Submitted:
07 October 2024
Posted:
08 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
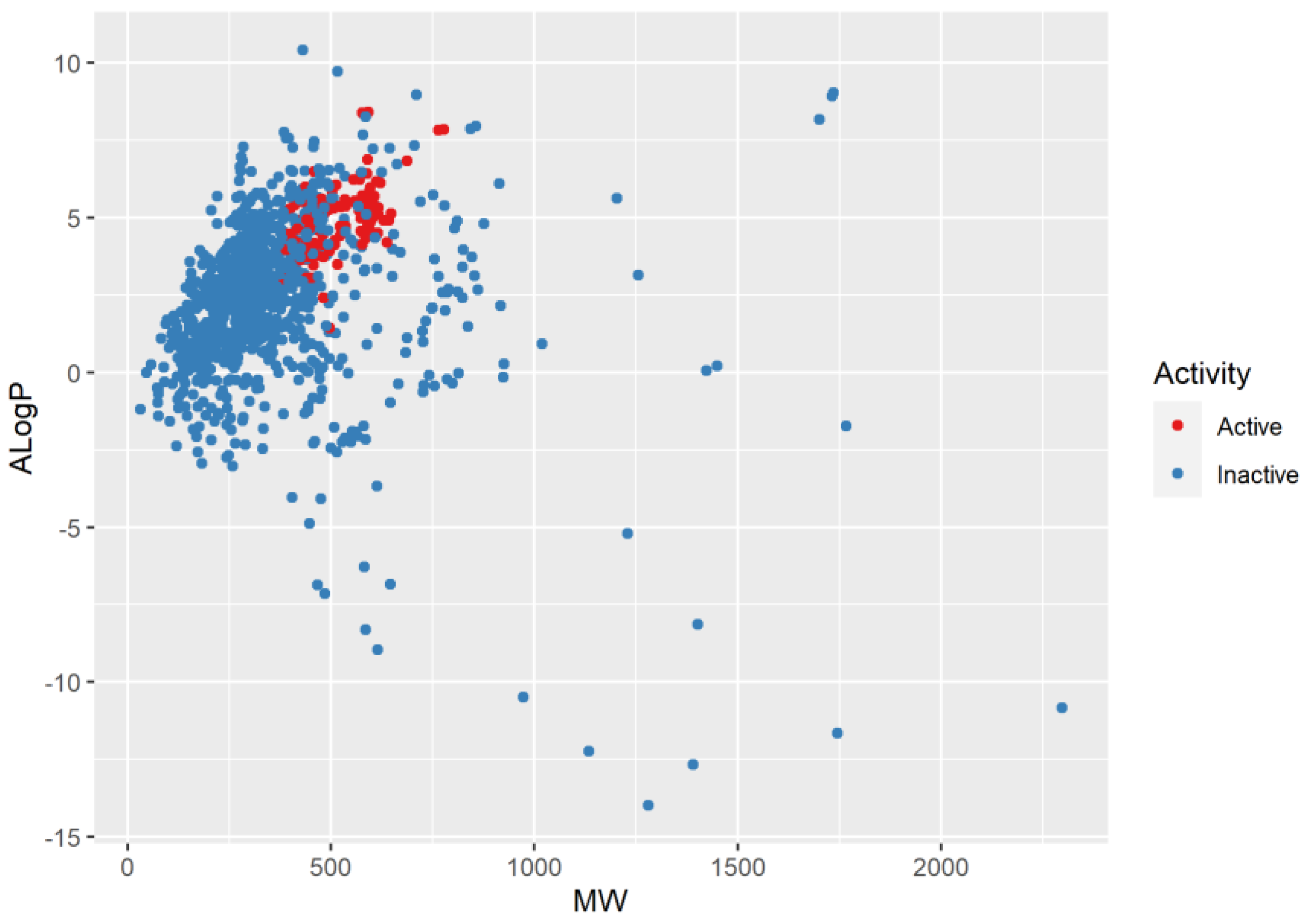
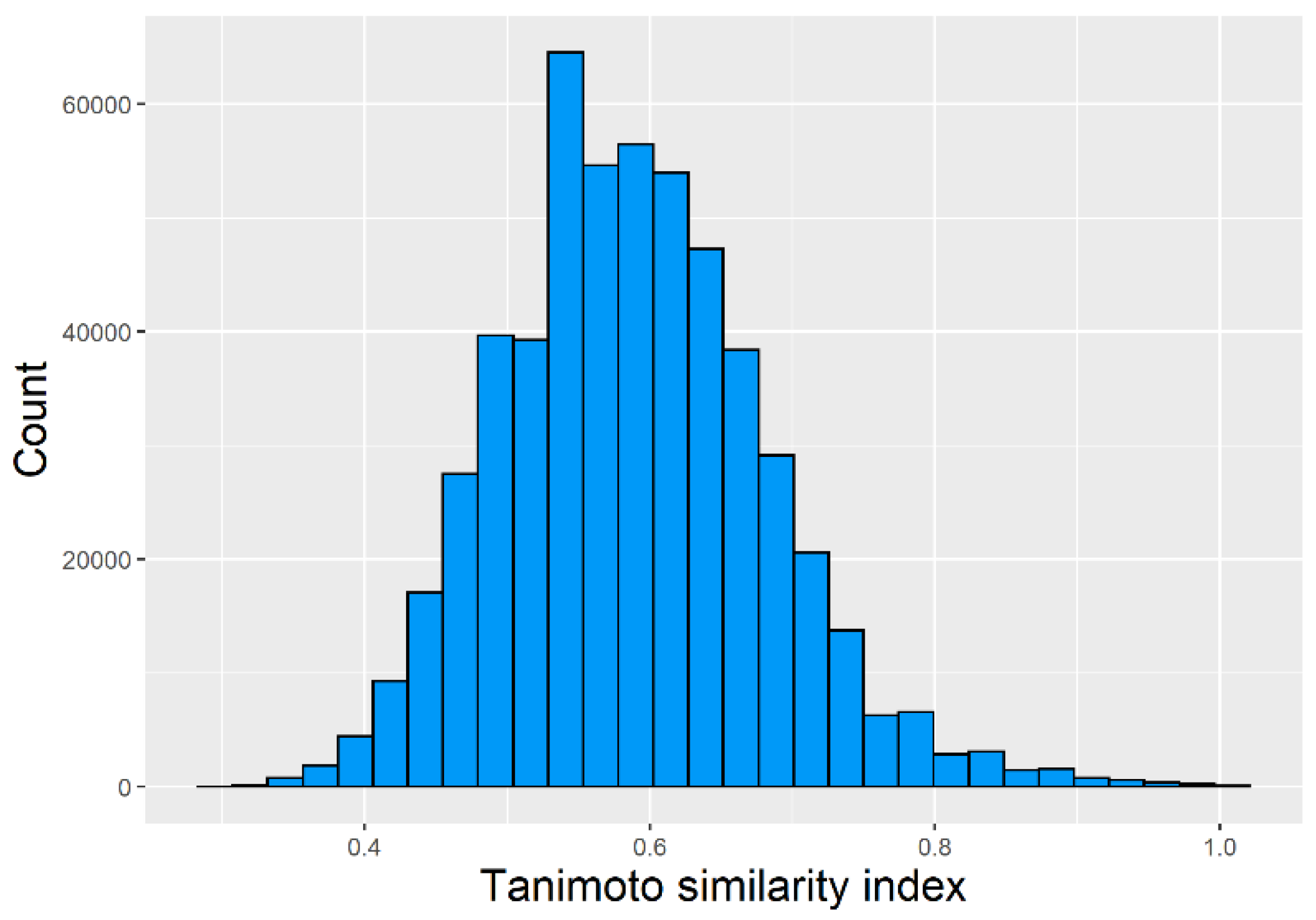
2.1. Chemical Space Distribution and Diversity of the Compounds in the Training Data Set
2.1. Regression Models and Their Performance
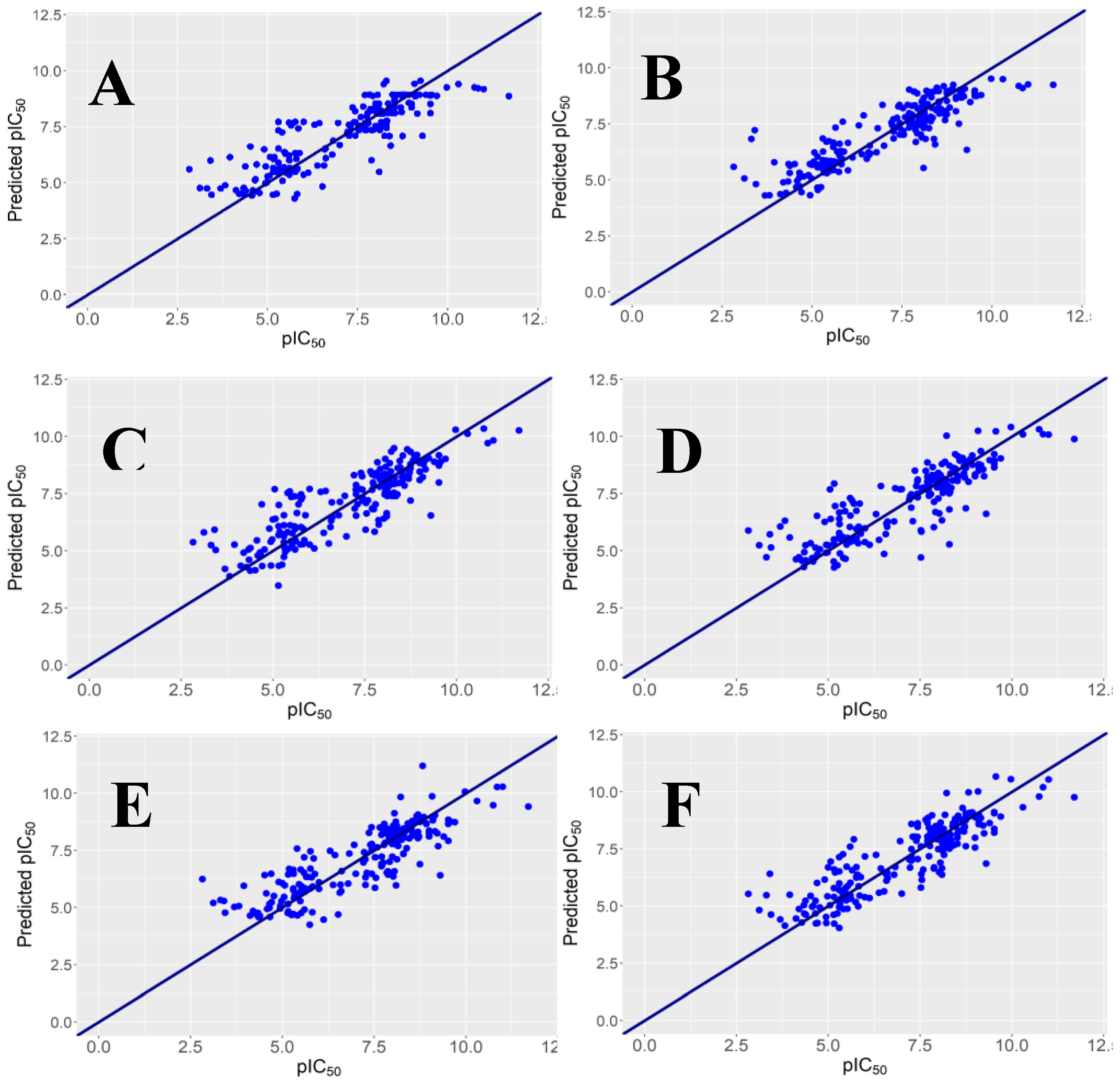
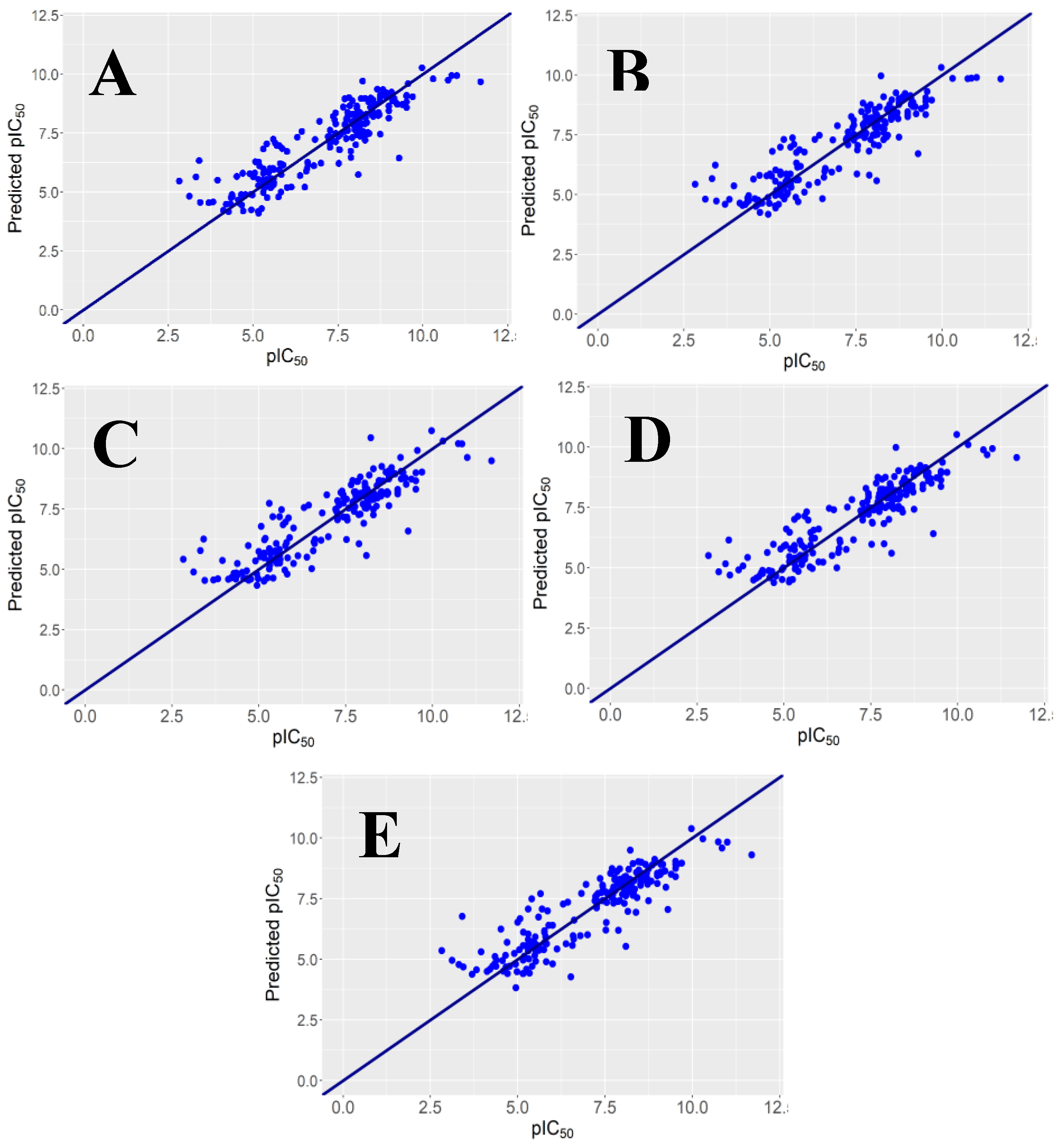
y-Randomization
Descriptors Useful for HMGCo-A Inhibition Prediction
Virtual Screening of a Data Set of Natural Compounds
Use Case Example for Herbal Extracts
3. Discussion
4. Materials and Methods
4.1. Data Set
Molecular Fingerprint Calculation
Chemical Space Distribution and Diversity
Feature Selection, Model Building and Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Toth, P.P.; Banach, M. Statins: Then and Now. Methodist DeBakey Cardiovascular Journal 2019, 15, 23. [Google Scholar] [CrossRef] [PubMed]
- Adhyaru, B.B.; Jacobson, T.A. Safety and Efficacy of Statin Therapy. Nat Rev Cardiol 2018, 15, 757–769. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.M.; DeBose-Boyd, R.A. Posttranslational Regulation of HMG CoA Reductase, the Rate-Limiting Enzyme in Synthesis of Cholesterol. Annu. Rev. Biochem. 2021, 90, 659–679. [Google Scholar] [CrossRef] [PubMed]
- Almeida, S.O.; Budoff, M. Effect of Statins on Atherosclerotic Plaque. Trends in Cardiovascular Medicine 2019, 29, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Arefieva, T.I.; Filatova, A.Yu.; Potekhina, A.V.; Shchinova, A.M. Immunotropic Effects and Proposed Mechanism of Action for 3-Hydroxy-3-Methylglutaryl-Coenzyme A Reductase Inhibitors (Statins). Biochemistry Moscow 2018, 83, 874–889. [Google Scholar] [CrossRef]
- Saeedi Saravi, S.S.; Saeedi Saravi, S.S.; Arefidoust, A.; Dehpour, A.R. The Beneficial Effects of HMG-CoA Reductase Inhibitors in the Processes of Neurodegeneration. Metab Brain Dis 2017, 32, 949–965. [Google Scholar] [CrossRef]
- Sodero, A.O.; Barrantes, F.J. Pleiotropic Effects of Statins on Brain Cells. Biochimica et Biophysica Acta (BBA) - Biomembranes 2020, 1862, 183340. [Google Scholar] [CrossRef]
- Stine, J.E.; Guo, H.; Sheng, X.; Han, X.; Schointuch, M.N.; Gilliam, T.P.; Gehrig, P.A.; Zhou, C.; Bae-Jump, V.L. The HMG-CoA Reductase Inhibitor, Simvastatin, Exhibits Anti-Metastatic and Anti-Tumorigenic Effects in Ovarian Cancer. Oncotarget 2016, 7, 946–960. [Google Scholar] [CrossRef]
- Ahmadi, M.; Amiri, S.; Pecic, S.; Machaj, F.; Rosik, J.; Łos, M.J.; Alizadeh, J.; Mahdian, R.; Da Silva Rosa, S.C.; Schaafsma, D.; et al. Pleiotropic Effects of Statins: A Focus on Cancer. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 2020, 1866, 165968. [Google Scholar] [CrossRef]
- Bahrami, A.; Bo, S.; Jamialahmadi, T.; Sahebkar, A. Effects of 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase Inhibitors on Ageing: Molecular Mechanisms. Ageing Research Reviews 2020, 58, 101024. [Google Scholar] [CrossRef]
- Zhou, H.; Xie, Y.; Baloch, Z.; Shi, Q.; Huo, Q.; Ma, T. The Effect of Atorvastatin, 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase Inhibitor (HMG-CoA), on the Prevention of Osteoporosis in Ovariectomized Rabbits. J Bone Miner Metab 2017, 35, 245–254. [Google Scholar] [CrossRef] [PubMed]
- De La Cruz, J.A.; Mihos, C.G.; Horvath, S.A.; Santana, O. The Pleiotropic Effects of Statins in Endocrine Disorders. EMIDDT 2019, 19, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Climent, E.; Benaiges, D.; Pedro-Botet, J. Hydrophilic or Lipophilic Statins? Front. Cardiovasc. Med. 2021, 8, 687585. [Google Scholar] [CrossRef]
- Montastruc, J. Rhabdomyolysis and Statins: A Pharmacovigilance Comparative Study between Statins. Brit J Clinical Pharma 2023, 89, 2636–2638. [Google Scholar] [CrossRef]
- Ma, M.-M.; Xu, Y.-Y.; Sun, L.-H.; Cui, W.-J.; Fan, M.; Zhang, S.; Liu, L.; Wu, L.-Z.; Li, L.-C. Statin-Associated Liver Dysfunction and Muscle Injury: Epidemiology, Mechanisms, and Management Strategies. International Journal of General Medicine 2024, 2055–2063. [Google Scholar] [CrossRef]
- Clarke, A.T.; Johnson, P.C.D.; Hall, G.C.; Ford, I.; Mills, P.R. High Dose Atorvastatin Associated with Increased Risk of Significant Hepatotoxicity in Comparison to Simvastatin in UK GPRD Cohort. PLoS ONE 2016, 11, e0151587. [Google Scholar] [CrossRef]
- Thakker, D.; Nair, S.; Pagada, A.; Jamdade, V.; Malik, A. Statin Use and the Risk of Developing Diabetes: A Network Meta-analysis. Pharmacoepidemiology and Drug 2016, 25, 1131–1149. [Google Scholar] [CrossRef] [PubMed]
- Sinyavskaya, L.; Gauthier, S.; Renoux, C.; Dell’Aniello, S.; Suissa, S.; Brassard, P. Comparative Effect of Statins on the Risk of Incident Alzheimer Disease. Neurology 2018, 90. [Google Scholar] [CrossRef]
- Hirota, T.; Fujita, Y.; Ieiri, I. An Updated Review of Pharmacokinetic Drug Interactions and Pharmacogenetics of Statins. Expert Opinion on Drug Metabolism & Toxicology 2020, 16, 809–822. [Google Scholar] [CrossRef]
- Zhang, X.; Xing, L.; Jia, X.; Pang, X.; Xiang, Q.; Zhao, X.; Ma, L.; Liu, Z.; Hu, K.; Wang, Z.; et al. Comparative Lipid-Lowering/Increasing Efficacy of 7 Statins in Patients with Dyslipidemia, Cardiovascular Diseases, or Diabetes Mellitus: Systematic Review and Network Meta-Analyses of 50 Randomized Controlled Trials. Cardiovascular Therapeutics 2020, 2020, 1–21. [Google Scholar] [CrossRef]
- Leelananda, S.P.; Lindert, S. Computational Methods in Drug Discovery. Beilstein Journal of Organic Chemistry 2016, 12, 2694–2718. [Google Scholar] [CrossRef] [PubMed]
- Khan, P.M.; Roy, K. Current Approaches for Choosing Feature Selection and Learning Algorithms in Quantitative Structure–Activity Relationships (QSAR). Expert Opinion on Drug Discovery 2018, 13, 1075–1089. [Google Scholar] [CrossRef]
- Grisoni, F.; Ballabio, D.; Todeschini, R.; Consonni, V. Molecular Descriptors for Structure–Activity Applications: A Hands-On Approach. In Computational Toxicology; Nicolotti, O., Ed.; Methods in Molecular Biology; Springer New York: New York, NY, 2018; Volume 1800, pp. 3–53. ISBN 978-1-4939-7898-4. [Google Scholar]
- Sato, A.; Miyao, T.; Jasial, S.; Funatsu, K. Comparing Predictive Ability of QSAR/QSPR Models Using 2D and 3D Molecular Representations. J Comput Aided Mol Des 2021, 35, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Rajathei, D.M.; Parthasarathy, S.; Selvaraj, S. Combined QSAR Model and Chemical Similarity Search for Novel HMG-CoA Reductase Inhibitors for Coronary Heart Disease. Current Computer-Aided Drug Design 2020, 16, 473–485. [Google Scholar] [CrossRef]
- Moorthy, N.H.N.; Cerqueira, N.M.; Ramos, M.J.; Fernandes, P.A. Ligand Based Analysis on HMG-CoA Reductase Inhibitors. Chemometrics and Intelligent Laboratory Systems 2015, 140, 102–116. [Google Scholar] [CrossRef]
- Samizo, S.; Kaneko, H. Predictive Modeling of HMG-CoA Reductase Inhibitory Activity and Design of New HMG-CoA Reductase Inhibitors. ACS Omega 2023, 8, 27247–27255. [Google Scholar] [CrossRef]
- Zang, Y.; Li, Y.; Yin, Y.; Chen, S.; Kai, Z. Discovery and Quantitative Structure–Activity Relationship Study of Lepidopteran HMG-CoA Reductase Inhibitors as Selective Insecticides. Pest Management Science 2017, 73, 1944–1952. [Google Scholar] [CrossRef]
- Oliveira, M.A.; Araújo, R.D.C.M.U.; Lopes, C.D.C.; De Oliveira, B.G. In Silico Studies Combining QSAR Models, DFT-Based Reactivity Descriptors and Docking Simulations of Phthalimide Congeners with Hypolipidemic Activity. Orbital: Electron. J. Chem. 2021, 13, 188–199. [Google Scholar] [CrossRef]
- Choudhary, M.I.; Naheed, S.; Jalil, S.; Alam, J.M.; Atta-ur-Rahman. Effects of Ethanolic Extract of Iris Germanica on Lipid Profile of Rats Fed on a High-Fat Diet. Journal of Ethnopharmacology 2005, 98, 217–220. [Google Scholar] [CrossRef]
- Naylor, M.R.; Ly, A.M.; Handford, M.J.; Ramos, D.P.; Pye, C.R.; Furukawa, A.; Klein, V.G.; Noland, R.P.; Edmondson, Q.; Turmon, A.C.; et al. Lipophilic Permeability Efficiency Reconciles the Opposing Roles of Lipophilicity in Membrane Permeability and Aqueous Solubility. J. Med. Chem. 2018, 61, 11169–11182. [Google Scholar] [CrossRef]
- De, P.; Kar, S.; Ambure, P.; Roy, K. Prediction Reliability of QSAR Models: An Overview of Various Validation Tools. Arch Toxicol 2022, 96, 1279–1295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S. Partial Dependence of Breast Tumor Malignancy on Ultrasound Image Features Derived from Boosted Trees. J. Electron. Imaging 2010, 19, 023004. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC 15 – Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.D.; Martin, Y.C. The Information Content of 2D and 3D Structural Descriptors Relevant to Ligand-Receptor Binding. Journal of Chemical Information and Computer Sciences 1997, 37, 1–9. [Google Scholar] [CrossRef]
- Verma, J.; Khedkar, V.M.; Coutinho, E.C. 3D-QSAR in Drug Design-a Review. Current topics in medicinal chemistry 2010, 10, 95–115. [Google Scholar] [CrossRef]
- Hadni, H.; Elhallaoui, M. 2D and 3D-QSAR, Molecular Docking and ADMET Properties in Silico Studies of Azaaurones as Antimalarial Agents. New Journal of Chemistry 2020, 44, 6553–6565. [Google Scholar] [CrossRef]
- Fan, T.; Sun, G.; Zhao, L.; Cui, X.; Zhong, R. QSAR and Classification Study on Prediction of Acute Oral Toxicity of N-Nitroso Compounds. IJMS 2018, 19, 3015. [Google Scholar] [CrossRef]
- Gramatica, P.; Papa, E. QSAR Modeling of Bioconcentration Factor by Theoretical Molecular Descriptors. QSAR Comb. Sci. 2003, 22, 374–385. [Google Scholar] [CrossRef]
- Abreu, R.M.V.; Ferreira, I.C.F.R.; Queiroz, M.J.R.P. QSAR Model for Predicting Radical Scavenging Activity of Di(Hetero)Arylamines Derivatives of Benzo[b]Thiophenes. European Journal of Medicinal Chemistry 2009, 44, 1952–1958. [Google Scholar] [CrossRef]
- Sharma, S.; Prabhakar, Y.S.; Singh, P.; Sharma, B.K. QSAR Study about ATP-Sensitive Potassium Channel Activation of Cromakalim Analogues Using CP-MLR Approach. European Journal of Medicinal Chemistry 2008, 43, 2354–2360. [Google Scholar] [CrossRef]
- Fernández, M.; Caballero, J. QSAR Modeling of Matrix Metalloproteinase Inhibition by N-Hydroxy-α-Phenylsulfonylacetamide Derivatives. Bioorganic & Medicinal Chemistry 2007, 15, 6298–6310. [Google Scholar] [CrossRef]
- Kadam, R.U.; Roy, N. Cluster Analysis and Two-Dimensional Quantitative Structure-Activity Relationship (2D-QSAR) of Pseudomonas Aeruginosa Deacetylase LpxC Inhibitors. Bioorg Med Chem Lett 2006, 16, 5136–5143. [Google Scholar] [CrossRef] [PubMed]
- Seraj, K.; Asadollahi-Baboli, M. In Silico Evaluation of 5-Hydroxypyrazoles as LSD1 Inhibitors Based on Molecular Docking Derived Descriptors. Journal of Molecular Structure 2019, 1179, 514–524. [Google Scholar] [CrossRef]
- Adhikari, N.; Banerjee, S.; Baidya, S.K.; Ghosh, B.; Jha, T. Ligand-Based Quantitative Structural Assessments of SARS-CoV-2 3CLpro Inhibitors: An Analysis in Light of Structure-Based Multi-Molecular Modeling Evidences. Journal of Molecular Structure 2022, 1251, 132041. [Google Scholar] [CrossRef]
- Kumar, V.; Gupta, M.K.; Singh, G.; Prabhakar, Y.S. CP-MLR/PLS Directed QSAR Study on the Glutaminyl Cyclase Inhibitory Activity of Imidazoles: Rationales to Advance the Understanding of Activity Profile. Journal of Enzyme Inhibition and Medicinal Chemistry 2013, 28, 515–522. [Google Scholar] [CrossRef]
- De Melo, E.B. Multivariate SAR/QSAR of 3-Aryl-4-Hydroxyquinolin-2(1H)-One Derivatives as Type I Fatty Acid Synthase (FAS) Inhibitors. European Journal of Medicinal Chemistry 2010, 45, 5817–5826. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, X.; Chen, J. Quantitative Structure–Property Relationship of Distribution Coefficients of Organic Compounds. SAR and QSAR in Environmental Research 2020, 31, 585–596. [Google Scholar] [CrossRef]
- Stone, B.; Sapper, E. Machine Learning for the Design and Development of Biofilm Regulators 2018.
- Ishfaq, M.; Aamir, M.; Ahmad, F.; M Mebed, A.; Elshahat, S. Machine Learning-Assisted Prediction of the Biological Activity of Aromatase Inhibitors and Data Mining to Explore Similar Compounds. ACS Omega 2022, 7, 48139–48149. [Google Scholar] [CrossRef] [PubMed]
- Lavado, G.J.; Baderna, D.; Carnesecchi, E.; Toropova, A.P.; Toropov, A.A.; Dorne, J.L.C.M.; Benfenati, E. QSAR Models for Soil Ecotoxicity: Development and Validation of Models to Predict Reproductive Toxicity of Organic Chemicals in the Collembola Folsomia Candida. Journal of Hazardous Materials 2022, 423, 127236. [Google Scholar] [CrossRef]
- Yu, X. Global Classification Models for Predicting Acute Toxicity of Chemicals towards Daphnia Magna. Environmental Research 2023, 238, 117239. [Google Scholar] [CrossRef]
- Ghasemi, J.B.; Zohrabi, P.; Khajehsharifi, H. Quantitative Structure–Activity Relationship Study of Nonpeptide Antagonists of CXCR2 Using Stepwise Multiple Linear Regression Analysis. Monatsh Chem 2010, 141, 111–118. [Google Scholar] [CrossRef]
- Matias, M.; Campos, G.; Santos, A.O.; Falcão, A.; Silvestre, S.; Alves, G. Synthesis, in Vitro Evaluation and QSAR Modelling of Potential Antitumoral 3,4-Dihydropyrimidin-2-(1H)-Thiones. Arabian Journal of Chemistry 2019, 12, 5086–5102. [Google Scholar] [CrossRef]
- Shekhawat, N.; Singh, P. CP-MLR/PLS Directed Structure-Activity Study in Modeling of the Aggrecanase-1 Inhibitory Activity of Biphenylsulfonamides. Indian Journal of Chemistry 2024, 63, 315–324. [Google Scholar] [CrossRef]
- Worachartcheewan, A.; Nantasenamat, C.; Prachayasittikul, S.; Aiemsaard, A.; Prachayasittikul, V. Towards the Design of 3-Aminopyrazole Pharmacophore of Pyrazolopyridine Derivatives as Novel Antioxidants. Med Chem Res 2017, 26, 2699–2706. [Google Scholar] [CrossRef]
- Mansouri, K.; Ringsted, T.; Ballabio, D.; Todeschini, R.; Consonni, V. Quantitative Structure–Activity Relationship Models for Ready Biodegradability of Chemicals. J. Chem. Inf. Model. 2013, 53, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-H.; Xia, Z.-N.; Yan, L.; Liu, S.-S. Prediction of Placental Barrier Permeability: A Model Based on Partial Least Squares Variable Selection Procedure. Molecules 2015, 20, 8270–8286. [Google Scholar] [CrossRef]
- Lei, B.; Li, J.; Lu, J.; Du, J.; Liu, H.; Yao, X. Rational Prediction of the Herbicidal Activities of Novel Protoporphyrinogen Oxidase Inhibitors by Quantitative Structure−Activity Relationship Model Based on Docking-Guided Active Conformation. J. Agric. Food Chem. 2009, 57, 9593–9598. [Google Scholar] [CrossRef]
- De, P.; Roy, K. QSAR and QSAAR Modeling of Nitroimidazole Sulfonamide Radiosensitizers: Application of Small Dataset Modeling. Struct Chem 2021, 32, 631–642. [Google Scholar] [CrossRef]
- Cañizares-Carmenate, Y.; Mena-Ulecia, K.; MacLeod Carey, D.; Perera-Sardiña, Y.; Hernández-Rodríguez, E.W.; Marrero-Ponce, Y.; Torrens, F.; Castillo-Garit, J.A. Machine Learning Approach to Discovery of Small Molecules with Potential Inhibitory Action against Vasoactive Metalloproteases. Mol Divers 2022, 26, 1383–1397. [Google Scholar] [CrossRef]
- Hasegawa, K.; Funatsu, K. Advanced PLS Techniques in Chemoinformatics Studies. Current computer-aided drug design 2010, 6, 103–127. [Google Scholar] [CrossRef]
- Speck-Planche, A.; Cordeiro, M. Computer-Aided Discovery in Antimicrobial Research: In Silico Model for Virtual Screening of Potent and Safe Anti-Pseudomonas Agents. CCHTS 2015, 18, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Noorizadeh, H. Linear and Nonlinear Quantitative Structure Linear Retention Indices Relationship Models for Essential Oils. Eurasian Journal of Analytical Chemistry 2013, 8. [Google Scholar]
- Sharma, S.; Sharma, B.K.; Pilania, P.; Singh, P.; Prabhakar, Y.S. Modeling of the Growth Hormone Secretagogue Receptor Antagonistic Activity Using Chemometric Tools. Journal of Enzyme Inhibition and Medicinal Chemistry 2009, 24, 1024–1033. [Google Scholar] [CrossRef] [PubMed]
- Jahan, A.; Sharma, B.K.; Sharma, V.D. Quantitative Structure-Activity Relationship Study on the MMP-13 Inhibitory Activity of Fused Pyrimidine Derivatives Possessing a 1, 2, 4-Triazol-3-Yl Group as a ZBG. GSC Biological and Pharmaceutical Sciences 2021, 16, 251–265. [Google Scholar] [CrossRef]
- Xuan, Y.; Zhou, Y.; Yue, Y.; Zhang, N.; Sun, G.; Fan, T.; Zhao, L.; Zhong, R. Identification of Potential Natural Product Derivatives as CK2 Inhibitors Based on GA-MLR QSAR Modeling, Synthesis and Biological Evaluation. Medicinal Chemistry Research 2024, 1–14. [Google Scholar] [CrossRef]
- Duchowicz, P.R.; Talevi, A.; Bellera, C.; Bruno-Blanch, L.E.; Castro, E.A. Application of Descriptors Based on Lipinski’s Rules in the QSPR Study of Aqueous Solubilities. Bioorganic & medicinal chemistry 2007, 15, 3711–3719. [Google Scholar] [CrossRef]
- Choudhary, M.; Deshpande, S.; Sharma, B. CP-MLR Directed QSAR Rationales for the 1-Aryl Sulfonyl Tryptamines as 5-HT6 Receptor Ligands. British Journal of Pharmaceutical Research 2015, 8, 1–17. [Google Scholar] [CrossRef]
- Meena, D.K.; Sharma, B.K.; Parihar, R. Quantitative Structure-Activity Relationship Study on the CDK2 Inhibitory Activity of 6-Substituted 2-Arylaminopurines. GSC Biological and Pharmaceutical Sciences 2022, 20, 107–119. [Google Scholar] [CrossRef]
- Raghuraj, P.; Afsar, J.; Kishore, S.B. CP-MLR Derived QSAR Rationales for the PPARy Agonistic Activity of the Pyridyloxybenzene-Acylsulfonamide Derivatives. GSC Biological and Pharmaceutical Sciences 2020, 12, 273–285. [Google Scholar] [CrossRef]
- Sharma, B.K.; Sarbhai, K.; Singh, P. A Rationale for the Activity Profile of Arylpiperazinylthioalkyls as 5-HT1A-Serotonin and A1-Adrenergic Receptor Ligands. European Journal of Medicinal Chemistry 2010, 45, 1927–1934. [Google Scholar] [CrossRef]
- Santos Cruz, D.; Santos Castilho, M. 2D QSAR Studies on Series of Human Beta-Secretase (BACE-1) Inhibitors. Medicinal Chemistry 2014, 10, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Dolatabadi, M.; Nekoei, M.; Banaei, A. Prediction of Antibacterial Activity of Pleuromutilin Derivatives by Genetic Algorithm–Multiple Linear Regression (GA–MLR). Monatsh Chem 2010, 141, 577–588. [Google Scholar] [CrossRef]
- Ojha, P.K.; Roy, K. Chemometric Modeling of Odor Threshold Property of Diverse Aroma Components of Wine. RSC Adv. 2018, 8, 4750–4760. [Google Scholar] [CrossRef] [PubMed]
- Antypenko, L.M.; Kovalenko, S.I.; Los’, T.S.; Rebec’, O.L. Synthesis and Characterization of Novel N -(Phenyl, Benzyl, Hetaryl)-2-([1,2,4]Triazolo[1,5- c ]Quinazolin-2-ylthio)Acetamides by Spectral Data, Antimicrobial Activity, Molecular Docking and QSAR Studies. Journal of Heterocyclic Chem 2017, 54, 1267–1278. [Google Scholar] [CrossRef]
- Abreu, R.M.V.; Ferreira, I.C.F.R.; Calhelha, R.C.; Lima, R.T.; Vasconcelos, M.H.; Adega, F.; Chaves, R.; Queiroz, M.-J.R.P. Anti-Hepatocellular Carcinoma Activity Using Human HepG2 Cells and Hepatotoxicity of 6-Substituted Methyl 3-Aminothieno[3,2-b]Pyridine-2-Carboxylate Derivatives: In Vitro Evaluation, Cell Cycle Analysis and QSAR Studies. European Journal of Medicinal Chemistry 2011, 46, 5800–5806. [Google Scholar] [CrossRef]
- Nembri, S.; Grisoni, F.; Consonni, V.; Todeschini, R. In Silico Prediction of Cytochrome P450-Drug Interaction: QSARs for CYP3A4 and CYP2C9. International Journal of Molecular Sciences 2016, 17, 914. [Google Scholar] [CrossRef]
- Huang, J.; Ma, G.; Muhammad, I.; Cheng, Y. Identifying P-Glycoprotein Substrates Using a Support Vector Machine Optimized by a Particle Swarm. J. Chem. Inf. Model. 2007, 47, 1638–1647. [Google Scholar] [CrossRef]
- Zhao, Z.; Cui, J.; Yin, Y.; Zhang, H.; Liu, Y.; Zeng, R.; Fang, C.; Kai, Z.; Wang, Z.; Wu, F. Synthesis and Biological Evaluation of Gem-Difluoromethylenated Statin Derivatives as Highly Potent HMG-CoA Reductase Inhibitors. Chin. J. Chem. 2016, 34, 801–808. [Google Scholar] [CrossRef]
- Andrade-Ochoa, S.; Correa-Basurto, J.; Rodríguez-Valdez, L.M.; Sánchez-Torres, L.E.; Nogueda-Torres, B.; Nevárez-Moorillón, G.V. In Vitro and in Silico Studies of Terpenes, Terpenoids and Related Compounds with Larvicidal and Pupaecidal Activity against Culex Quinquefasciatus Say (Diptera: Culicidae). Chemistry Central Journal 2018, 12, 53. [Google Scholar] [CrossRef]
- Scotti, M.T.; Scotti, L.; Ishiki, H.M.; Peron, L.M.; De Rezende, L.; Do Amaral, A.T. Variable-Selection Approaches to Generate QSAR Models for a Set of Antichagasic Semicarbazones and Analogues. Chemometrics and Intelligent Laboratory Systems 2016, 154, 137–149. [Google Scholar] [CrossRef]
- Galvez-Llompart, M.; Hierrezuelo, J.; Blasco, M.; Zanni, R.; Galvez, J.; De Vicente, A.; Pérez-García, A.; Romero, D. Targeting Bacterial Growth in Biofilm Conditions: Rational Design of Novel Inhibitors to Mitigate Clinical and Food Contamination Using QSAR. Journal of Enzyme Inhibition and Medicinal Chemistry 2024, 39, 2330907. [Google Scholar] [CrossRef] [PubMed]
- Seth, A.; Roy, K. QSAR Modeling of Algal Low Level Toxicity Values of Different Phenol and Aniline Derivatives Using 2D Descriptors. Aquatic Toxicology 2020, 228, 105627. [Google Scholar] [CrossRef] [PubMed]
- Stanton, D.T.; Baker, J.R.; McCluskey, A.; Paula, S. Development and Interpretation of a QSAR Model for in Vitro Breast Cancer (MCF-7) Cytotoxicity of 2-Phenylacrylonitriles. J Comput Aided Mol Des 2021, 35, 613–628. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.C.; Sharma, S. Molecular Modeling Study of Uracil-Based Hydroxamic Acids-Containing Histone Deacetylase Inhibitors. Arabian Journal of Chemistry 2019, 12, 2206–2215. [Google Scholar] [CrossRef]
- Jovanović, M.; Radan, M.; Čarapić, M.; Filipović, N.; Nikolic, K.; Crevar, M. Application of Parallel Artificial Membrane Permeability Assay Technique and Chemometric Modeling for Blood–Brain Barrier Permeability Prediction of Protein Kinase Inhibitors. Future Medicinal Chemistry 2024, 16, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Baba, H.; Takahara, J.; Yamashita, F.; Hashida, M. Modeling and Prediction of Solvent Effect on Human Skin Permeability Using Support Vector Regression and Random Forest. Pharmaceutical Research 2015, 32, 3604–3617. [Google Scholar] [CrossRef]
- Li, Y.; Fan, T.; Ren, T.; Zhang, N.; Zhao, L.; Zhong, R.; Sun, G. Ecotoxicological Risk Assessment of Pesticides against Different Aquatic and Terrestrial Species: Using Mechanistic QSTR and iQSTTR Modelling Approaches to Fill the Toxicity Data Gap. Green Chem. 2024, 26, 839–856. [Google Scholar] [CrossRef]
- Catherene Tomy, P.; Mohan, C.G. Chemical Space Navigation by Machine Learning Models for Discovering Selective MAO-B Enzyme Inhibitors for Parkinson’s Disease. Artificial Intelligence Chemistry 2023, 1, 100012. [Google Scholar] [CrossRef]
- Sun, G.; Fan, T.; Sun, X.; Hao, Y.; Cui, X.; Zhao, L.; Ren, T.; Zhou, Y.; Zhong, R.; Peng, Y. In Silico Prediction of O6-Methylguanine-DNA Methyltransferase Inhibitory Potency of Base Analogs with QSAR and Machine Learning Methods. Molecules 2018, 23, 2892. [Google Scholar] [CrossRef]
- Jamaludin, R.; Ibrahim, N.A.; Maarof, H. Development of Structure-Activity Modelling of Carboxamides Compounds for Aedes Aegypti Repellents. Journal of Advanced Research Design 2017, 35, 26–32. [Google Scholar]
- Erzincan, P.; Saçan, M.T.; Yüce-Dursun, B.; Danış, Ö.; Demir, S.; Erdem, S.S.; Ogan, A. QSAR Models for Antioxidant Activity of New Coumarin Derivatives. SAR and QSAR in Environmental Research 2015, 26, 721–737. [Google Scholar] [CrossRef] [PubMed]
- Żołnowska, B.; Sławiński, J.; Brzozowski, Z.; Kawiak, A.; Belka, M.; Zielińska, J.; Bączek, T.; Chojnacki, J. Synthesis, Molecular Structure, Anticancer Activity, and QSAR Study of N-(Aryl/Heteroaryl)-4-(1H-Pyrrol-1-Yl)Benzenesulfonamide Derivatives. IJMS 2018, 19, 1482. [Google Scholar] [CrossRef] [PubMed]
- Stasiak, J.; Koba, M.; Gackowski, M.; Baczek, T. Chemometric Analysis for the Classification of Some Groups of Drugs with Divergent Pharmacological Activity on the Basis of Some Chromatographic and Molecular Modeling Parameters. CCHTS 2018, 21, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Jeličić, M.-L.; Kovačić, J.; Cvetnić, M.; Mornar, A.; Amidžić Klarić, D. Antioxidant Activity of Pharmaceuticals: Predictive QSAR Modeling for Potential Therapeutic Strategy. Pharmaceuticals 2022, 15, 791. [Google Scholar] [CrossRef]
- Mukherjee, R.K.; Kumar, V.; Roy, K. Chemometric Modeling of Plant Protection Products (PPPs) for the Prediction of Acute Contact Toxicity against Honey Bees (A. Mellifera): A 2D-QSAR Approach. Journal of Hazardous Materials 2022, 423, 127230. [Google Scholar] [CrossRef]
- He, J.; Peng, T.; Yang, X.; Liu, H. Development of QSAR Models for Predicting the Binding Affinity of Endocrine Disrupting Chemicals to Eight Fish Estrogen Receptor. Ecotoxicology and Environmental Safety 2018, 148, 211–219. [Google Scholar] [CrossRef]
- Yuan, J.; Yu, S.; Gao, S.; Gan, Y.; Zhang, Y.; Zhang, T.; Wang, Y.; Yang, L.; Shi, J.; Yao, W. Predicting the Biological Activities of Triazole Derivatives as SGLT2 Inhibitors Using Multilayer Perceptron Neural Network, Support Vector Machine, and Projection Pursuit Regression Models. Chemometrics and Intelligent Laboratory Systems 2016, 156, 166–173. [Google Scholar] [CrossRef]
- Daghighi, A.; Casanola-Martin, G.M.; Timmerman, T.; Milenković, D.; Lučić, B.; Rasulev, B. In Silico Prediction of the Toxicity of Nitroaromatic Compounds: Application of Ensemble Learning QSAR Approach. Toxics 2022, 10, 746. [Google Scholar] [CrossRef]
- Gregori-Puigjané, E.; Mestres, J. SHED: Shannon Entropy Descriptors from Topological Feature Distributions. J. Chem. Inf. Model. 2006, 46, 1615–1622. [Google Scholar] [CrossRef]
- Mauri, A.; Bertola, M. Alvascience: A New Software Suite for the QSAR Workflow Applied to the Blood–Brain Barrier Permeability. IJMS 2022, 23, 12882. [Google Scholar] [CrossRef]
- Athista, M.; Hariharan, V.; Namratha, K.; Pavankumar, G.; Perciya, J.L.; Sunkar, S. Computational Identification of Natural Compounds as Potential Inhibitors for HMGCoA Reductase. Current Trends in Biotechnology and Pharmacy 2023, 17, 1457–1485. [Google Scholar]
- Cuccioloni, M.; Bonfili, L.; Mozzicafreddo, M.; Cecarini, V.; Scuri, S.; Cocchioni, M.; Nabissi, M.; Santoni, G.; Eleuteri, A.M.; Angeletti, M. Mangiferin Blocks Proliferation and Induces Apoptosis of Breast Cancer Cells via Suppression of the Mevalonate Pathway and by Proteasome Inhibition. Food Funct. 2016, 7, 4299–4309. [Google Scholar] [CrossRef] [PubMed]
- Min, S.-W.; Kim, D.-H. Kakkalide and Irisolidone: HMG-CoA Reductase Inhibitors Isolated from the Flower of Pueraria Thunbergiana. Biological & Pharmaceutical Bulletin 2007, 30, 1965–1968. [Google Scholar] [CrossRef]
- Gaulton, A.; Hersey, A.; Nowotka, M.; Bento, A.P.; Chambers, J.; Mendez, D.; Mutowo, P.; Atkinson, F.; Bellis, L.J.; Cibrián-Uhalte, E.; et al. The ChEMBL Database in 2017. Nucleic Acids Res 2017, 45, D945–D954. [Google Scholar] [CrossRef]
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. DataWarrior: An Open-Source Program For Chemistry Aware Data Visualization And Analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024. [Google Scholar]
- Cao, D.-S.; Xiao, N.; Xu, Q.-S.; Chen, A.F. Rcpi: R/Bioconductor Package to Generate Various Descriptors of Proteins, Compounds and Their Interactions. Bioinformatics 2015, 31, 279–281. [Google Scholar] [CrossRef] [PubMed]
- RStudio Team RStudio: Integrated Development Environment for R; RStudio, PBC.: Boston, MA, 2021.
- Ballabio, D.; Grisoni, F.; Consonni, V.; Todeschini, R. Integrated QSAR Models to Predict Acute Oral Systemic Toxicity. Mol. Inf. 2019, 38, 1800124. [Google Scholar] [CrossRef]
- Tomberg, A.; Boström, J. Can Easy Chemistry Produce Complex, Diverse, and Novel Molecules? Drug Discovery Today 2020, 25, 2174–2181. [Google Scholar] [CrossRef]
- Gao, K.; Nguyen, D.D.; Sresht, V.; Mathiowetz, A.M.; Tu, M.; Wei, G.-W. Are 2D Fingerprints Still Valuable for Drug Discovery? Phys. Chem. Chem. Phys. 2020, 22, 8373–8390. [Google Scholar] [CrossRef]
- Mauri, A. alvaDesc: A Tool to Calculate and Analyze Molecular Descriptors and Fingerprints. In Ecotoxicological QSARs; Roy, K., Ed.; Methods in Pharmacology and Toxicology; Springer US: New York, NY, 2020; pp. 801–820. ISBN 978-1-07-160149-5. [Google Scholar]
- Shi, J.; Zhao, G.; Wei, Y. Computational QSAR Model Combined Molecular Descriptors and Fingerprints to Predict HDAC1 Inhibitors. Med Sci (Paris) 2018, 34, 52–58. [Google Scholar] [CrossRef]
- Boudergua, S.; Alloui, M.; Belaidi, S.; Al Mogren, M.M.; Ellatif Ibrahim, U.A.A.; Hochlaf, M. QSAR Modeling and Drug-Likeness Screening for Antioxidant Activity of Benzofuran Derivatives. Journal of Molecular Structure 2019, 1189, 307–314. [Google Scholar] [CrossRef]
- Meyer, D.; Buchta, C. Proxy: Distance and Similarity Measures; 2021.
- Remeseiro, B.; Bolon-Canedo, V. A Review of Feature Selection Methods in Medical Applications. Computers in Biology and Medicine 2019, 112, 103375. [Google Scholar] [CrossRef] [PubMed]
- Concu, R.; Cordeiro, M.N.D.S. On the Relevance of Feature Selection Algorithms While Developing Non-Linear QSARs. In Ecotoxicological QSARs; Roy, K., Ed.; Methods in Pharmacology and Toxicology; Springer US: New York, NY, 2020; pp. 177–194. ISBN 978-1-07-160149-5. [Google Scholar]
- Lang, M.; Binder, M.; Richter, J.; Schratz, P.; Pfisterer, F.; Coors, S.; Au, Q.; Casalicchio, G.; Kotthoff, L.; Bischl, B. Mlr3: A Modern Object-Oriented Machine Learning Framework in R. Journal of Open Source Software 2019. [Google Scholar] [CrossRef]
- Zuber, V.; Strimmer, K. Care: High-Dimensional Regression and CAR Score Variable Selection; 2021.
- Kursa, M.B. Praznik: High Performance Information-Based Feature Selection. SoftwareX 2021, 16, 100819. [Google Scholar] [CrossRef]
- Zawadzki, Z.; Kosinski, M. FSelectorRcpp: “Rcpp” Implementation of “FSelector” Entropy-Based Feature Selection Algorithms with a Sparse Matrix Support; 2021.
- Hutson, G. FeatureTerminatoR: Feature Selection Engine to Remove Features with Minimal Predictive Power; 2021.
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of Statistical Software 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Milborrow, S.; Hastie, T.; Tibshirani, R. Earth: Multivariate Adaptive Regression Splines; 2024.
- Schliep, K.; Hechenbichler, K. Kknn: Weighted k-Nearest Neighbors; 2016.
- Beygelzimer, A.; Kakadet, S.; Langford, J.; Arya, S.; Mount, D.; Li, S. FNN: Fast Nearest Neighbor Search Algorithms and Applications; 2024.
- Kuhn, M.; Quinlan, R. Cubist: Rule- And Instance-Based Regression Modeling; 2024.
- Hornik, K.; Buchta, C.; Zeileis, A. Open-Source Machine Learning: R Meets Weka. Computational Statistics 2009, 24, 225–232. [Google Scholar] [CrossRef]
- Wright, M.N.; Ziegler, A. Ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. Journal of Statistical Software 2017, 77, 1–17. [Google Scholar] [CrossRef]
- Hothorn, T.; Hornik, K.; Zeileis, A. Unbiased Recursive Partitioning: A Conditional Inference Framework. Journal of Computational and Graphical Statistics 2006, 15, 651–674. [Google Scholar] [CrossRef]
- Zeileis, A. Object-Oriented Computation of Sandwich Estimators. J. Stat. Soft. 2006, 16. [Google Scholar] [CrossRef]
- Hothorn, T.; Hornik, K.; Wiel, M.A.V.D.; Zeileis, A. Implementing a Class of Permutation Tests: The Coin Package. J. Stat. Soft. 2008, 28. [Google Scholar] [CrossRef]
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F. E1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien; 2023.
- Helleputte, T.; Paul, J.; Gramme, P. LiblineaR: Linear Predictive Models Based on the LIBLINEAR C/C++ Library; 2024.
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K.; Mitchell, R.; Cano, I.; Zhou, T.; et al. Xgboost: Extreme Gradient Boosting; 2023.
- Ridgeway, G.; Developers, G.B.M. Gbm: Generalized Boosted Regression Models; 2024.
- Sparapani, R.; Spanbauer, C.; McCulloch, R. Nonparametric Machine Learning and Efficient Computation with Bayesian Additive Regression Trees: The BART R Package. Journal of Statistical Software 2021, 97, 1–66. [Google Scholar] [CrossRef]
- Majumdar, S.; Basak, S.C. Beware of External Validation! - A Comparative Study of Several Validation Techniques Used in QSAR Modelling. CAD 2018, 14, 284–291. [Google Scholar] [CrossRef]
- Feng, D. agRee: Various Methods for Measuring Agreement; 2020.
- Lin, L.I. A Concordance Correlation Coefficient to Evaluate Reproducibility. Biometrics 1989, 45, 255–268. [Google Scholar] [CrossRef]
- Chirico, N.; Gramatica, P. Real External Predictivity of QSAR Models: How To Evaluate It? Comparison of Different Validation Criteria and Proposal of Using the Concordance Correlation Coefficient. J. Chem. Inf. Model. 2011, 51, 2320–2335. [Google Scholar] [CrossRef]
- Gramatica, P. Principles of QSAR Modeling: Comments and Suggestions From Personal Experience. International Journal of Quantitative Structure-Property Relationships 2020, 5, 61–97. [Google Scholar] [CrossRef]
- Rücker, C.; Rücker, G.; Meringer, M. Y-Randomization and Its Variants in QSPR/QSAR. J. Chem. Inf. Model. 2007, 47, 2345–2357. [Google Scholar] [CrossRef] [PubMed]
- Biecek, P. DALEX: Explainers for Complex Predictive Models in R. Journal of Machine Learning Research 2018, 19, 1–5. [Google Scholar]
- Molnar, C.; Bischl, B.; Casalicchio, G. Iml: An R Package for Interpretable Machine Learning. JOSS 2018, 3, 786. [Google Scholar] [CrossRef]
- Hajalsiddig, T.T.H.; Osman, A.B.M.; Saeed, A.E.M. 2D-QSAR Modeling and Molecular Docking Studies on 1 H -Pyrazole-1-Carbothioamide Derivatives as EGFR Kinase Inhibitors. ACS Omega 2020, 5, 18662–18674. [Google Scholar] [CrossRef]
- Gotti, M.; Kuhn, M. Applicable: A Compilation of Applicability Domain Methods; 2022.
- Cortes, D. Isotree: Isolation-Based Outlier Detection; 2023.
- Rutz, A.; Sorokina, M.; Galgonek, J.; Mietchen, D.; Willighagen, E.; Gaudry, A.; Graham, J.G.; Stephan, R.; Page, R.; Vondrášek, J.; et al. The LOTUS Initiative for Open Knowledge Management in Natural Products Research. eLife 2022, 11, e70780. [Google Scholar] [CrossRef]
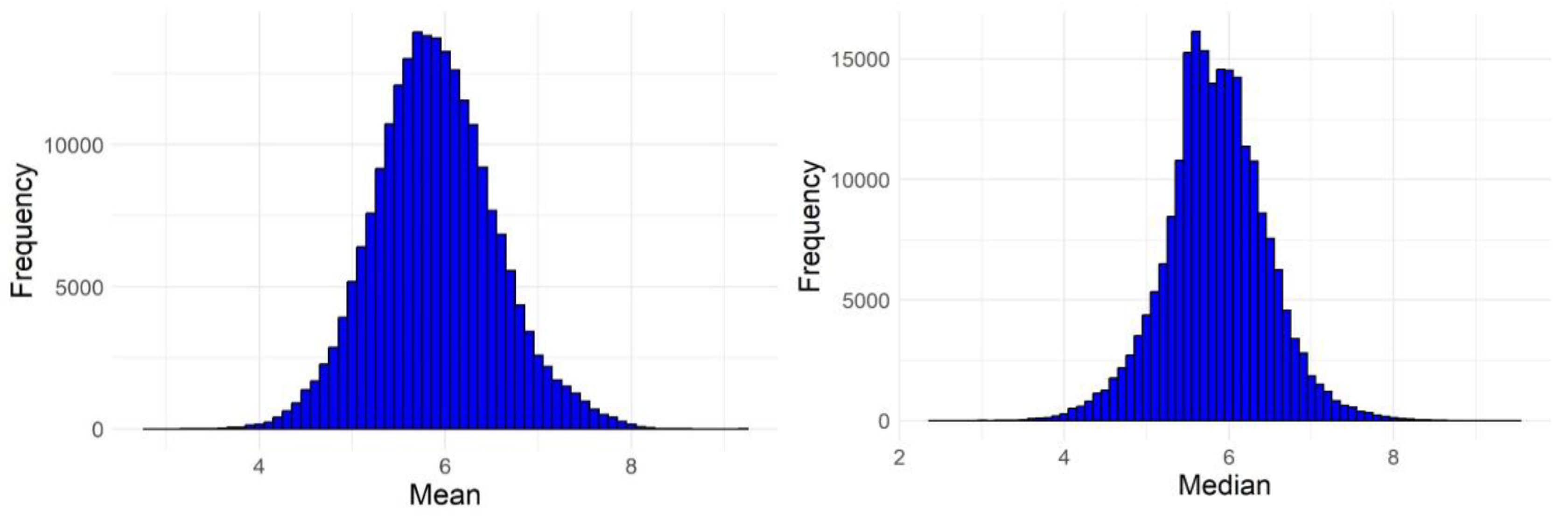
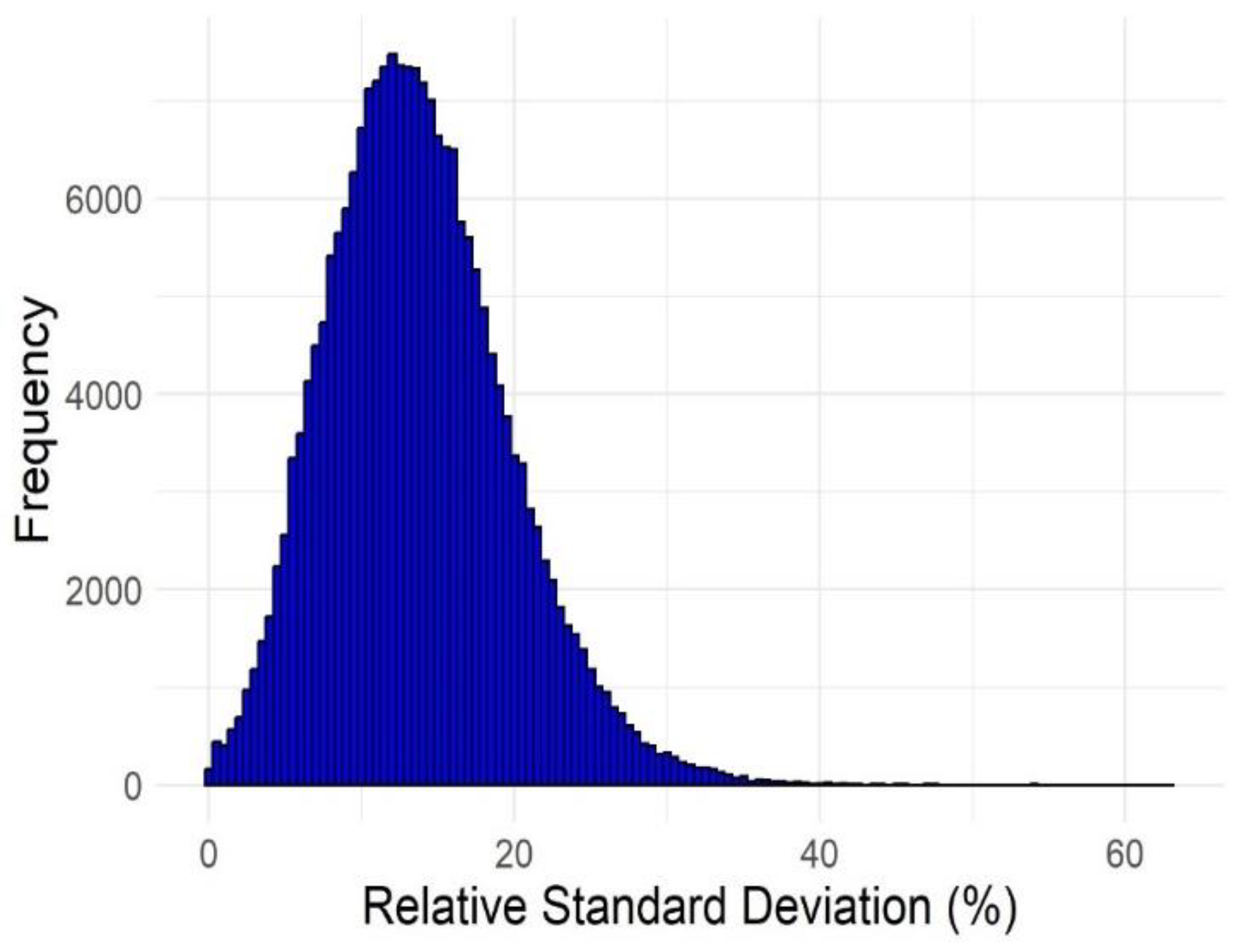
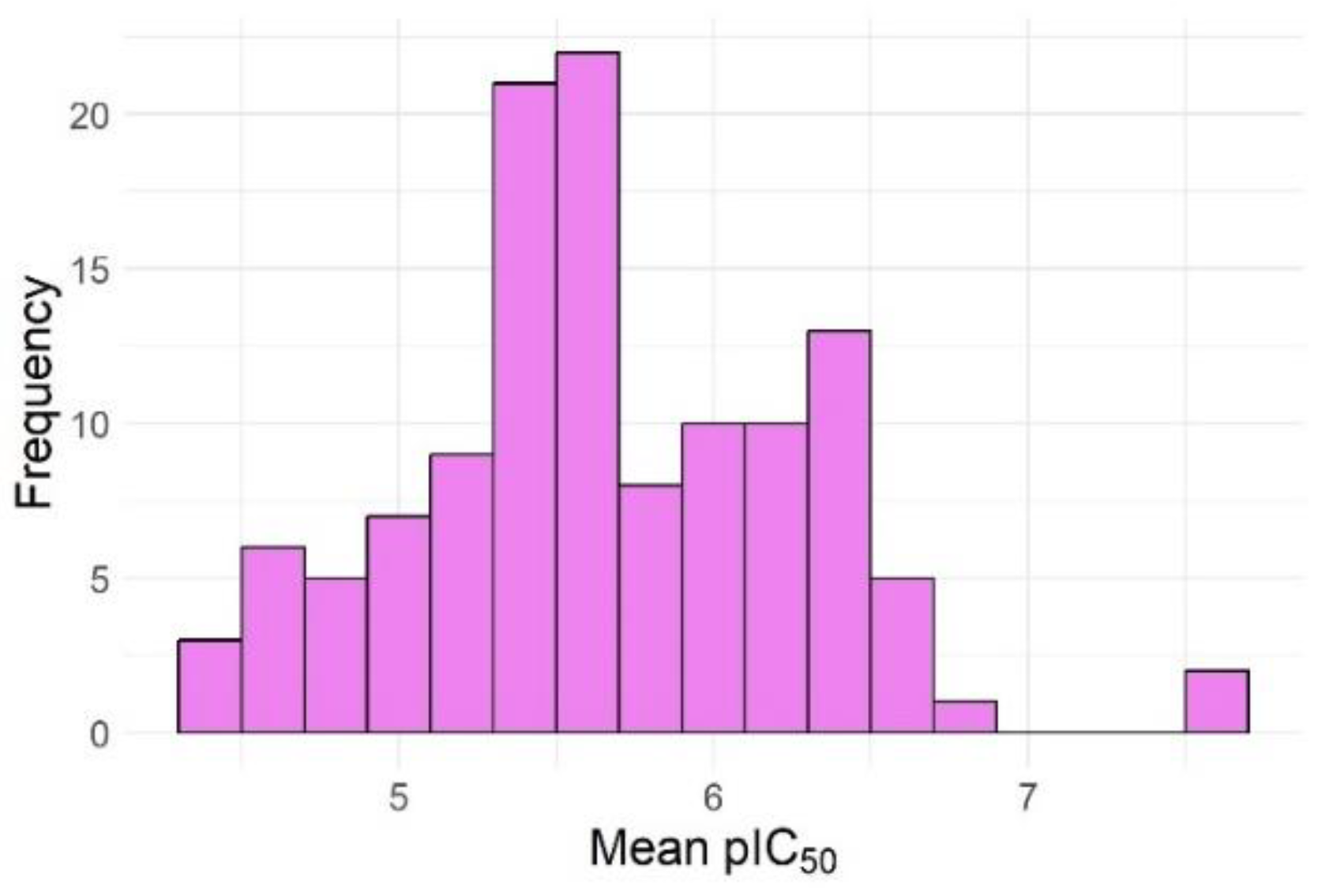
| No. of failures | Active compounds | Inactive compounds |
|---|---|---|
| 0 | 46 | 649 |
| 1 | 27 | 127 |
| 2 | 31 | 52 |
| 3 | 34 | 34 |
| 4 | 0 | 40 |
| 5 | 0 | 2 |
| No. | Regression algorithm | Descriptor set | Feature selection method | CCC (nested CV, n = 5) mean (s.d.) |
R2 (nested CV, n=5) mean (s.d.) |
RMSE (n=5) mean (s.d.) |
|---|---|---|---|---|---|---|
| 1 | Random forest (“ranger”) | MACCS | “cmim” | 0.837 0.840 0.846 0.833 0.825 0.836 (0.008) |
0.707 0.716 0.745 0.698 0.720 0.717 (0.018) |
0.872 0.867 0.835 0.884 0.877 0.867 (0.019) |
| 2 | XGboost | MACCS | Boruta | 0.848 0.861 0.840 0.850 0.848 0.849 (0.008) |
0.726 0.744 0.725 0.701 0.741 0.727 (0.017) |
0.848 0.821 0.873 0.870 0.835 0.849 (0.022) |
| 3 | Random forest (“ranger”) | MACCS | Boruta | 0.835 0.827 0.833 0.826 0.834 0.831 (0.004) |
0.712 0.696 0.722 0.679 0.707 0.702 (0.016) |
0.890 0.903 0.877 0.916 0.886 0.891 (0.018) |
| 4 | Support vector machines | MACCS | Boruta | 0.857 0.853 0.857 0.843 0.845 0.851 (0.007) |
0.743 0.740 0.754 0.708 0.738 0.737 (0.017) |
0.832 0.831 0.815 0.872 0.839 0.838 (0.021) |
| 5 | Gradient boosting machine (“GBM”) | Set2 | Boruta | 0.858 0.820 0.827 0.830 0.829 0.833 (0.015) |
0.752 0.681 0.702 0.696 0.667 0.700 (0.032) |
0.815 0.942 0.912 0.915 0.926 0.902 (0.050) |
| 6 | Support vector machines | Set2 | “jmim” | 0.840 0.841 0.850 0.839 0.841 0.842 (0.004) |
0.734 0.727 0.756 0.728 0.746 0.738 (0.012) |
0.854 0.850 0.824 0.849 0.838 0.843 (0.012) |
| 7 | BART | Set2 | Gaselect | 0.846 0.848 0.854 0.850 0.845 0.849 (0.004) |
0.730 0.739 0.745 0.733 0.689 0.727 (0.022) |
0.858 0.833 0.830 0.847 0.864 0.846 (0.015) |
| 8 | Random forest (“ranger”) | Set2 | Gaselect | 0.827 0.830 0.823 0.830 0.818 0.826 (0.005) |
0.733 0.730 0.708 0.742 0.723 0.727 (0.013) |
0.848 0.849 0.864 0.850 0.874 0.857 (0.011) |
| 9 | XGboost | Set2 | “jmim” | 0.832 0.843 0.832 0.830 0.823 0.832 (0.007) |
0.724 0.724 0.706 0.684 0.705 0.709 (0.017) |
0.868 0.852 0.885 0.890 0.901 0.879 (0.019) |
| 10 | BART | Set2 | Boruta | 0.867 0.825 0.825 0.834 0.829 0.836 (0.018) |
0.764 0.690 0.697 0.707 0.674 0.704 (0.034) |
0.797 0.929 0.917 0.901 0.919 0.893 (0.054) |
| 11 | Rule- and instance-cased regression | Set2 | Gaselect | 0.837 0.821 0.835 0.843 0.823 0.832 (0.009) |
0.724 0.707 0.717 0.727 0.660 0.707 (0.027) |
0.860 0.891 0.881 0.851 0.893 0.875 (0.019) |
| 12 | Support vector machines | Set2 | Gaselect | 0.849 0.853 0.840 0.856 0.851 0.850 (0.006) |
0.748 0.754 0.715 0.766 0.757 0.748 (0.020) |
0.821 0.804 0.846 0.804 0.819 0.819 (0.017) |
| 13 | Random forest (“ranger”) | Set3 | Gaselect | 0.812 0.832 0.826 0.826 0.823 0.824 (0.007) |
0.702 0.720 0.727 0.731 0.717 0.719 (0.011) |
0.873 0.841 0.860 0.862 0.876 0.862 (0.014) |
| 14 | BART | Set4 | “jmim” | 0.864 0.845 0.858 0.853 0.852 0.854 (0.007) |
0.751 0.710 0.742 0.731 0.730 0.733 (0.015) |
0.821 0.888 0.844 0.858 0.856 0.853 (0.024) |
| 15 | Weighted k-Nearest Neighbor | Set4 | Boruta | 0.826 0.854 0.865 0.848 0.858 0.850 (0.015) |
0.690 0.740 0.739 0.709 0.737 0.723 (0.022) |
0.923 0.846 0.821 0.874 0.858 0.864 (0.038) |
| 16 | BART | Set4 | Gaselect | 0.856 0.847 0.845 0.846 0.859 0.851 (0.006) |
0.743 0.726 0.715 0.719 0.745 0.730 (0.014) |
0.833 0.865 0.871 0.877 0.848 0.859 (0.018) |
| 17 | XGboost | Set4 | “jmim” | 0.835 0.832 0.856 0.830 0.846 0.840 (0.011) |
0.701 0.702 0.750 0.705 0.737 0.719 (0.022) |
0.857 0.896 0.825 0.902 0.844 0.865 (0.033) |
| 18 | Random forest (“ranger”) | Set4 | Boruta | 0.831 0.846 0.857 0.855 0.847 0.847 (0.010) |
0.702 0.748 0.754 0.749 0.738 0.738 (0.021) |
0.861 0.835 0.796 0.829 0.848 0.834 (0.024) |
| 19 | Rule- and instance-cased regression | Set4 | Gaselect | 0.851 0.813 0.841 0.842 0.837 0.837 (0.014) |
0.726 0.655 0.721 0.715 0.688 0.701 (0.030) |
0.859 0.947 0.882 0.879 0.896 0.893 (0.033) |
| 20 | BART | Set4 | Boruta | 0.856 0.870 0.868 0.872 0.877 0.869 (0.008) |
0.743 0.757 0.743 0.760 0.768 0.754 (0.011) |
0.833 0.814 0.836 0.805 0.800 0.818 (0.016) |
| 21 | XGboost | Set4 | Boruta | 0.850 0.849 0.850 0.855 0.843 0.849 (0.004) |
0.737 0.747 0.734 0.738 0.711 0.733 (0.013) |
0.848 0.834 0.838 0.842 0.893 0.851 (0.024) |
| Ensemble algorithm | CCC (nested CV) | R2 (nested cross-validation) | RMSE (nested cross-validation) |
|---|---|---|---|
| Support vector machines | 0.893 | 0.798 | 0.730 |
| BART | 0.888 | 0.789 | 0.745 |
| KKNN | 0.887 | 0.789 | 0.750 |
| Random forests | 0.889 | 0.794 | 0.739 |
| Xgboost | 0.883 | 0.784 | 0.760 |
| Model whose features were randomized | CCC (nested CV, n = 20) mean (s.d.) |
Rr2 (nested CV, n=20) mean (s.d.) |
RMSE (n=20) mean (s.d.) |
Rp2 (for the corresponding model) |
|---|---|---|---|---|
| Model 19 in Table 2 | 0.047 (0.055) | -0.220 (0.077) | 1.804 (0.045) | 0.803 |
| Model 17 in Table 2 | -0.007 (0.034) | -0.113 (0.068) | 1.731 (0.027) | 0.773 |
| Model 20 in Table 2 | 0.078 (0.060) | -0.056 (0.040) | 1.685 (0.032) | 0.781 |
| MACCS Key | Structural Pattern | Association |
|---|---|---|
| 62 | "A$A!A$A" (any atom – ring bond – any atom – chain bond – any atom – ring bond – any atom) | Positive |
| 85 | CN(C)C (a closed ring formed by a C-N-C chain) | Positive |
| 105 | "A$A($A)$A" (aromatic atom – substructure – aromatic atom) | Negative |
| 22 | Three-membered ring system (3M ring) | Relatively strongly negative |
| 65 | Carbon and nitrogen united by an aromatic query bond | Positive |
| 145 | 6M RING > 1 (more than one six-member rings) | Positive |
| 89 | OAAAO (two oxygen atoms connected by three other atoms) | Positive |
| 97 | NAAAO (a nitrogen atom connected by a sequence of four single bonds to an oxygen atom) | Weakly negative |
| 107 | XA(A)A (where X is a halogen and A any atom) | Weakly positive |
| 42 | F (a fluorine atom) | Weakly positive |
| Descriptor | Correlation coefficient (for other descriptors) | Correlated Descriptors | Activity Relationship |
|---|---|---|---|
| MATS3e (Moran autocorrelation of lag 3 weighted by Sanderson electronegativity) |
r = 0.846 |
MATS3s (Moran autocorrelation of lag 3 weighted by I-state) |
Negative values → higher activity |
| SpMax_B(p) (Leading eigenvalue from Burden matrix weighted by polarizability) |
r >0.91 r>0.80 |
SpDiam_B(p) (Diameter from Burden matrix weighted by polarizability) SpMax1_Bh(p) (Leading eigenvalue n. 1 of Burden matrix weighted by polarizability) piPC06 (molecular multiple path count of order 6) SpDiam_B(v) ( spectral diameter from Burden matrix weighted by van der Waals volume) SpMax_B.v. |
Inverted U-shape |
| VE1sign_B(s) (Coefficient sum of the last eigenvector from Burden matrix weighted by I-State) |
N/A |
None |
Higher values → lower activity |
| SpMin1_Bh(e) (Smallest eigenvalue n. 1 of Burden matrix weighted by Sanderson electronegativity) |
r = 0.99 r>0.87 -0.80 |
SpMin1_Bh(i) (Smallest eigenvalue n. 1 of Burden matrix weighted by ionization potential) SpMin1_Bh(v) (Smallest eigenvalue n. 1 of Burden matrix weighted by van der Waals volume) SpMin1_Bh(p) (Smallest eigenvalue n. 1 of Burden matrix weighted by polarizability) WiA_D/Dt (average Wiener-like index from distance/detour matrix) |
Negative association with an asymmetric inverted U-shape |
| SM3_X (Spectral moment of order 3 from chi matrix) |
r > 0.90 r=0.81 |
nR03 (Number of 3-membered rings) D/Dtr03 (Distance/detour ring index of order 3) SRW03 (Self-returning walk count of order 3) SM5_X (Spectral moment of order 5 from chi matrix) B04[N-S] (Presence/absence of N – S at topological distance 4) B06[O-S] (Presence/absence of O – S at topological distance 6) F06[O-S] (Frequency of O – S at topological distance 6) |
Negative correlation with pIC50 |
| GATS5v (Geary autocorrelation of lag 5 weighted by van der Waals volume) |
r = -0.903 r = 0.80 |
MATS5p (Moran autocorrelation of lag 5 weighted by polarizability) GATS5p (Geary autocorrelation of lag 5 weighted by polarizability) |
Increasing values → higher activity |
| MATS1p (Moran autocorrelation of lag 1 weighted by polarizability) |
r = 0.93 r = 0.87 |
MATS1v (Moran autocorrelation of lag 1 weighted by van der Waals volume), MATS1i (Moran autocorrelation of lag 1 weighted by ionization potential) | Inverted U-shaped relationship with activity |
| JGI5 (Mean topological charge index of order 5) |
NA |
None |
Higher values → higher inhibitory activity |
| TI2_L (Second Mohar index from Laplace matrix) |
r > 0.8 for all but none > 0.9 |
MSD (Mean square distance index (Balaban)) AECC (Average eccentricity) DECC (Eccentric) ICR (Radial centric information index) MaxTD (Max topological distance) S3K (3-path Kier alpha-modified shape index) IDE (Mean information content on the distance equality) HVcpx (Graph vertex complexity index) WiA_Dz(Z) (Average Wiener-like index from Barysz matrix weighted by atomic number) SpPosA_Dz(Z) (Normalized spectral positive sum from Barysz matrix weighted by atomic number) SpMaxA_Dz(Z) (Normalized leading eigenvalue from Barysz matrix weighted by atomic number) SpMAD_Dz(Z) (Spectral mean absolute deviation from Barysz matrix weighted by atomic number) WiA_Dz(m) (Average Wiener-like index from Barysz matrix weighted by mass) SpPosA_Dz(m) (Normalized spectral positive sum from Barysz matrix weighted by mass) SpMaxA_Dz(m) (Normalized leading eigenvalue from Barysz matrix weighted by mass) SpMAD_Dz(m) (Spectral mean absolute deviation from Barysz matrix weighted by mass) WiA_Dz(v) (Average Wiener-like index from Barysz matrix weighted by van der Waals volume) SpPosA_Dz(v) (Normalized spectral positive sum from Barysz matrix weighted by van der Waals volume) SpMaxA_Dz(v) (Normalized leading eigenvalue from Barysz matrix weighted by van der Waals volume) SpMAD_Dz(v) (Spectral mean absolute deviation from Barysz matrix weighted by van der Waals volume) WiA_Dz(e) (Average Wiener-like index from Barysz matrix weighted by Sand |
Higher values → lower inhibitory activity |
| Descriptor | Correlated Descriptors | Correlation coefficient(s) | Activity Relationship |
|---|---|---|---|
| C-034 (R–CR..X) | nPyrroles (number of pyrrole rings), N-073 (Ar2NH / Ar3N / Ar2N-Al / R..N..R), SaasN (sum of aasN E-states), NaasN (number of atoms of type aasN) | R=0.89 – 0.90 |
Higher values → higher activity |
| SHED_AA (Shannon entropy descriptor, acceptor-acceptor) | SHED_DA (Shannon entropy descriptor, acceptor-acceptor) | r=0.91 | Lower values → higher activity |
| C-003 (a CHR3 group) | nCt (number of total tertiary C), nCrt (number of ring tertiary C) | r=0.88 - 0.99 | ≤3 → lower activity, 4 or 5 → higher activity |
| nCrt (number of ring tertiary C) | nCt, C-003, SpMin1_Bh(s) (smallest eigenvalue n. 1 of Burden matrix weighted by I-state) | 0.80 – 0.88 | 0 → higher activity, ≥1 → lower activity |
| CATS2D_04_AA (CATS2D Acceptor-Acceptor at lag 04) | F04[O-O] (Frequency of O – O at topological distance 4) | r=0.81 | ≥3 → Stronger activity |
| NsF (number of atoms of type sF, i.e. -F) | nF (number of fluorine atoms), nX (number of halogen atoms), P_VSA_e_6 (P_VSA-like on Sanderson electronegativity, bin 6), F-084 (F attached to C1(sp2)), SsF (sum of sF E-states), NsF (number of atoms of type sF), F01[C-F] (frequency of C – F at topological distance 1), F02[C-F] (frequency of C – F at topological distance 2), F03[C-F] (frequency of C – F at topological distance 3), F07[C-F] (frequency of C – F at topological distance ), F08[C-F] (frequency of C – F at topological distance 8) | r>0.9 or r=1.0 | Fluorinated → higher activity |
| CATS2D_04_DA (CATS2D Donor-Acceptor at lag 04) | CATS2D_04_DD, F04[O-O] | r > 0.80 | Higher values → slightly higher inhibition |
| SHED_AN (Shannon entropy descriptor, acceptor-negative) | SHED_DN, CATS2D_01_DN (CATS2D Donor-Negative at lag 01), CATS2D_00_NN (CATS2D Negative-Negative at lag 00, i.e. number of negative atoms) | r>0.90 | Higher values → slightly lower activities |
| CATS2D_02_AL (CATS2D acceptor-lipophilic at lag 02) | F04[O-O] | r = 0.84 | Higher values → slightly higher inhibition |
| CATS2D_09_DL (CATS2D Donor-Lipophilic at lag 09) | CATS2D_02_DL, CATS2D_07_DL, CATS2D_08_DL | r > 0.80 | Lower values → higher inhibitory activity |
| No. | Compound | IC50* (μM) | IC50** (μM) |
|---|---|---|---|
| Isoflavonoids | |||
| 1 | irigenin (5,7,3'-trihydroxy-6,4',5'-trimethoxyisoflavone) | 0.56 | 1.37 |
| 2 | tectoridin (shekanin; 4',5-dihydro-6-methoxy-7-(o-glucoside)isoflavone) | 0.84 | 0.72 |
| 3 | irisolidone (4'-O-methyltectorigenin) | 0.53 | 1.24 |
| 4 | iristectorin A | 0.89 | 0.82 |
| 5 | iristectorigenin B | 0.54 | 1.12 |
| 6 | homotectoridin | 0.87 | 0.70 |
| 7 | germanaism A | 0.52 | |
| 8 | irilone 4'-O-glucoside | 0.53 | 0.73 |
| 9 | germanaism B | 0.64 | 0.80 |
| 10 | germanaism A | 0.52 | 0.95 |
| 11 | Kakkalidone (irisolidone 7-O-beta-D-glucoside and its stereoisomers) | 0.59 | 0.75 |
| 12 | homotectoridin | 0.87 | |
| 13 | irisflorentin | 1.73 | 0.80 |
| 14 | pratensein 7-O-glucopyranoside | 2.08 | 0.82 |
| 15 | germanaism G | 2.34 | 0.82 |
| 16 | 3-(3-hydroxy-4,5-dimethoxyphenyl)-7-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxychromen-4-one | 1.24 | 0.69 |
| 17 | 5-hydroxy-3-(3-hydroxy-4,5-dimethoxyphenyl)-7-[(2R,3S,4R,5R,6S)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxychromen-4-one | 1.41 | 0.78 |
| 18 | germanaism D | 2.44 | 0.85 |
| flavonoids | |||
| 19 | isoswertiajaponin | 0.83 | 0.97 |
| 20 | swertisin (flavocommelitin, 6-C-glucopyranosyl-7-O-methylapigenin) | 1.24 | 0.84 |
| 21 | isoswertisin (isoflavocommelitin, 7-O-methylvitexin) | 1.07 | 0.85 |
| 22 | embigenin | 1.30 | 0.66 |
| terpenoids | |||
| 23 | iriflorentan (2Z-2-[(2R,3S,4S)-4-hydroxy-3-(hydroxymethyl)-2-(3-hydroxypropyl)-4-methyl-3-[(3E,5E)-4-methyl-6-[(1R,3S)-2,2,3-trimethyl-6-methylidenecyclohexyl]hexa-3,5-dienyl]cyclohexylidene]propanal) | 0.62 | 1.51 |
| 24 | germanical C (2-[4-hydroxy-3-(hydroxymethyl)-2-(3-hydroxypropyl)-4-methyl-3-[4-methyl-6-(2,5,6,6-tetramethylcyclohex-2-en-1-yl)hexa-3,5-dien-1-yl]cyclohexylidene]propanal) | 0.76 | 1.65 |
| 25 | irisgermanical B (2-[4-hydroxy-3-(hydroxymethyl)-2-(3-hydroxypropyl)-4-methyl-3-[4-methyl-6-(2,2,3-trimethyl-6-methylidenecyclohexyl)hexa-3,5-dien-1-yl]cyclohexylidene]propanal) | 0.62 | 1.51 |
| xanthonoids | |||
| 26 | mangiferin | 1.68 | 0.91 |
| 27 | irisxanthone | 1.84 | 0.98 |
| 28 | isomangiferin | 2.49 | 0.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





