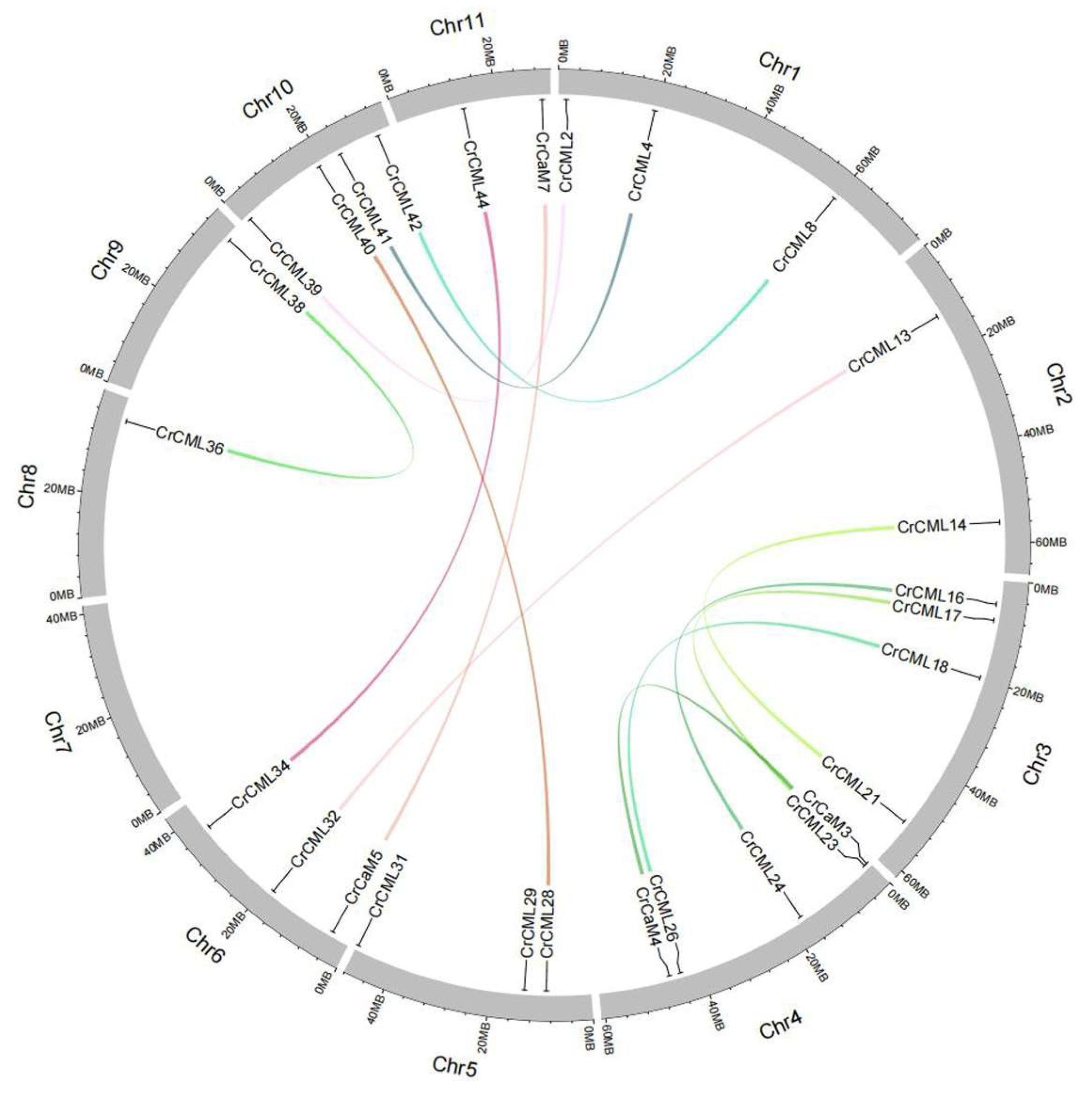
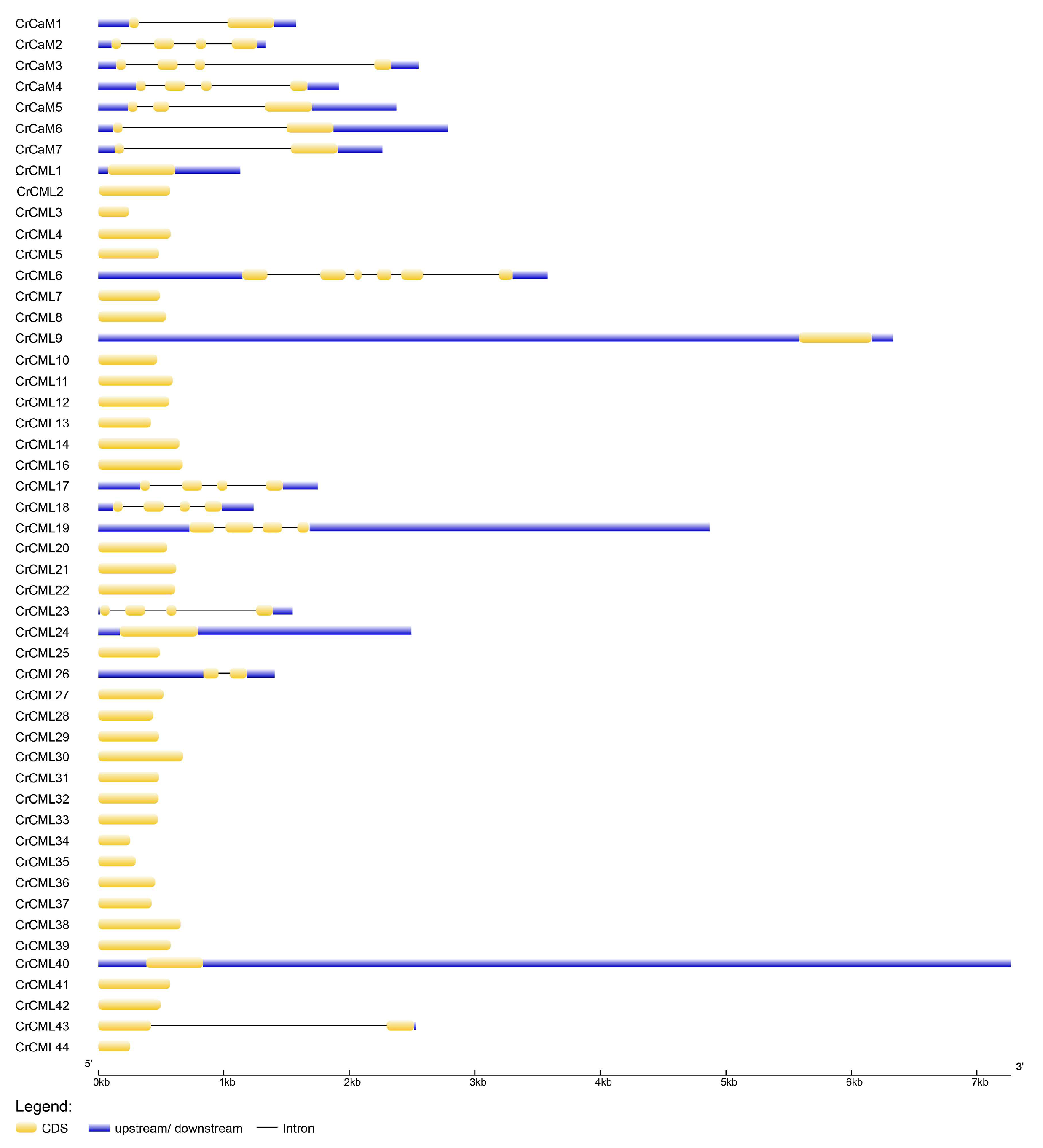
2.2. The Genes’ Localization and Analysis of the CrCaM/CrCML Genes Structure
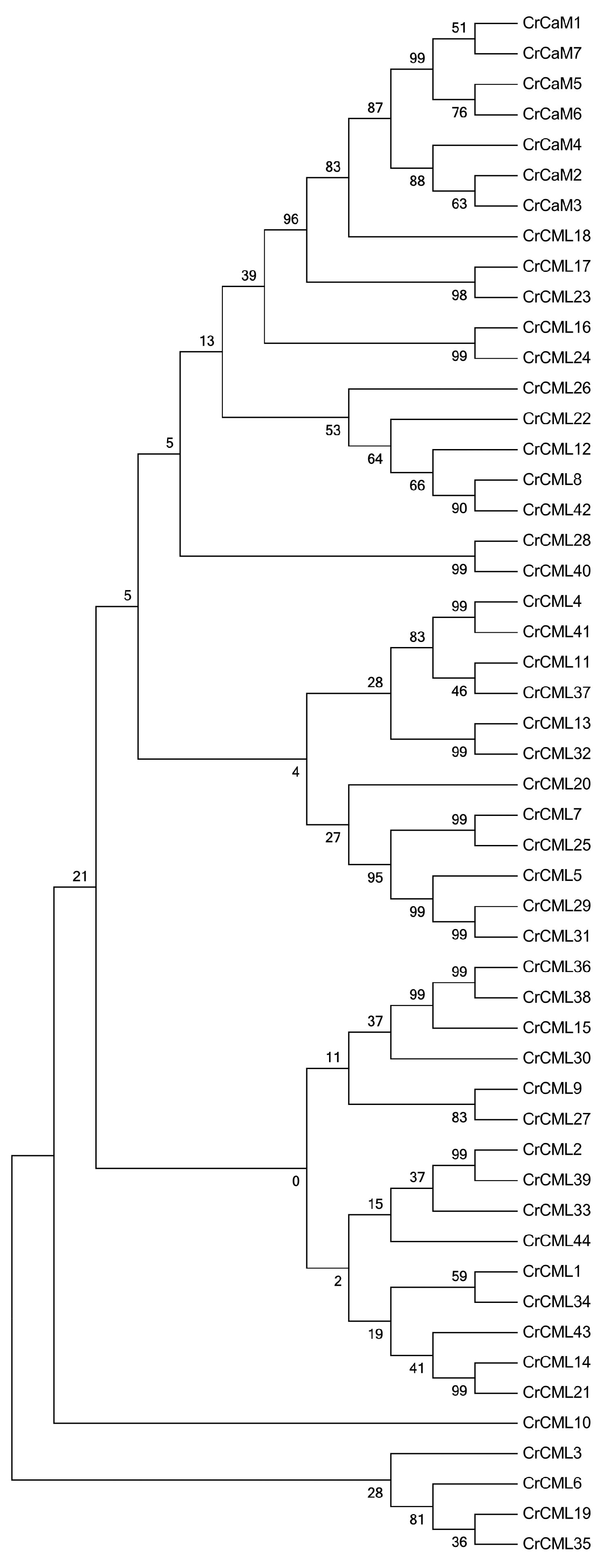
2.3. The Conserved Motifs of Proteins and Phylogenetic Analysis
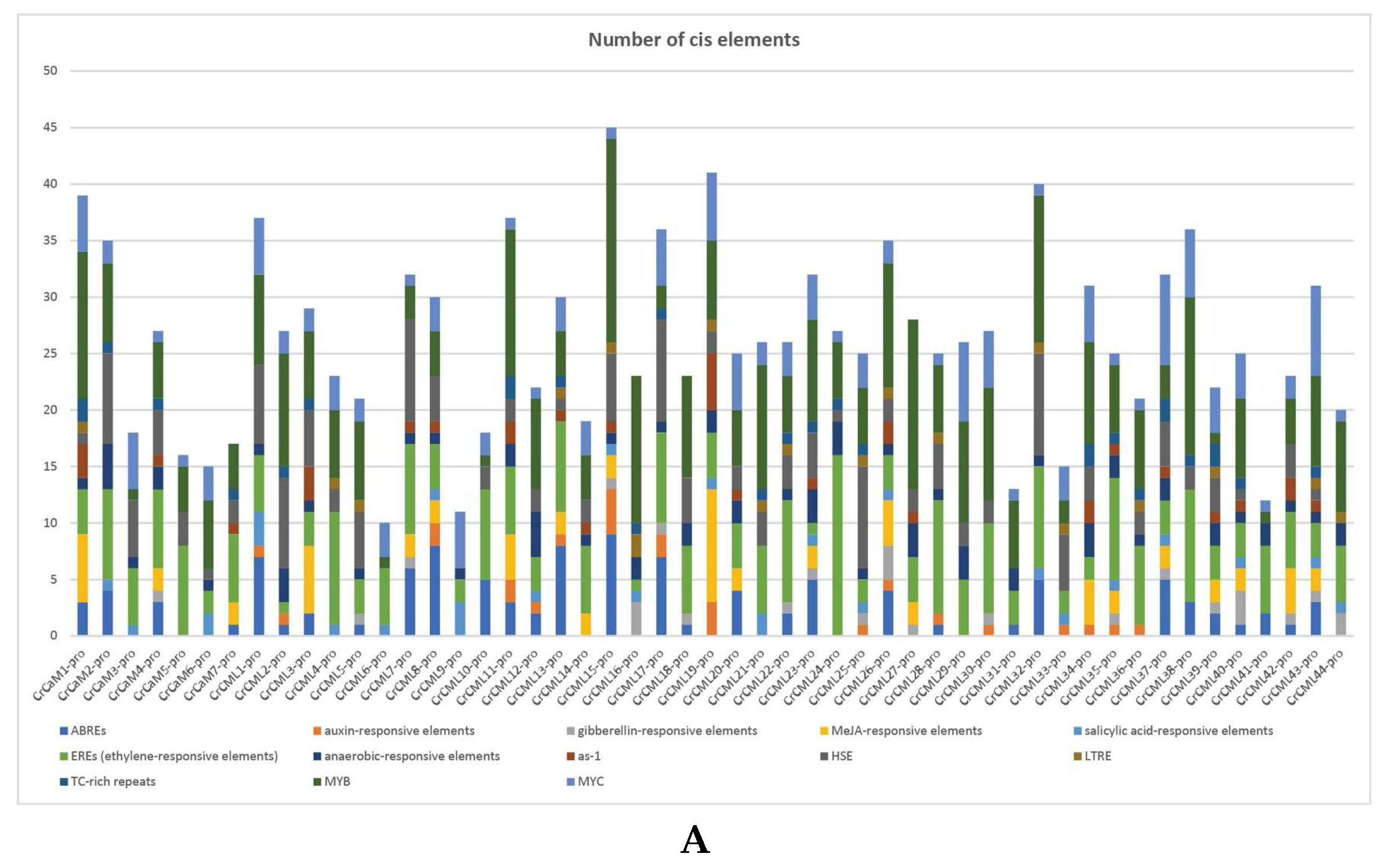
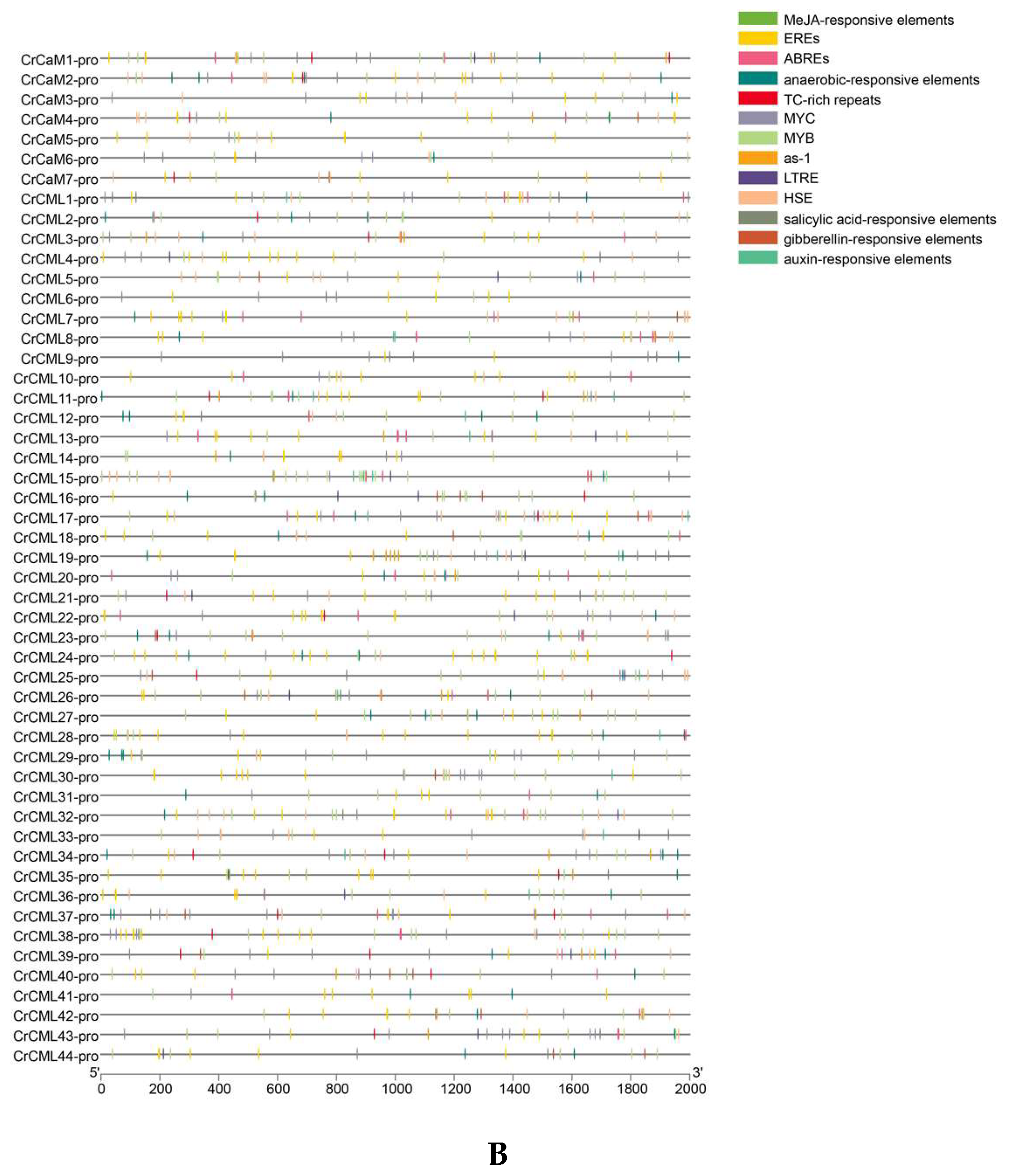
2.4. Abiotic Stress-Related Cis-Regulatory Elements (CEs) in the CrCaM/CrCML Promoters
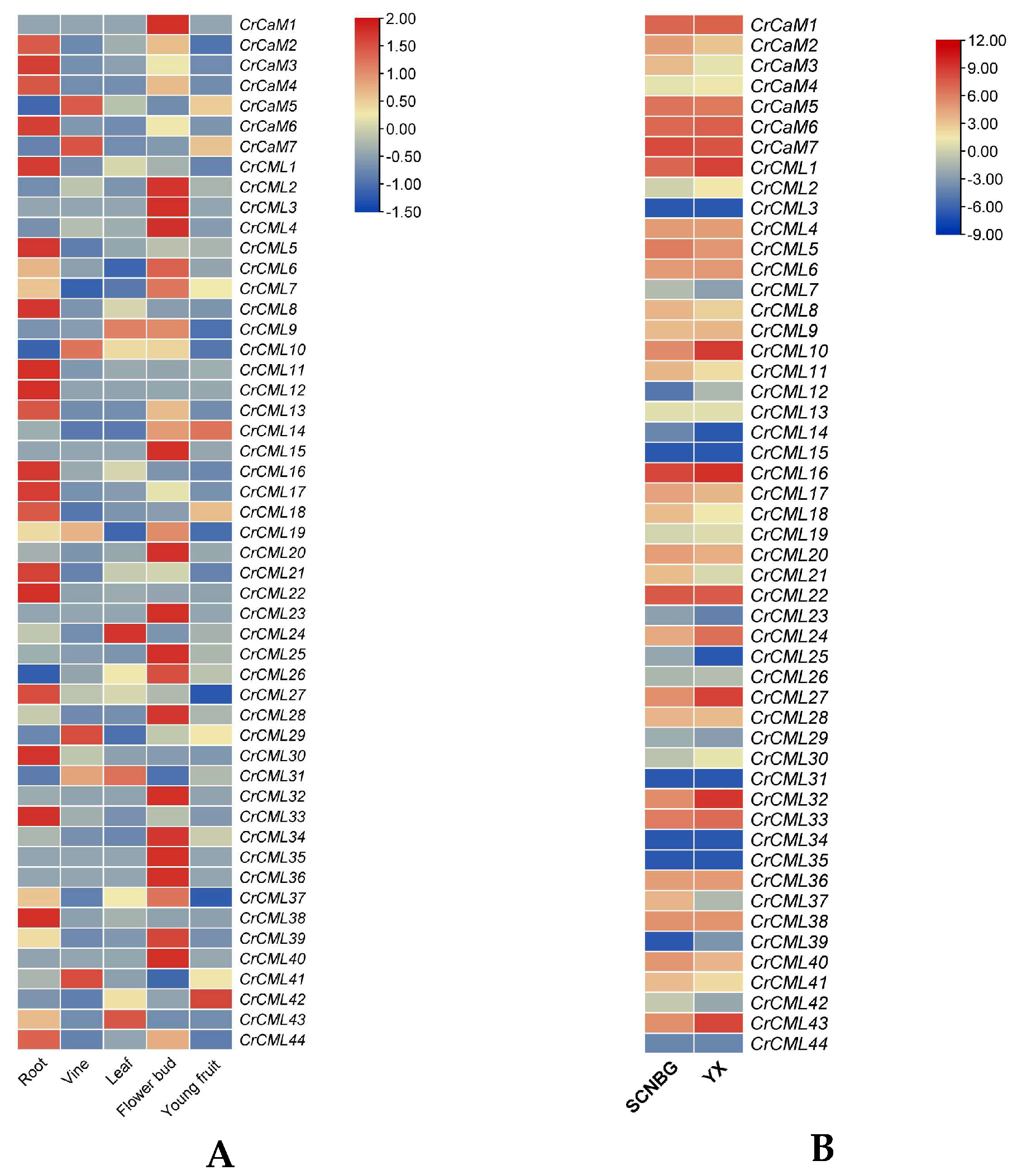
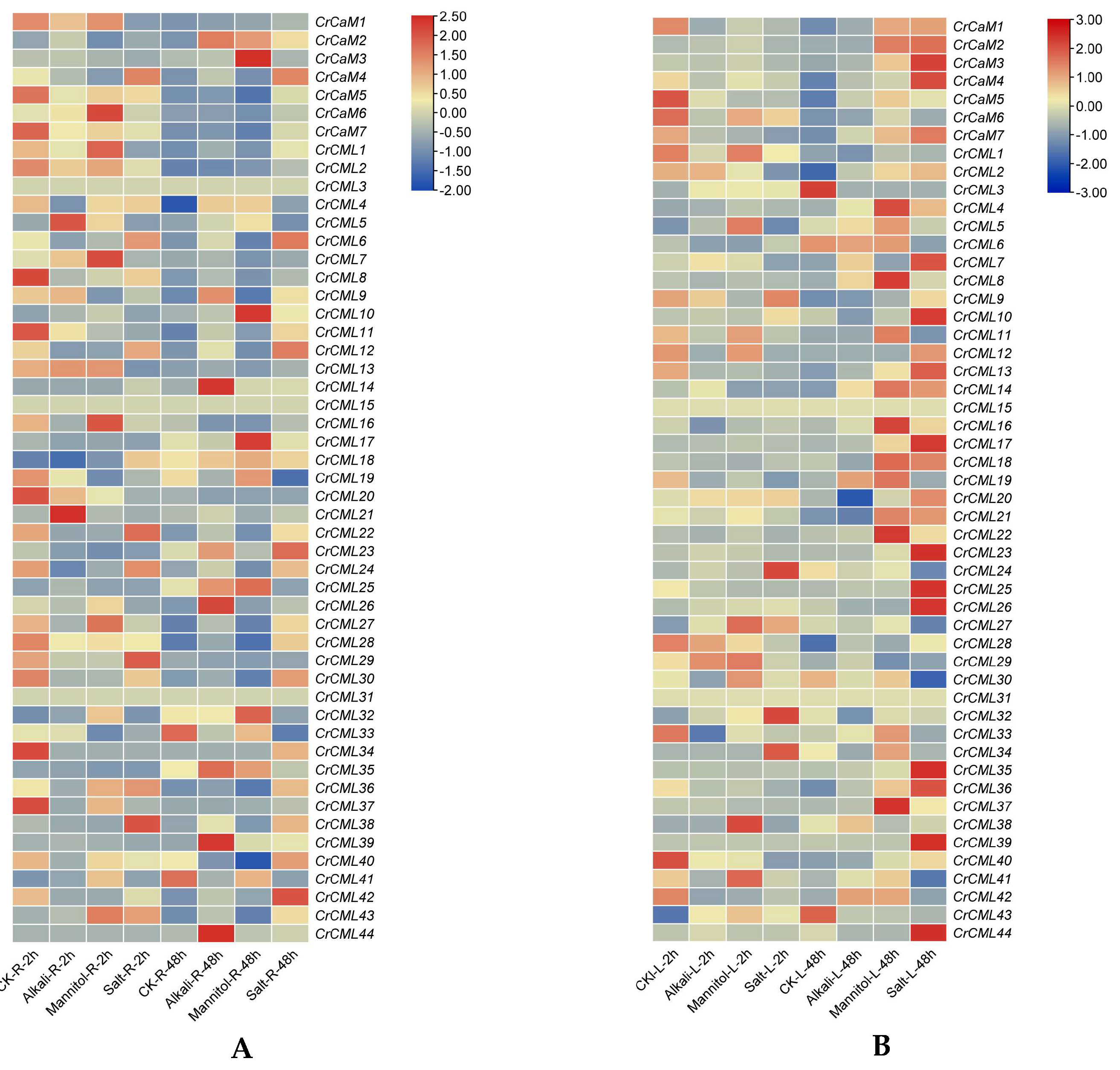
2.5. Tissue- and Habitat-Specific Expression Profiles of the CrCaMs/CrCMLs
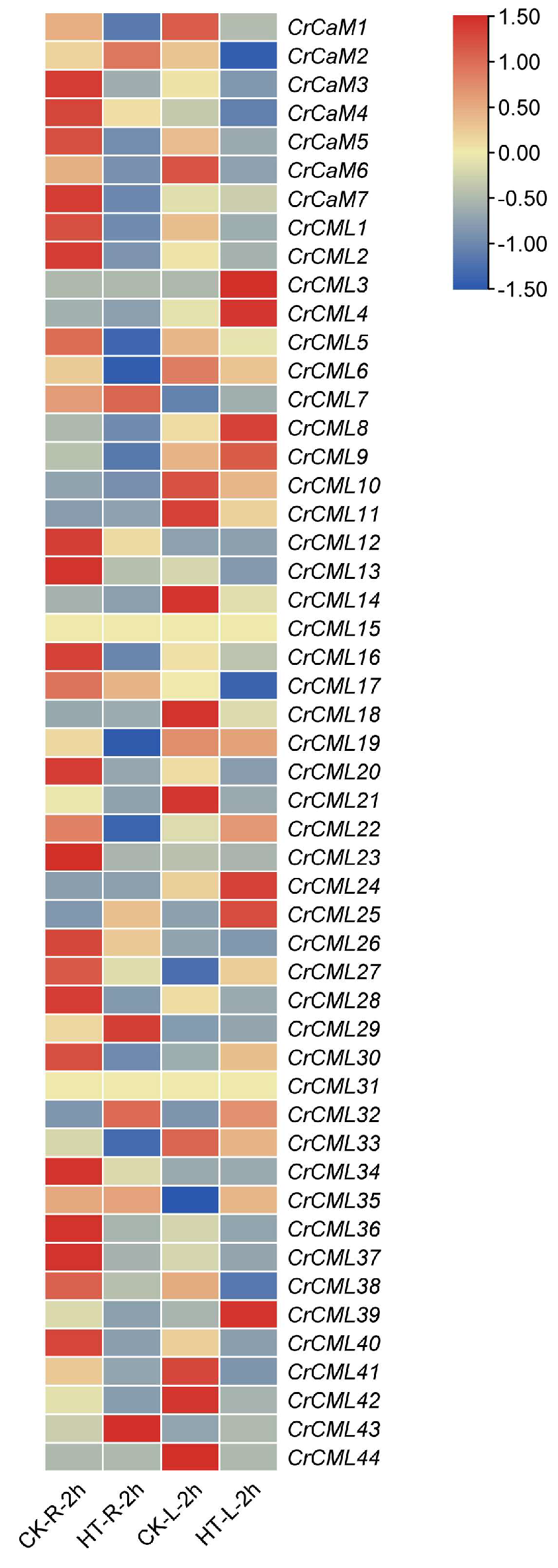
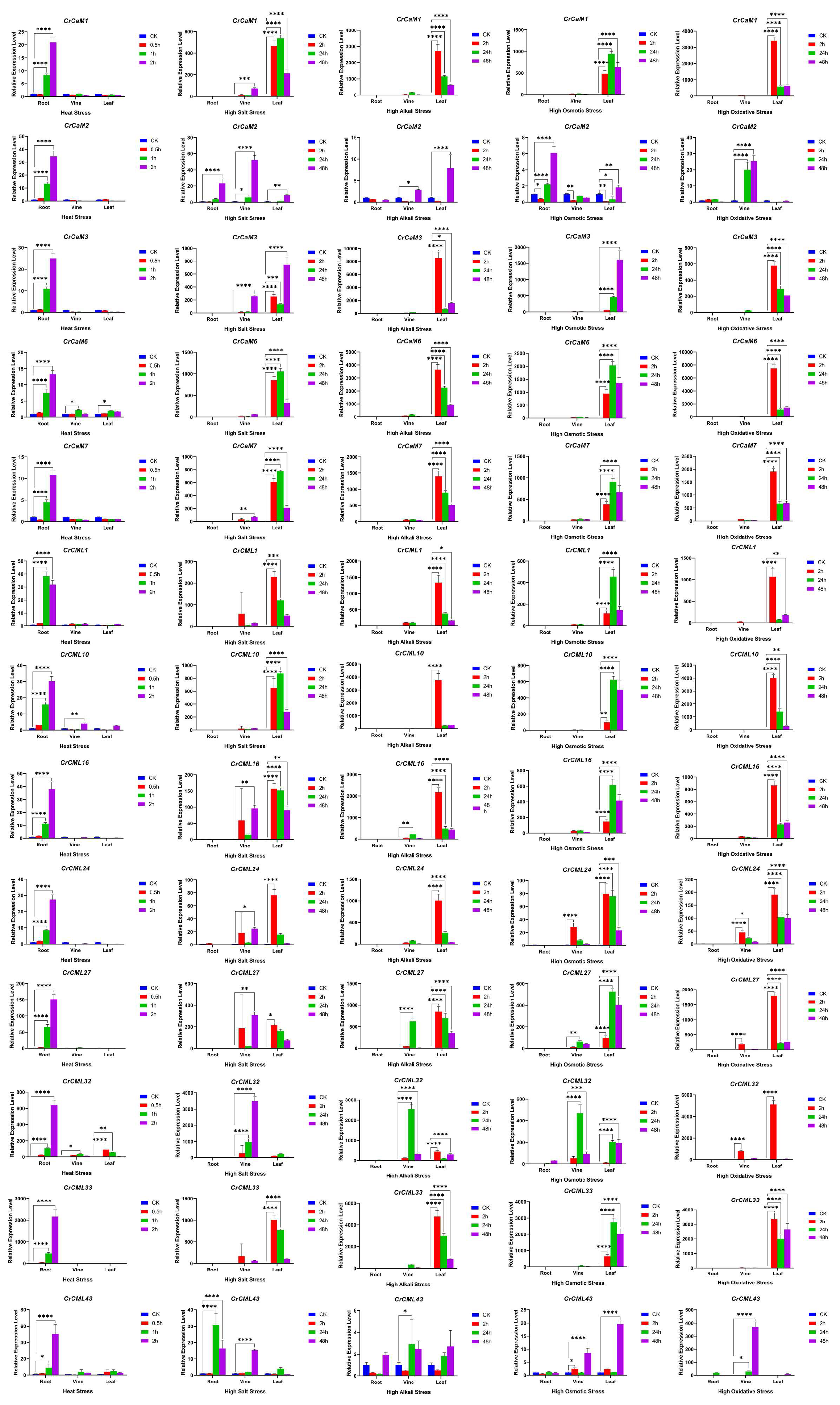
2.6. Expression Profile of the CrCaM/CrCML Genes in Response to Abiotic Stress
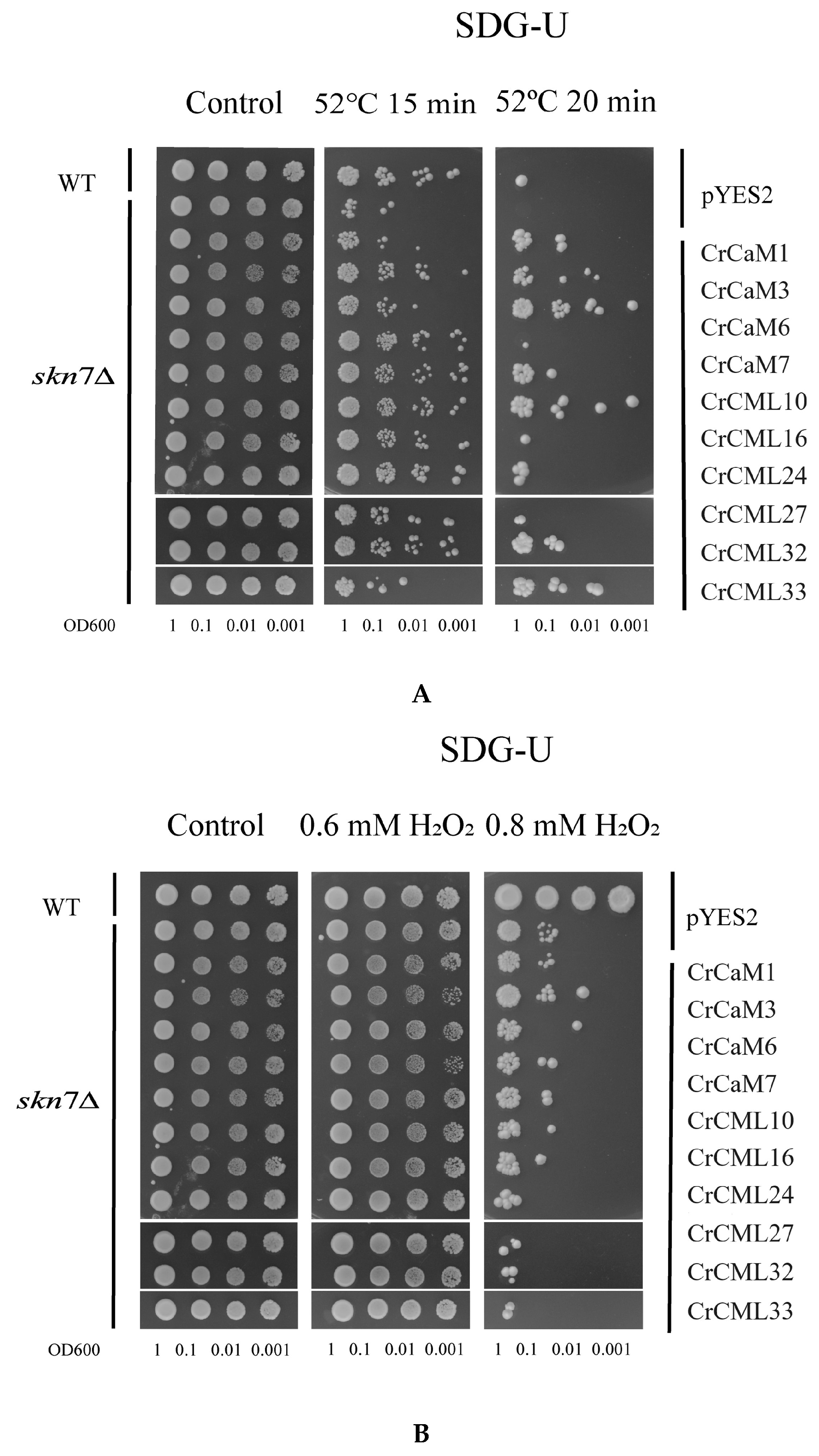
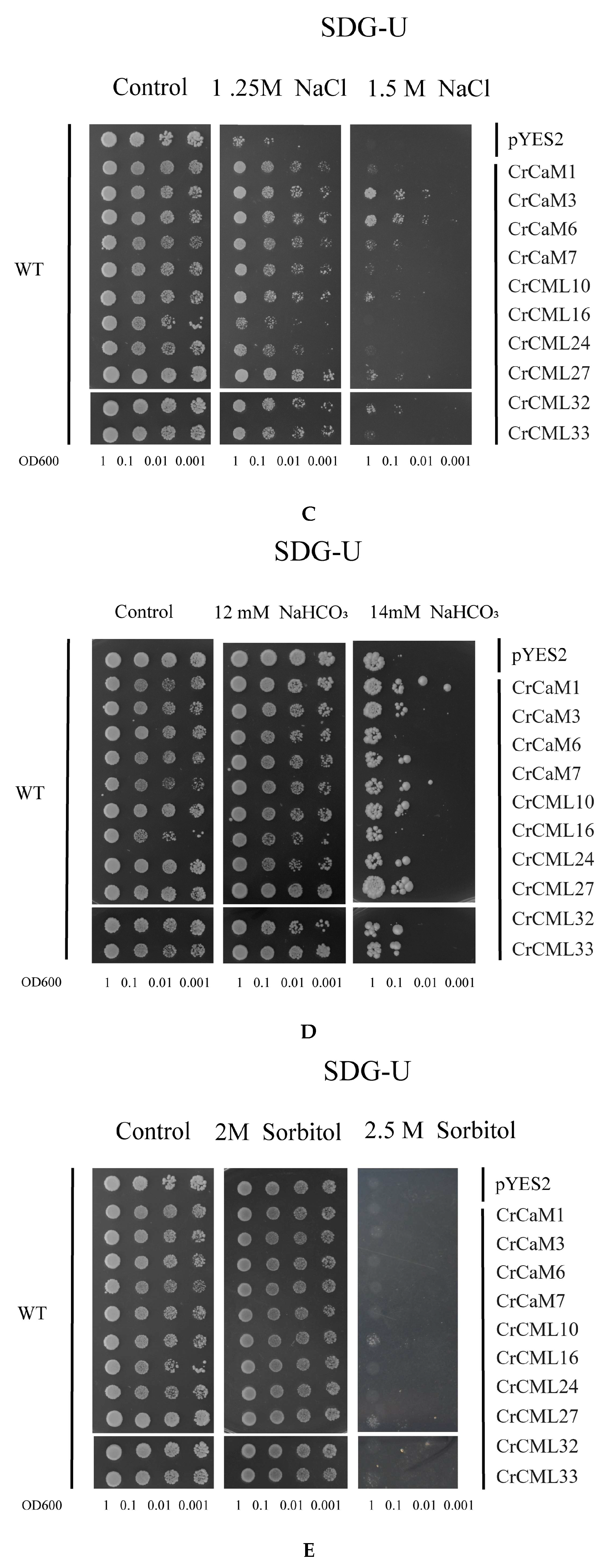
2.7. Functional Characterization of the CrCaMs/CrCMLs in Yeast
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Stress Treatments
4.2. Identification and Evolutionary Analyses of the CrCaM/CrCML Family in C. rosea
4.3. Chromosomal Location, Ka/Ks Calculation of the CrCaM/CrCML Genes, and Conserved Motifs of the Proteins
4.4. Cis-Regulatory Element Analysis of the CrCaM/CrCML Promoters
4.5. RNA-seq of the CrCaMs/CrCMLs in Different C. rosea Tissues or under Different Stress Treatments
4.6. Expression Pattern Analysis Using Quantitative Reverse Transcription (qRT)-PCR
4.7. Functional Identification Using a Yeast Expression System
4.8. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ranty, B.; Aldon, D.; Cotelle, V.; Galaud, J.P.; Thuleau, P.; Mazars, C. Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 2016, 7, 327. [Google Scholar] [CrossRef] [PubMed]
- McCormack, E.; Tsai, Y.C.; Braam, J. Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 2005, 10, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Dunand, C.; Snedden, W.; Galaud, J.P. CaM and CML emergence in the green lineage. Trends Plant Sci. 2015, 20, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Luan, S.; Kudla, J.; Rodriguez-Concepcion, M.; Yalovsky, S.; Gruissem, W. Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell 2002, 14, S389–400. [Google Scholar] [CrossRef]
- White, P.J.; Broadley, M.R. Calcium in plants. Ann. Bot. 2003, 92, 487–511. [Google Scholar] [CrossRef]
- Ketehouli, T.; Nguyen Quoc, V.H.; Dong, J.; Do, H.; Li, X.; Wang, F. Overview of the roles of calcium sensors in plants’ response to osmotic stress signalling. Funct. Plant Biol. 2022, 49, 589–599. [Google Scholar] [CrossRef]
- Chin, D.; Means, A.R. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000, 10, 322–328. [Google Scholar] [CrossRef]
- La-Verde, V.; Dominici, P.; Astegno, A. Towards understanding plant calcium signaling through calmodulin-like proteins: a biochemical and structural perspective. Int. J. Mol. Sci. 2018, 19, 1331. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Kumar, P.; Bae, H. Genomics and evolutionary aspect of calcium signaling event in calmodulin and calmodulin-like proteins in plants. BMC Plant Biol. 2017, 17, 38. [Google Scholar] [CrossRef]
- Virdi, A.S.; Singh, S.; Singh, P. Abiotic stress responses in plants: roles of calmodulin-regulated proteins. Front. Plant Sci. 2015, 6, 809. [Google Scholar] [CrossRef]
- He, M.; He, H.C.Q.; Ding, N.Z. Abiotic stresses: general defenses of land plants and chances for engineering multistress tolerance. Front. Plant Sci. 2018, 9, 1771. [Google Scholar] [CrossRef] [PubMed]
- McCormack, E.; Braam, J. Calmodulins and related potential calcium sensors of Arabidopsis. New Phytol. 2003, 159, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Boonburapong, B.; Buaboocha, T. Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biol. 2007, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Gao, L.; Yu, F.; Lü, S.; Yang, P. Evolution and diversification of CaM/CML gene family in green plants. Plant Physiol. Biochem. 2023, 202, 107922. [Google Scholar] [CrossRef]
- Huang, J.; Liu, N.; Ren, H.; Jian, S.G. Physiology and biochemical characteristics of Canavalia maritime under stress. J. Trop. Subtrop. Bot. 2019, 27, 157–163. [Google Scholar]
- Lin, R.; Zheng, J.; Pu, L.; Wang, Z.; Mei, Q.; Zhang, M.; Jian, S. Genome-wide identification and expression analysis of aquaporin family in Canavalia rosea and their roles in the adaptation to saline-alkaline soils and drought stress. BMC Plant Biol. 2021, 21, 333. [Google Scholar] [CrossRef]
- Jia, L.; Chu, H.; Wu, D.; Feng, M.; Zhao, L. Role of calmodulin in thermotolerance. Plant Signal Behav. 2014, 9, e28887. [Google Scholar] [CrossRef]
- Zeng, H.; Xu, L.; Singh, A.; Wang, H.; Du, L.; Poovaiah, B.W. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 2015, 6, 600. [Google Scholar] [CrossRef]
- Ranty, B.; Aldon, D.; Cotelle, V.; Galaud, J.P.; Thuleau, P.; Mazars, C. Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 2016, 7, 327. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Z.; Han, S.; Liu, P.; Sadeghnezhad, E.; Liu, M. Growth or survival: what is the role of calmodulin-like proteins in plant? Int. J. Biol. Macromol. 2023, 242(Pt1), 124733. [Google Scholar] [CrossRef]
- Shen, Q.; Fu, L.; Su, T.; Ye, L.; Huang, L.; Kuang, L.; Wu, L.; Wu, D.; Chen, Z.H.; Zhang, G. Calmodulin HvCaM1 negatively regulates salt tolerance via modulation of HvHKT1s and HvCAMTA4. Plant Physiol. 2020, 183, 1650–1662. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Xiong, X.; Arif, S.; Gao, L.; Zhao, L.; Shah, I.H.; Zhang, Y. A calmodulin-like CmCML13 from Cucumis melo improved transgenic Arabidopsis salt tolerance through reduced shoot’s Na+, and also improved drought resistance. Plant Physiol. Biochem. 2020, 155, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Chen, N.; Song, L.; Wang, D.; Cai, H.; Yao, L.; Li, X.; Guo, C. Alfalfa (Medicago sativa L.) MsCML46 gene encoding calmodulin-like protein confers tolerance to abiotic stress in tobacco. Plant Cell Rep 2021, 40, 1907–1922. [Google Scholar] [CrossRef] [PubMed]
- Jamra, G.; Agarwal, A.; Singh, N.; Sanyal, S.K.; Kumar, A.; Pandey, G.K. Ectopic expression of finger millet calmodulin confers drought and salinity tolerance in Arabidopsis thaliana. Plant Cell Rep. 2021, 40, 2205–2223. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, J.; Hou, Q.; Liu, Y.; Wang, J.; Deng, S. Isolation and functional characterization of a salt-responsive calmodulin-like gene MpCML40 from semi-mangrove Millettia pinnata. Int. J. Mol. Sci. 2021, 22, 3475. [Google Scholar] [CrossRef]
- Hau, B.; Symonds, K.; Teresinski, H.; Janssen, A.; Duff, L.; Smith, M.; Benidickson, K.; Plaxton, W.; Snedden, W.A. Arabidopsis calmodulin-like proteins CML13 and CML14 interact with calmodulin-binding transcriptional activators and function in salinity stress response. Plant Cell Physiol. 2024, 65, 282–300. [Google Scholar] [CrossRef]
- Raina, M.; Kumar, A.; Yadav, N.; Kumari, S.; Yusuf, M.A.; Mustafiz, A.; Kumar, D. StCaM2, a calcium binding protein, alleviates negative effects of salinity and drought stress in tobacco. Plant Mol Biol 2021, 106, 85–108. [Google Scholar] [CrossRef]
- Yang, M.; Zhou, B.; Song, Z.; Tan, Z.; Liu, R.; Luo, Y.; Guo, Z.; Lu, S. A calmodulin-like protein PvCML9 negatively regulates salt tolerance. Plant Physiol. Biochem. 2024, 210, 108642. [Google Scholar] [CrossRef]
- Xue, N.; Sun, M.; Gai, Z.; Bai, M.; Sun, J.; Sai, S.; Zhang, L. Genome-wide identification and expression analysis of calmodulin (CaM) and calmodulin-like (CML) genes in the brown algae Saccharina japonica. Plants (Basel) 2023, 12, 1934. [Google Scholar] [CrossRef]
- Gao, L.; Damaris, R.N.; Yu, F.; Yang, P.F. Genome-wide identification and expression analysis of CaM/CML gene family in sacred lotus (Nelumbo nucifera). Plant Mol. Biol. Rep. 2022, 40, 418–432. [Google Scholar] [CrossRef]
- Yang, M.; Chen, J.; Liu, T.; Xiang, L.; Zhou, B.F. Genome-wide identification and expression analysis of calmodulin-like gene family in Paspalums vaginatium revealed their role in response to salt and cold stress. Curr. Issues Mol. Biol. 2023, 45, 1693–1711. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhu, J.; Zhu, L.; Tang, Y.; Hao, Z.; Zhang, J.; Shi, J.; Cheng, T.; Lu, L. Genome-wide analyses of calmodulin and calmodulin-like proteins in the halophyte Nitraria sibirica reveal their involvement in response to salinity, drought and cold stress. Int. J. Biol. Macromol. 2023, 253 (Pt7)(Pt7), 127442. [Google Scholar] [CrossRef]
- Lin, R.; Zou, T.; Mei, Q.; Wang, Z.; Zhang, M.; Jian, S. Genome-wide analysis of the late embryogenesis abundant (LEA) and abscisic acid-, stress-, and ripening-induced (ASR) gene superfamily from Canavalia rosea and their roles in salinity/alkaline and drought tolerance. Int. J. Mol. Sci. 2021, 22, 4554. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Jian, S.; Wang, Z. Comprehensive analysis of the Hsp20 gene family in Canavalia rosea indicates its roles in the response to multiple abiotic stresses and adaptation to tropical coral islands. Int. J. Mol. Sci. 2022, 23, 6405. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, Z.; Jian, S. Functional characterization of heat shock factor (CrHsf) families provide comprehensive insight into the adaptive mechanisms of Canavalia rosea (Sw.) DC. to tropical coral islands. Int. J. Mol. Sci. 2022, 23, 12357. [Google Scholar] [CrossRef]
- Zou, T.; Lin, R.; Pu, L.; Mei, Q.; Wang, Z.; Jian, S.; Zhang, M. Genome-wide identification, structure characterization, expression pattern profiling, and substrate specificity of the metal tolerance protein family in Canavalia rosea (Sw.) DC. Plants (Basel) 2021, 10, 1340. [Google Scholar] [CrossRef]
- Zou, T.; Pu, L.; Lin, R.; Mo, H.; Wang, Z.; Jian, S.; Zhang, M. Roles of Canavalia rosea metallothioneins in metal tolerance and extreme environmental adaptation to tropical coral reefs. J. Plant Physiol. 2022, 268, 153559. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, Z.; Jian, S. Genome-wide identification and functional analysis of the GASA gene family responding to multiple stressors in Canavalia rosea. Genes (Basel) 2022, 13, 1988. [Google Scholar] [CrossRef]
- Ding, Q.; Liu, H.; Lin, R.; Wang, Z.; Jian, S.; Zhang, M. Genome-wide functional characterization of Canavalia rosea cysteine-rich trans-membrane module (CrCYSTM) genes to reveal their potential protective roles under extreme abiotic stress. Plant Physiol. Biochem. 2023, 200, 107786. [Google Scholar] [CrossRef]
- Goldtzvik, Y.; Sen, N.; Lam, S.D.; Orengo, C. Protein diversification through post-translational modifications, alternative splicing, and gene duplication. Curr. Opin. Struct. Biol. 2023, 81, 102640. [Google Scholar] [CrossRef]
- Das Laha, S.; Dutta, S.; Schäffner, A.R.; Das, M. Gene duplication and stress genomics in Brassicas: Current understanding and future prospects. J. Plant Physiol. 2020, 255, 153293. [Google Scholar] [CrossRef] [PubMed]
- Martins, T.S.; Da-Silva, C.J.; Shabala, S.; Striker, G.G.; Carvalho, I.R.; de Oliveira, A.C.B.; do Amarante, L. Understanding plant responses to saline waterlogging: insights from halophytes and implications for crop tolerance. Planta 2023, 259, 24. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; He, R.; Tian, C.Y.; Song, J. Utilization of halophytes in saline agriculture and restoration of contaminated salinized soils from genes to ecosystem: Suaeda salsa as an example. Mar. Pollut. Bull. 2023, 197, 115728. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Li, J.; Yang, Y.; Jiang, C.; Guo, Y. Designing salt stress-resilient crops: Current progress and future challenges. J. Integr. Plant Biol. 2024, 66, 303–329. [Google Scholar] [CrossRef]
- McAinsh, M.R.; Pittman, J.K. Shaping the calcium signature. New Phytol. 2009, 181, 275–294. [Google Scholar] [CrossRef]
- Sanders, D.; Brownlee, C.; Harper, J.F. Communicating with calcium. Plant Cell 1999, 11, 691–706. [Google Scholar] [CrossRef]
- Liao, J.; Deng, J.; Qin, Z.; Tang, J.; Shu, M.; Ding, C.; Liu, J.; Hu, C.; Yuan, M.; Huang, Y.; Yang, R.; Zhou, Y. Genome-wide identification and analyses of calmodulins and calmodulin-like proteins in Lotus japonicus. Front. Plant Sci. 2017, 8, 482. [Google Scholar] [CrossRef]
- Sun, Q.; Yu, S.; Guo, Z. Calmodulin-Like (CML) Gene family in Medicago truncatula: genome-wide identification, characterization and expression analysis. Int. J. Mol. Sci. 2020, 21, 7142. [Google Scholar] [CrossRef]
- Yadav, M.; Pandey, J.; Chakraborty, A.; Hassan, M.I.; Kundu, J.K.; Roy, A.; Singh, I.K.; Singh, A. A comprehensive analysis of calmodulin-Like proteins of Glycine max indicates their role in calcium signaling and plant defense against insect attack. Front. Plant Sci. 2022, 13, 817950. [Google Scholar] [CrossRef]
- Zhao, H.; Gao, Y.; Du, Y.; Du, J.; Han, Y. Genome-wide analysis of the CML gene family and its response to melatonin in common bean (Phaseolus vulgaris L.). Sci. Rep. 2023, 13, 1196. [Google Scholar] [CrossRef]
- Sun, Q.; Huang, R.; Zhu, H.; Sun, Y.; Guo, Z. A novel Medicago truncatula calmodulin-like protein (MtCML42) regulates cold tolerance and flowering time. Plant J. 2021, 108, 1069–1082. [Google Scholar] [CrossRef] [PubMed]
- Fu, N.; Wang, L.; Han, X.; Yang, Q.; Zhang, Y.; Tong, Z.; Zhang, J. Genome-wide identification and expression analysis of calmodulin and calmodulin-like genes, revealing CaM3 and CML13 participating in drought stress in Phoebe bournei. Int. J. Mol. Sci. 2023, 25, 545. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]


| Name | Locus | Protein length | Major amino acids (aa, %) | Mw (kDa) | PI | II | AI | GRAVY | Disordered aa (%) | WoLF_PSORT | Plant-PLoc |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CrCaM1 | 03T007739 | 149 | D(12.8%),E(12.8%),L(7.4%) | 16.85 | 4.11 | 24.52 | 69.40 | -0.619 | 32.89 | nucl: 4, mito: 4, extr: 3, cyto: 2 | Endoplasmic reticulum |
| CrCaM2 | 04T011067 | 171 | E(14.0%),D(9.9%),L(8.8%) | 19.68 | 4.05 | 38.15 | 84.33 | -0.337 | 30.99 | cyto: 6, chlo: 3, nucl: 3, plas: 1 | Chloroplast |
| CrCaM3 | 04T011068 | 150 | E(15.3%),D(12.0%),L(9.3%) | 17.04 | 3.89 | 46.07 | 89.00 | -0.331 | 25.33 | cyto: 8, chlo: 2, extr: 1, E.R.: 1, cysk: 1 | Nucleus |
| CrCaM4 | 04T013039 | 150 | E(14.7%),D(12.7%),L(8.0%) | 17.03 | 4.03 | 35.84 | 81.20 | -0.487 | 31.33 | chlo: 5, cyto: 4.5, cyto_nucl: 3.33333, extr: 3 | Endoplasmic reticulum |
| CrCaM5 | 06T017463 | 190 | D(11.1%),D(10.5%),L(10.0%) | 21.59 | 4.14 | 33.12 | 80.58 | -0.268 | 42.11 | chlo: 6, plas: 2, nucl: 1.5, cysk_nucl: 1.5, cyto: 1, mito: 1, vacu: 1 | Chloroplast |
| CrCaM6 | 07T020185 | 149 | E(13.4%),D(12.1%),L(7.4%) | 16.86 | 4.12 | 25.82 | 69.40 | -0.619 | 32.89 | nucl: 4, mito: 4, extr: 3, cyto: 2 | Endoplasmic reticulum |
| CrCaM7 | 11T029359 | 149 | D(12.8%),E(12.8%),A(7.4%),L(7.4%) | 16.83 | 4.11 | 23.23 | 70.07 | -0.602 | 32.89 | cyto: 4, mito: 4, nucl: 3, extr: 2 | Endoplasmic reticulum |
| CrCML1 | 01T000055 | 177 | N(10.7%),D(10.7%),E(8.5%),G(8.5%) | 19.60 | 4.23 | 39.80 | 59.49 | -0.808 | 58.19 | nucl: 6, chlo: 4, mito: 2, extr: 2 | Nucleus |
| CrCML2 | 01T000134 | 187 | D(8.2%),S(8.2%),E(9.1%),G(9.1%) | 20.52 | 4.30 | 45.03 | 75.56 | -0.396 | 55.61 | chlo: 10, mito: 4 | Nucleus |
| CrCML3 | 01T001229 | 81 | E(12.3%),D(11.1%),L(11.1%) | 9.21 | 4.62 | 41.03 | 85.43 | -0.593 | 22.22 | cyto: 5, nucl: 3.5, chlo: 3, cysk_nucl: 2.5, extr: 1 | Cytoplasm |
| CrCML4 | 01T001582 | 191 | S(10.5%),D(9.9%),G(8.4%) | 21.71 | 5.12 | 49.75 | 56.65 | -0.700 | 65.97 | mito: 9, nucl: 4 | Chloroplast |
| CrCML5 | 01T001986 | 160 | L(11.2%),D(8.1%),G(8.1%) | 17.62 | 4.21 | 35.31 | 83.56 | -0.124 | 38.75 | nucl: 5.5, nucl_plas: 4, cyto: 3.5, cyto_E.R.: 2.5, mito: 2, plas: 1.5 | Nucleus |
| CrCML6 | 01T002018 | 288 | E(11.1%),L(9.7%),F(8.7%) | 33.26 | 4.97 | 42.59 | 76.56 | -0.316 | 42.01 | cyto: 5, chlo: 3, plas: 3, E.R.: 2 | Chloroplast |
| CrCML7 | 01T002045 | 163 | L(12.3%),A(11.0%),E(9.2%) | 18.13 | 4.66 | 39.69 | 84.54 | -0.315 | 43.56 | plas: 5, nucl_plas: 5, nucl: 3, cyto: 2, mito: 2, chlo: 1 | Nucleus |
| CrCML8 | 01T002566 | 179 | L(11.2%),E(10.6%),N(7.8%) | 20.41 | 4.37 | 46.73 | 87.65 | -0.046 | 42.46 | chlo: 11, cyto: 1, extr: 1 | Chloroplast |
| CrCML9 | 01T002767 | 193 | D(9.3%),K(9.3%),S(8.8%) | 22.32 | 8.70 | 39.56 | 60.57 | -0.627 | 60.10 | nucl: 7.5, cyto_nucl: 4.5, chlo: 4, mito: 2 | Cytoplasm |
| CrCML10 | 01T003442 | 155 | G(14.2%),D(12.9%),L(7.7%) | 16.86 | 4.81 | 31.39 | 66.71 | -0.414 | 72.90 | cyto: 6, mito: 4, chlo: 2, nucl: 1 | Chloroplast |
| CrCML11 | 02T004181 | 197 | D(10.7%),L(9.6%),S(9.6%) | 21.89 | 4.55 | 41.87 | 72.79 | -0.406 | 63.45 | nucl: 7, cyto: 3, mito: 2, extr: 2 | Cytoplasm |
| CrCML12 | 02T004464 | 187 | G(10.7%),L(10.7%),E(10.2%) | 20.98 | 4.69 | 36.33 | 78.13 | -0.415 | 45.99 | cyto: 11, nucl: 1, mito: 1 | Cytoplasm |
| CrCML13 | 02T004981 | 139 | L(14.4%),E(10.8%),D(9.4%) | 16.00 | 4.34 | 27.74 | 80.58 | -0.491 | 34.53 | nucl: 5, mito: 5, chlo: 3 | Cytoplasm |
| CrCML14 | 02T006679 | 214 | L(9.8%),S(9.8%),D(9.3%) | 24.15 | 5.18 | 32.02 | 76.50 | -0.309 | 47.20 | chlo: 11, nucl: 1, mito: 1 | Cytoplasm |
| CrCML15 | 03T007802 | 152 | D(11.8%),G(10.5%),E(9.9%) | 17.03 | 4.32 | 15.81 | 73.09 | -0.447 | 32.24 | chlo: 4, extr: 4, nucl: 2, cyto: 2, mito: 2 | Cytoplasm |
| CrCML16 | 03T007833 | 223 | S(13.9%),D(9.4%),V(9.0%) | 24.55 | 4.63 | 48.90 | 73.32 | -0.526 | 60.54 | chlo: 5, cyto: 5, nucl: 2, mito: 1 | Chloroplast |
| CrCML17 | 03T008082 | 148 | E(14.2%),D(10.1%),I(9.5%) | 17.00 | 4.14 | 38.60 | 84.39 | -0.443 | 27.03 | cyto: 13 | Chloroplast |
| CrCML18 | 03T009031 | 149 | E(16.1%),D(10.7%),L(10.7%) | 16.93 | 4.02 | 44.45 | 83.09 | -0.448 | 38.26 | cyto_nucl: 5.83333, chlo: 4, nucl: 4, cyto: 3.5, cyto_E.R.: 2.83333 | Chloroplast |
| CrCML19 | 03T009705 | 220 | L(10.5%),E(8.6%),I(8.6%) | 25.58 | 5.69 | 27.67 | 86.00 | -0.378 | 41.82 | cyto: 10, mito: 2, chlo: 1 | Chloroplast |
| CrCML20 | 03T010270 | 182 | L(12.1%),D(8.8%),E(8.8%) | 20.31 | 4.37 | 39.87 | 88.41 | -0.234 | 41.21 | mito: 7.5, chlo_mito: 7, chlo: 5.5 | Nucleus |
| CrCML21 | 03T010292 | 206 | L(10.2%),S(10.2%),D(9.2%) | 23.42 | 5.47 | 33.93 | 79.95 | -0.242 | 30.10 | chlo: 10, extr: 2, cyto: 1 | Chloroplast |
| CrCML22 | 03T010654 | 203 | E(11.3%),F(10.8%),S(10.3%) | 23.75 | 4.75 | 52.45 | 72.96 | -0.467 | 38.92 | nucl: 9, cyto: 2, extr: 2 | Chloroplast |
| CrCML23 | 04T011122 | 148 | E(13.5%),D(12.8%),L(8.8%) | 17.08 | 4.05 | 24.54 | 88.85 | -0.420 | 32.43 | cyto: 6, cyto_nucl: 6, chlo: 4, extr: 1 | Cytoplasm |
| CrCML24 | 04T011931 | 206 | S(10.7%),A(9.7%),D(9.2%),G(9.2%) | 22.30 | 4.62 | 36.41 | 71.55 | -0.270 | 66.02 | nucl: 7, mito: 4, chlo: 2 | Chloroplast |
| CrCML25 | 04T012182 | 163 | L(13.5%),E(9.8%),A(9.2%) | 18.29 | 4.53 | 42.13 | 91.66 | -0.279 | 30.06 | nucl_plas: 5.5, nucl: 5, plas: 4, cyto: 2, mito: 2 | Chloroplast |
| CrCML26 | 04T012874 | 84 | E(15.5%),V(11.9%),L(8.3%) | 9.87 | 4.38 | 59.01 | 92.62 | -0.230 | 23.81 | cyto: 5, extr: 5, nucl: 2, chlo: 1 | Chloroplast |
| CrCML27 | 05T014580 | 172 | K(11.6%),S(10.5%),E(9.3%),G(9.3%) | 19.08 | 5.18 | 26.33 | 58.37 | -0.505 | 47.09 | cyto: 7, nucl: 3, chlo: 2, mito: 2 | Nucleus |
| CrCML28 | 05T014903 | 145 | E(11.0%),L(10.3%),D(9.0%) | 16.53 | 4.86 | 31.69 | 84.76 | -0.403 | 35.86 | nucl: 4, cyto: 3, plas: 2, chlo: 1, mito: 1, extr: 1, cysk: 1 | Chloroplast |
| CrCML29 | 05T015286 | 160 | L(13.8%),A(8.8%),G(8.8%) | 17.49 | 4.41 | 27.70 | 95.75 | -0.022 | 40.00 | plas: 5, nucl_plas: 4.5, cyto: 4, nucl: 2, chlo: 1, mito: 1 | Chloroplast |
| CrCML30 | 05T015406 | 224 | E(12.1%),L(9.8%),K(9.8%) | 25.17 | 4.83 | 44.01 | 88.75 | -0.214 | 50.00 | chlo: 9, extr: 3, nucl: 1 | Cytoplasm |
| CrCML31 | 05T017030 | 160 | L(13.8%),A(9.4%),D(8.8%),G(8.8%) | 17.59 | 4.30 | 32.93 | 90.31 | 0.057 | 42.50 | chlo: 10, mito: 2, nucl: 1 | Chloroplast |
| CrCML32 | 06T018151 | 159 | L(15.1%),D(10.7%),E(10.7%) | 18.21 | 4.31 | 46.05 | 86.42 | -0.359 | 43.40 | nucl: 5, cyto: 5, chlo: 2, mito: 1 | Cytoplasm |
| CrCML33 | 06T018267 | 157 | L(17.2%),E(12.1%),G(10.2%) | 16.96 | 4.10 | 25.21 | 108.09 | -0.061 | 33.76 | cyto: 10.5, cyto_E.R.: 6.33333, E.R._vacu: 1.33333 | Nucleus |
| CrCML34 | 06T018865 | 84 | D(11.9%),E(10.7%),A(9.5%) | 9.31 | 4.21 | 20.60 | 67.38 | -0.568 | 26.19 | nucl: 5, cyto: 3, mito: 3, chlo: 1, plas: 1 | Mitochondrion |
| CrCML35 | 06T019154 | 98 | G(9.2%),A(8.2%),K(8.2%) | 11.20 | 8.79 | 20.00 | 81.63 | -0.587 | 61.22 | mito: 8.5, chlo_mito: 6, chlo: 2.5, nucl: 1.5, cyto_nucl: 1.5 | Chloroplast |
| CrCML36 | 08T022893 | 150 | D(12.7%),G(10.7%),E(10.0%) | 16.99 | 4.34 | 26.75 | 72.07 | -0.516 | 29.33 | nucl: 4, chlo: 3, mito: 3, cyto: 2, extr: 2 | Cytoplasm |
| CrCML37 | 09T023581 | 141 | E(14.9%),L(12.1%),G(9.9%) | 16.14 | 4.34 | 46.10 | 75.39 | -0.365 | 29.08 | nucl: 6, chlo: 4, cyto: 2, mito: 1 | Chloroplast |
| CrCML38 | 09T025115 | 218 | D(9.6%),L(9.6%),I(8.7%) | 24.88 | 4.74 | 41.15 | 88.03 | -0.259 | 50.00 | chlo: 12, mito: 1 | Nucleus |
| CrCML39 | 10T025518 | 191 | S(12.0%),E(11.5%),D(8.9%) | 21.41 | 4.58 | 46.26 | 69.37 | -0.468 | 46.07 | chlo: 7, mito: 5, nucl: 2 | Nucleus |
| CrCML40 | 10T026298 | 149 | D(11.4%),L(9.4%),E(7.4%) | 16.89 | 4.70 | 34.32 | 80.54 | -0.436 | 59.06 | cyto: 9, cyto_nucl: 8, nucl: 3 | Cytoplasm |
| CrCML41 | 10T026655 | 190 | S(12.6%),D(10.5%),L(10.5%) | 21.90 | 4.77 | 51.14 | 70.32 | -0.670 | 61.05 | mito: 7.5, chlo_mito: 5.5, nucl: 4, chlo: 2.5 | Chloroplast |
| CrCML42 | 10T027366 | 165 | E(11.5%),F(9.1%),V(8.5%) | 19.30 | 4.82 | 45.76 | 83.15 | -0.259 | 24.85 | extr: 4, vacu: 3, E.R.: 3, golg: 2, chlo: 1 | Chloroplast |
| CrCML43 | 11T028665 | 210 | S(12.9%),K(10.0%),G(8.1%),L(8.1%) | 23.37 | 7.63 | 35.58 | 78.86 | -0.304 | 47.14 | nucl: 11, cyto: 3 | Nucleus |
| CrCML44 | 11T028666 | 84 | A(10.7%),D(10.7%),E(10.7%) | 9.31 | 4.37 | 14.27 | 74.40 | -0.425 | 35.71 | cyto: 8, nucl: 2, mito: 2, chlo: 1 | Peroxisome |
| Duplicated pair | Duplicate Type | Ka | Ks | Ka/Ks | P-Value(Fisher) | Positive Selection |
|---|---|---|---|---|---|---|
| CrCaM3-CrCaM4 | Segmental | 0.12532 | 0.542019 | 0.231209 | 6.07E-09 | No |
| CrCaM5-CrCaM7 | Segmental | 0.00565853 | 0.507362 | 0.0111528 | 7.70E-24 | No |
| CrCML2-CrCML39 | Segmental | 0.224798 | 1.46238 | 0.153721 | 4.27E-20 | No |
| CrCML4-CrCML41 | Segmental | 0.147676 | 0.735966 | 0.200656 | 3.32E-14 | No |
| CrCML8-CrCML42 | Segmental | 0.280668 | 0.761875 | 0.368392 | 5.48E-06 | No |
| CrCML13-CrCML32 | Segmental | 0.200534 | 0.803216 | 0.249664 | 1.16E-08 | No |
| CrCML14-CrCML21 | Segmental | 0.199284 | 0.538055 | 0.370378 | 1.25E-06 | No |
| CrCML16-CrCML24 | Segmental | 0.250644 | 0.787603 | 0.318237 | 3.62E-10 | No |
| CrCML17-CrCML23 | Segmental | 0.157841 | 0.707805 | 0.223001 | 2.98E-10 | No |
| CrCML18-CrCML26 | Segmental | 0.137486 | 0.583507 | 0.235619 | 1.30E-08 | No |
| CrCML28-CrCML40 | Segmental | 0.0930564 | 0.609897 | 0.152577 | 2.73E-13 | No |
| CrCML29-CrCML31 | Segmental | 0.0905119 | 0.792094 | 0.114269 | 8.83E-20 | No |
| CrCML34-CrCML44 | Segmental | 0.0645313 | 0.529471 | 0.121879 | 1.74E-08 | No |
| CrCML36-CrCML38 | Segmental | 0.112235 | 1.05912 | 0.105971 | 1.19E-19 | No |
| CrCaM2-CrCaM3 | Tandem | \ | \ | \ | \ | \ |
| CrCML43-CrCML44 | Tandem | \ | \ | \ | \ | \ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





