None of the currently available medications for
Naegleria fowleri targets organism’s glucose metabolism [
2,
6]. Hence, it is imperative to investigate the activity of existing treatments on the glucose metabolism of both pathogen and host before we looked for potential drugs. Multiple drugs have been used to treat
Naegleria fowleri infections. To identify the effectiveness of these drugs against glucokinase enzymes derived from both pathogen (PDBID:6DA0) and its human homolog (PDBID:3FGU), present drugs that are used for treatment such as azithromycin, amphotericin b, Miltefosine, fluconazole, ketoconazole were docked against both glucokinase protein. The structures of the drugs were retrieved from PubChem and ChemSpider. The binding affinities of the drugs to the enzymes revealed that these drugs may have a key role to play in negatively affecting carbohydrate metabolism.
Supplementary Figure 2. , and
Supplementary Figure 4. show the binding of mentioned drugs around nGLK and hGLK respectively.
Table 3.
Screening of established drugs that are used to treat Naegleria fowleri against nGLK and hGLK.
| Name of drug |
Binding affinity for N.F(kcal/mol) |
Binding affinity for Human |
| Azithromycin |
-7.0 |
-7.4 |
| Amphotericin B |
-7.6 |
-8.5 |
| Miltefosine |
-4.1 |
-5.5 |
| Fluconazole |
-7.4 |
-6.6 |
| ketoconazole |
-8.2 |
-8.8 |
| Rifampin |
-9.2 |
-8.3 |
| Dexamethasone |
-7.9 |
-7.6 |
2.3.1. Activity of Azithromycin against nGLK and hGLK
Azithromycin is an FDA-approved drug for the treatment of various bacterial infections. It is a derivative of erythromycin and is administered orally. It prevents protein synthesis by binding to 23S rRNA and inhibits the assembly of bacterial 50S ribosomal subunit [
19]. It is used to treat opportunistic infections including pneumonia, acute sinus, bronchitis, throat, and tonsil infection. There are side effects associated with the use of azithromycin [
20,
21,
22]. The docking of azithromycin against glucokinase gave a moderate binding affinity of -7kcal/mol. This indicates the use of azithromycin for the treatment of
Naegleria fowleri could have been affecting the carbohydrate metabolism of the pathogen by affecting glucokinase.
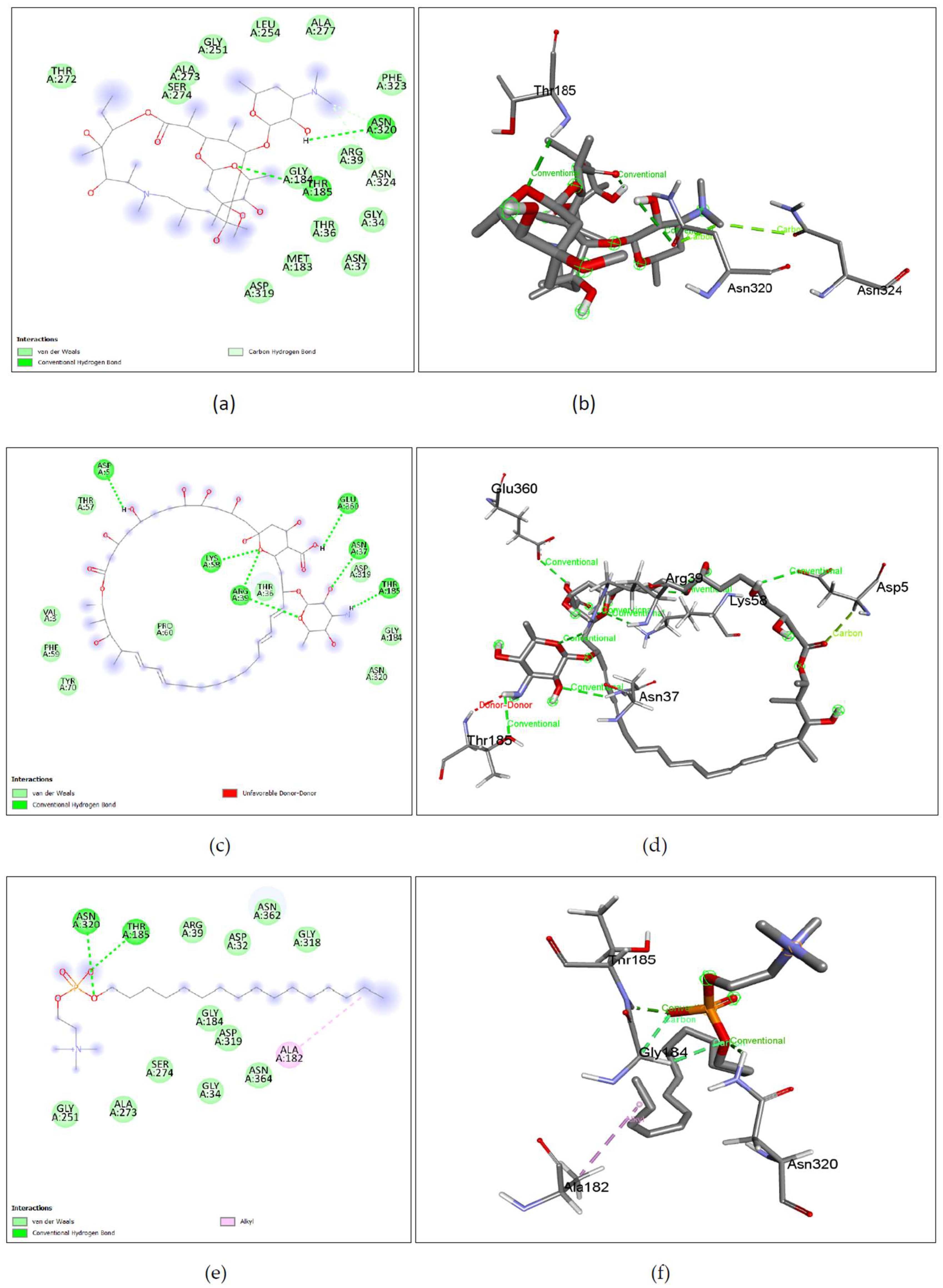
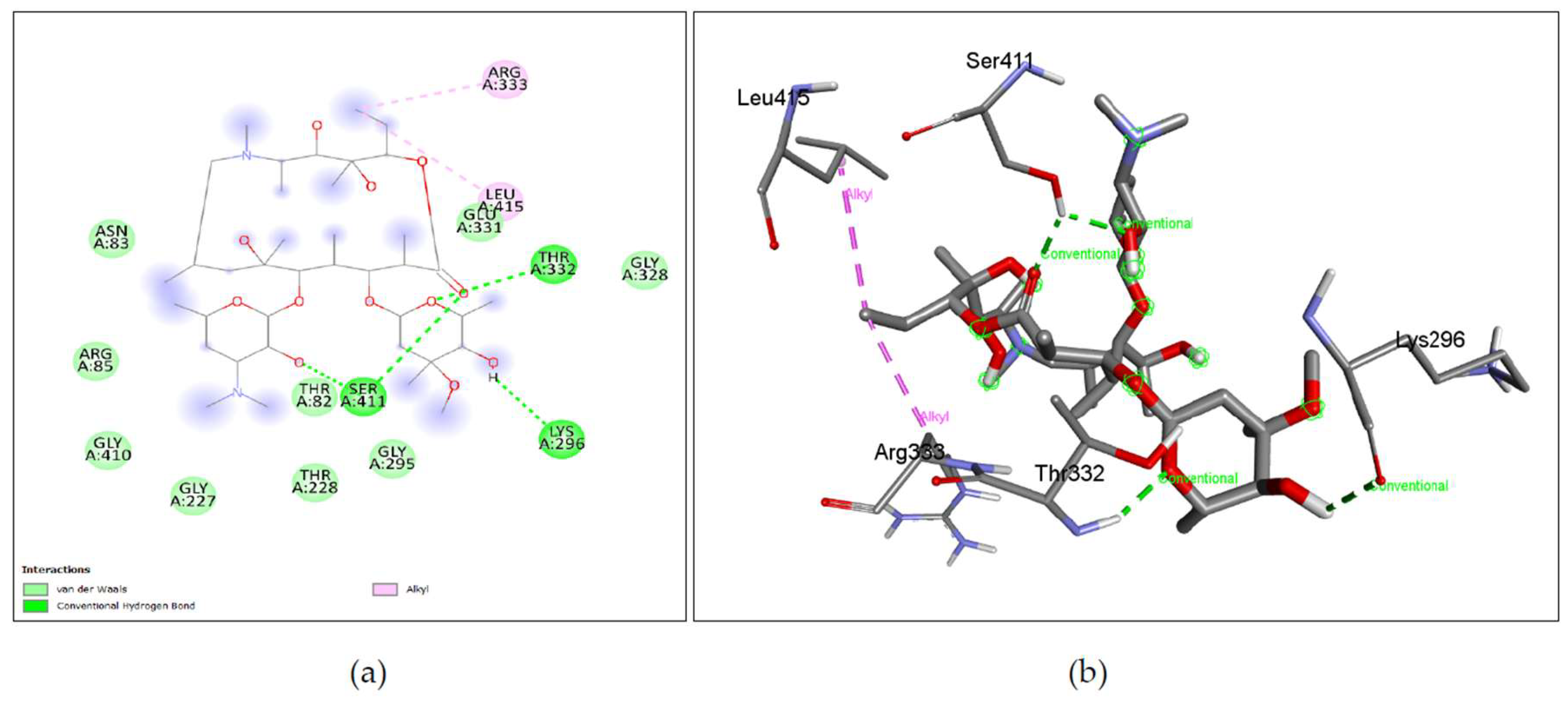
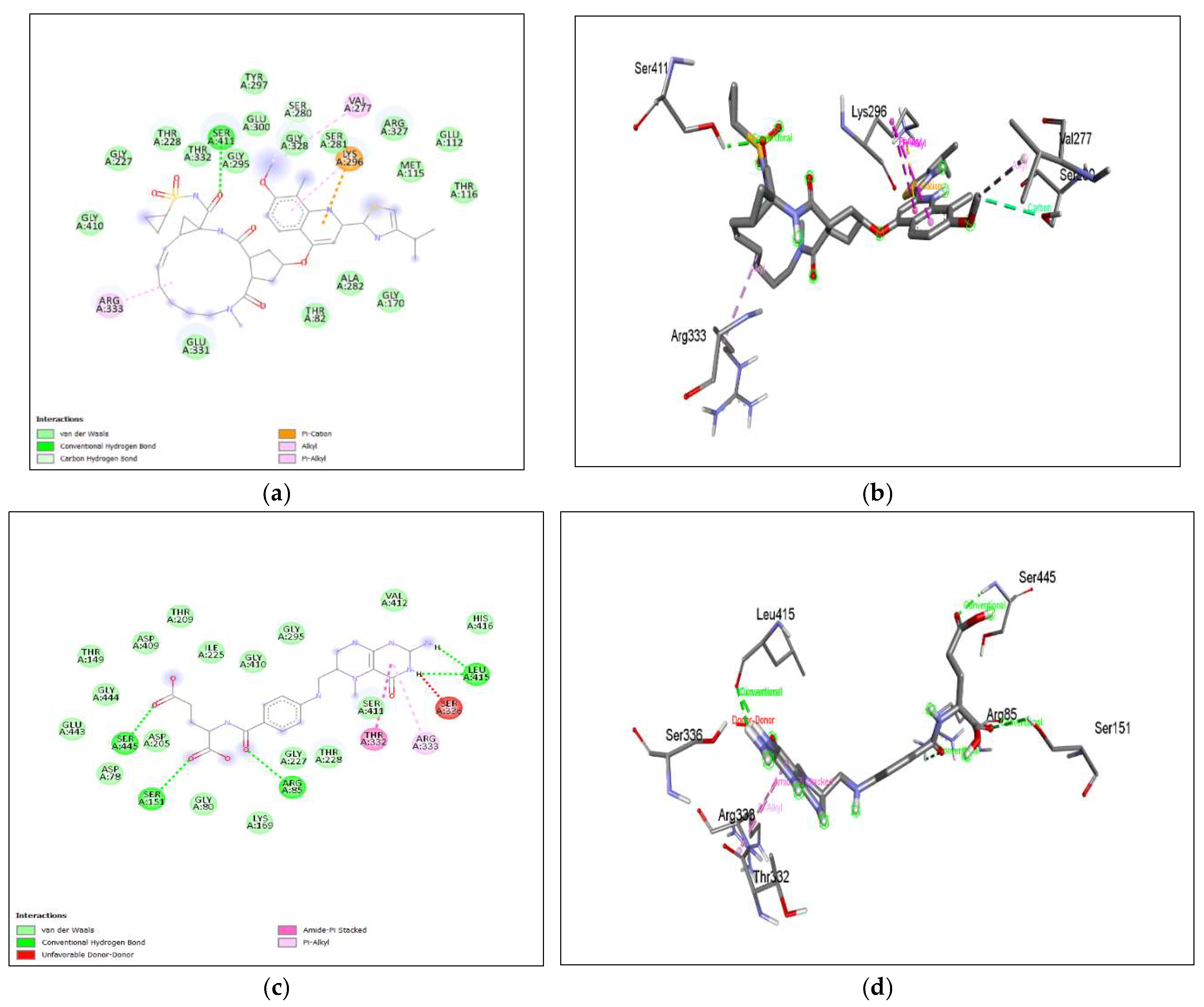
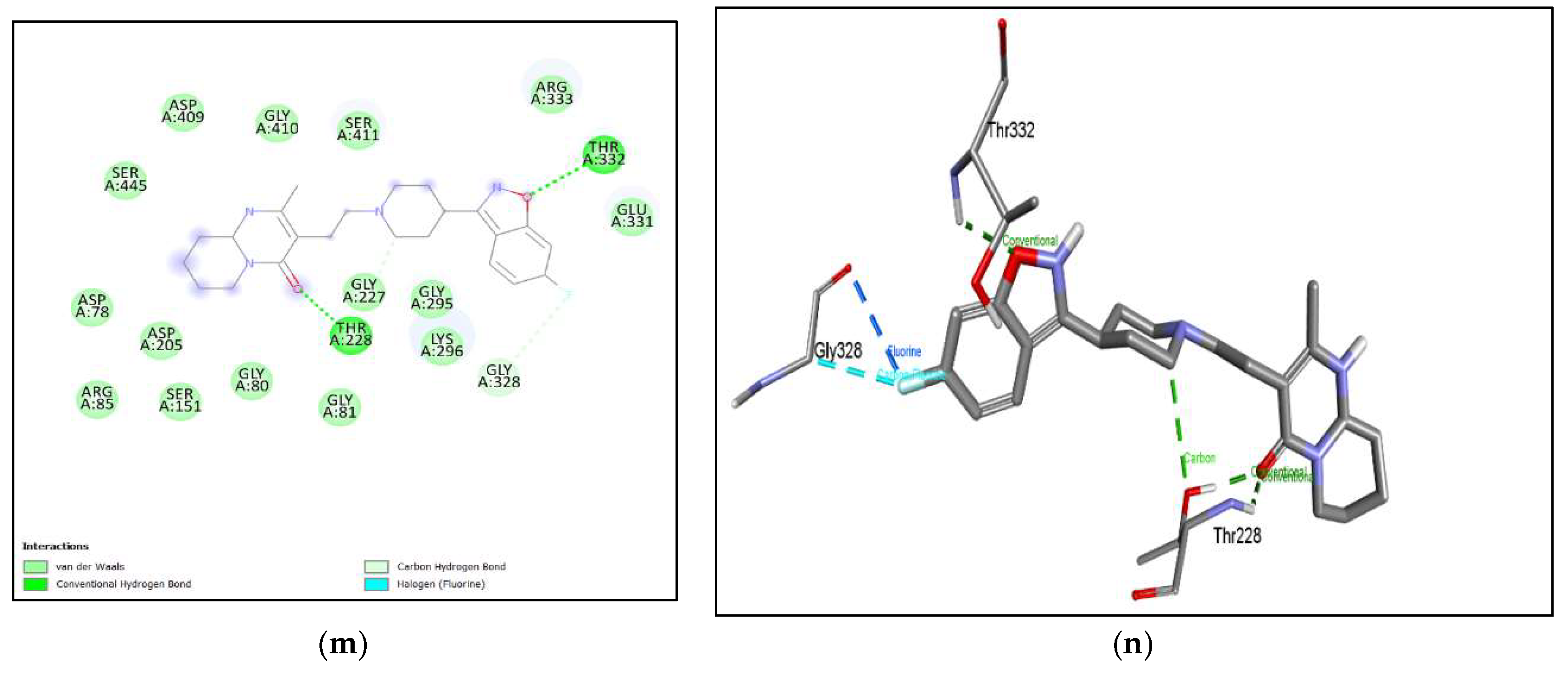
The binding affinity of the drug with the human homolog of glucokinase also gave a moderate binding affinity of -7.4kcalmol, which is even higher than the binding to pathogen glucokinase, indicating that this drug could be involved in affecting human carbohydrate metabolism. As shown in Figure 7. (a), (b) Azithromycin shows binding with nGLK around THR272, SER274, ALA273, GLY251, LEU254, ALA277, PHE323, ARG39, GLY34, ASN37, THR36, MET183, ASP319 and shows binding at ASN320, ASN324 via carbon hydrogen bond and THR185 via conventional hydrogen bond. Also, Figure 8. (a), (b) shows Azithromycin binding with hGLK around ASN83, ARG85, GLY410, GLY227, THR228, GLY295, THR82, GLU331, GLY328 and interacts with SER411, LYS296, THR332 via conventional hydrogen bond and LEU415, ARG333 via alkyl bond. Supplementary Figure 3. (a) and Supplementary Figure 5. (a) show amino acids of nGLK and hGLK within 5 A of azithromycin.
Figure 7.
Lig-plots and 3D images showing interactions between Target nGLK and well-known Naegleria fowleri drugs. (a) Azithromycin interacts with nGLK through Van Der Waals interactions (light green), carbon hydrogen bond (dark green), and conventional hydrogen bond (pale green). (b) Azithromycin interactions visualized in 3D. (c) Amphotericin B interacts with nGLK through Van Der Waals interactions (light green), and conventional hydrogen bond (dark green). (d) Amphotericin B interactions visualized in 3D. (e) Miltefosine interacts with nGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), and alkyl interactions (pale pink). (f) Miltefosine interactions visualized in 3D. (g) Fluconazole interacts with nGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), halogen interaction(blue), unfavorable bond(red), and pi-alkyl (pale pink). (h) Fluconazole interactions visualized in 3D. (i) Ketoconazole interacts with nGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), carbon hydrogen bond (pale green), pi-anion (gold), pi-alkyl (pale pink), and alkyl (pale pink). (j) Ketoconazole interactions visualized in 3D. (k) Rifampin interacts with nGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), carbon hydrogen bond (pale green), and attractive charge (gold) interaction. (l) Rifampin interactions visualized in 3D. (m) Dexamethasone interacts with nGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green). (n) Dexamethasone interactions visualized in 3D.
Figure 7.
Lig-plots and 3D images showing interactions between Target nGLK and well-known Naegleria fowleri drugs. (a) Azithromycin interacts with nGLK through Van Der Waals interactions (light green), carbon hydrogen bond (dark green), and conventional hydrogen bond (pale green). (b) Azithromycin interactions visualized in 3D. (c) Amphotericin B interacts with nGLK through Van Der Waals interactions (light green), and conventional hydrogen bond (dark green). (d) Amphotericin B interactions visualized in 3D. (e) Miltefosine interacts with nGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), and alkyl interactions (pale pink). (f) Miltefosine interactions visualized in 3D. (g) Fluconazole interacts with nGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), halogen interaction(blue), unfavorable bond(red), and pi-alkyl (pale pink). (h) Fluconazole interactions visualized in 3D. (i) Ketoconazole interacts with nGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), carbon hydrogen bond (pale green), pi-anion (gold), pi-alkyl (pale pink), and alkyl (pale pink). (j) Ketoconazole interactions visualized in 3D. (k) Rifampin interacts with nGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), carbon hydrogen bond (pale green), and attractive charge (gold) interaction. (l) Rifampin interactions visualized in 3D. (m) Dexamethasone interacts with nGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green). (n) Dexamethasone interactions visualized in 3D.
Figure 8.
Lig-plots showing interactions between Human glucokinase hGLK and well-known Naegleria fowleri drugs. (a) Azithromycin interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), and alkyl interactions. (b) Azithromycin interactions visualized in 3D. (c) Azithromycin B interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), and unfavorable bond (red). (d) Amphotericin B interactions visualized in 3D. (e) Miltefosine interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), attractive charge (yellow), carbon hydrogen bond (pale green), alkyl interactions (pale pink), and unfavorable bond (red). (f) Miltefosine interactions visualized in 3D. (g) Fluconazole interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), pi-sigma interactions (purple), and pi-alkyl interactions (pale pink). (h) Fluconazole interactions visualized in 3D. (i) Ketoconazole interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), pi-sigma interactions (purple), alkyl (pale pink), and pi-alkyl interactions (pale pink). (j) Ketoconazole interactions visualized in 3D. (k)) Rifampin interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), and pi-cation interactions (yellow). (l) Rifampin interactions visualized in 3D. (m) Dexamethasone interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), and halogen interactions (blue). (n) Dexamethasone interactions visualied in 3D.
Figure 8.
Lig-plots showing interactions between Human glucokinase hGLK and well-known Naegleria fowleri drugs. (a) Azithromycin interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), and alkyl interactions. (b) Azithromycin interactions visualized in 3D. (c) Azithromycin B interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), and unfavorable bond (red). (d) Amphotericin B interactions visualized in 3D. (e) Miltefosine interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), attractive charge (yellow), carbon hydrogen bond (pale green), alkyl interactions (pale pink), and unfavorable bond (red). (f) Miltefosine interactions visualized in 3D. (g) Fluconazole interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), pi-sigma interactions (purple), and pi-alkyl interactions (pale pink). (h) Fluconazole interactions visualized in 3D. (i) Ketoconazole interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), pi-sigma interactions (purple), alkyl (pale pink), and pi-alkyl interactions (pale pink). (j) Ketoconazole interactions visualized in 3D. (k)) Rifampin interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), and pi-cation interactions (yellow). (l) Rifampin interactions visualized in 3D. (m) Dexamethasone interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), and halogen interactions (blue). (n) Dexamethasone interactions visualied in 3D.
2.3.2. Activity of Amphotericin B against nGLK and hGLK
Amphotericin B, also known as Fungizone is an FDA-approved antifungal drug used to treat histoplasmosis, cryptococcosis, and opportunistic infections due to HIV. Amphotericin B binds to ergosterol, found on the fungal and
Naegleria fowleri membrane. It is reported to induce immune cell stimulation[
18,
23]. The binding affinity of amphotericin B against glucokinase was -7.6kcal/mol. This score suggests that the binding is moderately strong and could mean that the drug could reduce the efficiency of the glucokinase activity of the pathogen. Amphotericin B has an even higher binding affinity of -8.5kcal/mol to the human homolog of glucokinase, which indicates that the drug could be affecting the carbohydrate metabolism of the host. This result confirm studies that have shown marked difference in carbohydrate metabolism caused by amphotericin B [
24,
25]. Previously, our study had identified sterol 24-C methyltransferase inhibitors that could inhibit ergosterol biosynthesis in
N. gruberi [
10]. Amphotericin is also known to inhibit the ergosterol pathway which is essential for fungi. N.
fowleri also requires ergosterol in its cell membrane[
26]. This docking result could indicate that amphotericin could inhibit both lipid and carbohydrate metabolism. The administration of this drug is limited due to its severe acute and chronic toxicity leading to nausea, vomiting, fever, hypoxia, hypertension, and nephrotoxicity[
23]. As shown in
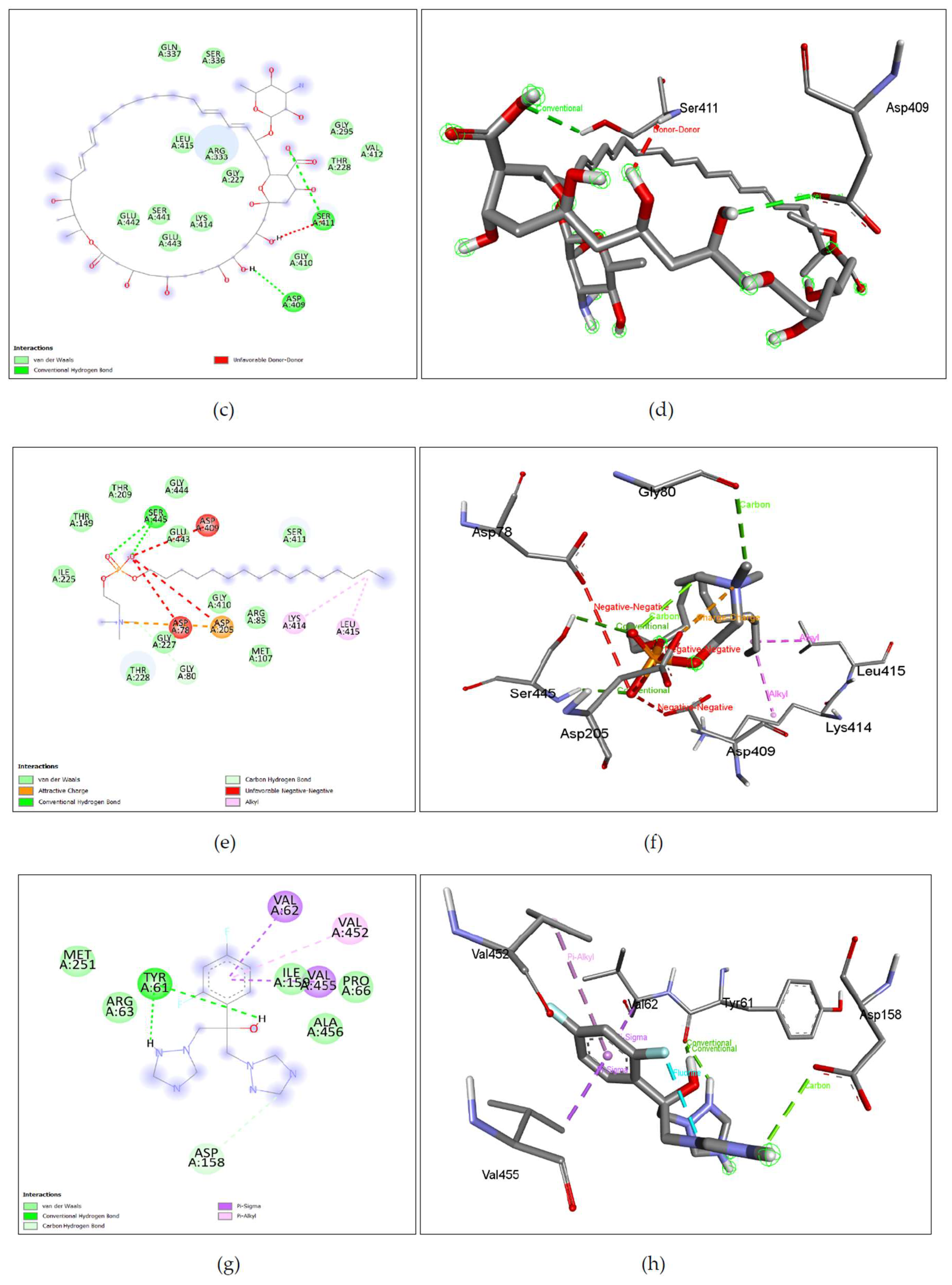
Figure 7. (c), (d) Amphotericin B shows binding with nGLK around VAL3, PHE59, TYR70, PRO60, ASN320, GLY184, ASP319, THR57 and interacts with ASP5, LYS58, ARG39, ASN37, GLU360, THR185 via Conventional hydrogen bond, and THR185 via unfavorable interactions. Also,
Figure 8. (c), (d) shows amphotericin B binding with human glucokinase around GLN337, SER336, GLY295, VAL412, THR228, GLY410, GLY227, ARG333, LEU415, LYS414, GLU443, GLU442, SER441 and interacts with SER411, ASP 409 via conventional hydrogen bond and SER411 via unfavorable donor-donor interaction.
Supplementary Figure 3. (b) and
Supplementary Figure 5. (b) show amino acids of nGLK and hGLK within 5 A of amphotericin.
2.3.3. Activity of Miltefosine against nGLK and hGLK
Miltefosine is an antimicrobial drug that was originally used against cancer where it interfered with lipid-dependent cell signaling and induced apoptosis. Miltefosine is an ester of phosphocholine compound and has anti-inflammatory, anti-fungal, antiprotozoal, and antiviral effects [
7,
27]. It modulates cell membrane permeability, lipid composition, signal transduction, and phospholipid metabolism. It is known to be an inhibitor of MAPK and induces apoptosis by balancing MAPK and JNK pathways[
7,
27,
28]. It also modulates the immune response. The docking of miltefosine against nGLK gave a low binding score of -4.7kcal/mol. It also bound to the hGLK with an affinity of -5.5kcal/mol, indicating this could be affecting carbohydrate metabolism in the host. It is known to be teratogenic, impair fertility and cause fetal death below recommended doses in animals (IMPAVIDO). The administration of these drugs has potentially lasting side effects on patients. It is administered to young adults for the treatment of
Naegleria infections. As shown in
Figure 7. (e), (f), Mitefosine shows binding with nGLK at ARG39, ASP32, GLY318, ASN362, GLY184, ASP319, GLY34, ASN364, SER274, ALA273, GLY251 and shows interaction with ASN320, THR185 via conventional hydrogen bond and ALA182 via Alkyl interaction against the pathogen’s glucokinase. Also,
Figure 8. (e), (f), shows Miltefosine binding with hGLK around THR149, THR209, GLY444, ILE225, SER411, GLY227, THR228, GLY410, ARG85, MET107, GLY80, GLY443 and interacts with SER445 via conventional hydrogen bond, ASP205 via attractive charge interactions, LYS414, LEU415 via alkyl interactions, GLY80 via carbon hydrogen bond and ASP78, ASP205, ASP409 via unfavorable interactions.
Supplementary Figure 3. (c) and
Supplementary Figure 5. (c) show amino acids of nGLK and hGLK within 5 A of miltefosine.
2.3.4. Activity of Fluconazole against nGLK and hGLK
Fluconazole is an FDA-approved antifungal drug used against esophagus, throat, vaginal infection, meningitis, and other opportunistic infections. It works against superficial fungal infections [
29]. It is a synthetic triazole acting against fungal sterol demethylation. It had a moderate binding affinity of -7.4 kcal/mol against glucokinase. The drug may affect the glucokinase enzyme negatively. The drug has a binding affinity of -6.6kcal/mol when docked against hGLK. It is a highly selective inhibitor for the fungal ergosterol synthesis enzyme, lanosterol 14-α demethylase. It is known to cause hallucinations and paranoia and carries a risk of severe hepatic toxicity
[30]. As shown in
Figure 7. (g), (h), Fluconazole shows binding with nGLK around ASN364, LEU135, SER138, PHE363, ILE142, GLY318, ASP319, MET183, TYR361, LEU187, GLY184, ARG39, THR185, ASN37, GLY34 and interacts with ASP32, ALA182, GLY186 via conventional hydrogen bond and ALA317, ALA182 via pi-alkyl interactions and GLN357 via Halogen interaction. Also,
Figure 8. ((g), (h), shows Fluconazole binding with hGLK around MET251, ARG63, ALA456, PRO66, ILE159 and interacts with TYR61 via a conventional hydrogen bond, VAL455, VAL62 via pi-sigma bond, VAL452 via pi-alkyl and ASP158 via carbon hydrogen bond.
Supplementary Figure 3. (d) and
Supplementary Figure 5. (d) show amino acids of nGLK and hGLK within 5 A of fluconazole.
2.3.5. Activity of Ketoconazole against nGLK and hGLK
Ketoconazole also known as xolegel is a broad-spectrum fungicide for superficial fungal infections. It is used in immunosuppressed individuals. It inhibits ergosterol synthesis by interacting with 14-α-sterol demethylase. It is an imidazole with acute drug-induced liver toxicity and is not used for anti-fungal treatments [
31,
32]. Ketoconazole is a potent inhibitor of CYP3A4/5 and is known to affect the concentration of saxagalaptin, an orally available drug for type 2 diabetes [
33]. The binding affinity of ketoconazole against pathogen glucokinase was -8.2kcal/mol which is a good binding score. It could mean that the treatment administered to
Naegleria fowleri patients may have been inhibiting the glucokinase activity and affecting the pathogen negatively. Inhibition of the glucokinase enzyme in
Naegleria fowleri was shown to be detrimental to the growth of the pathogen. Ketoconazole also binds to the human homolog of glucokinase with a binding affinity of -8.8kcal/mol. As shown in
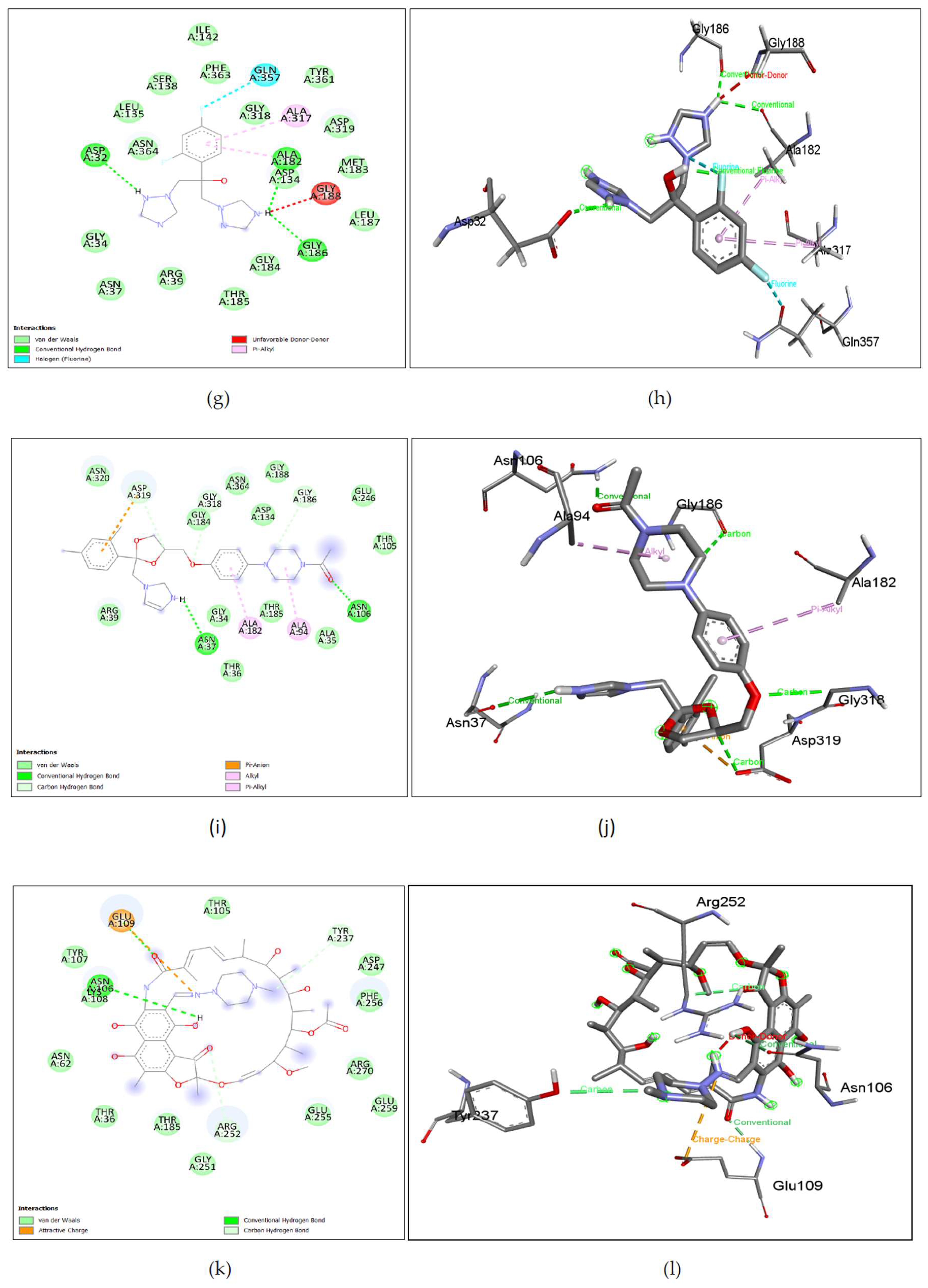
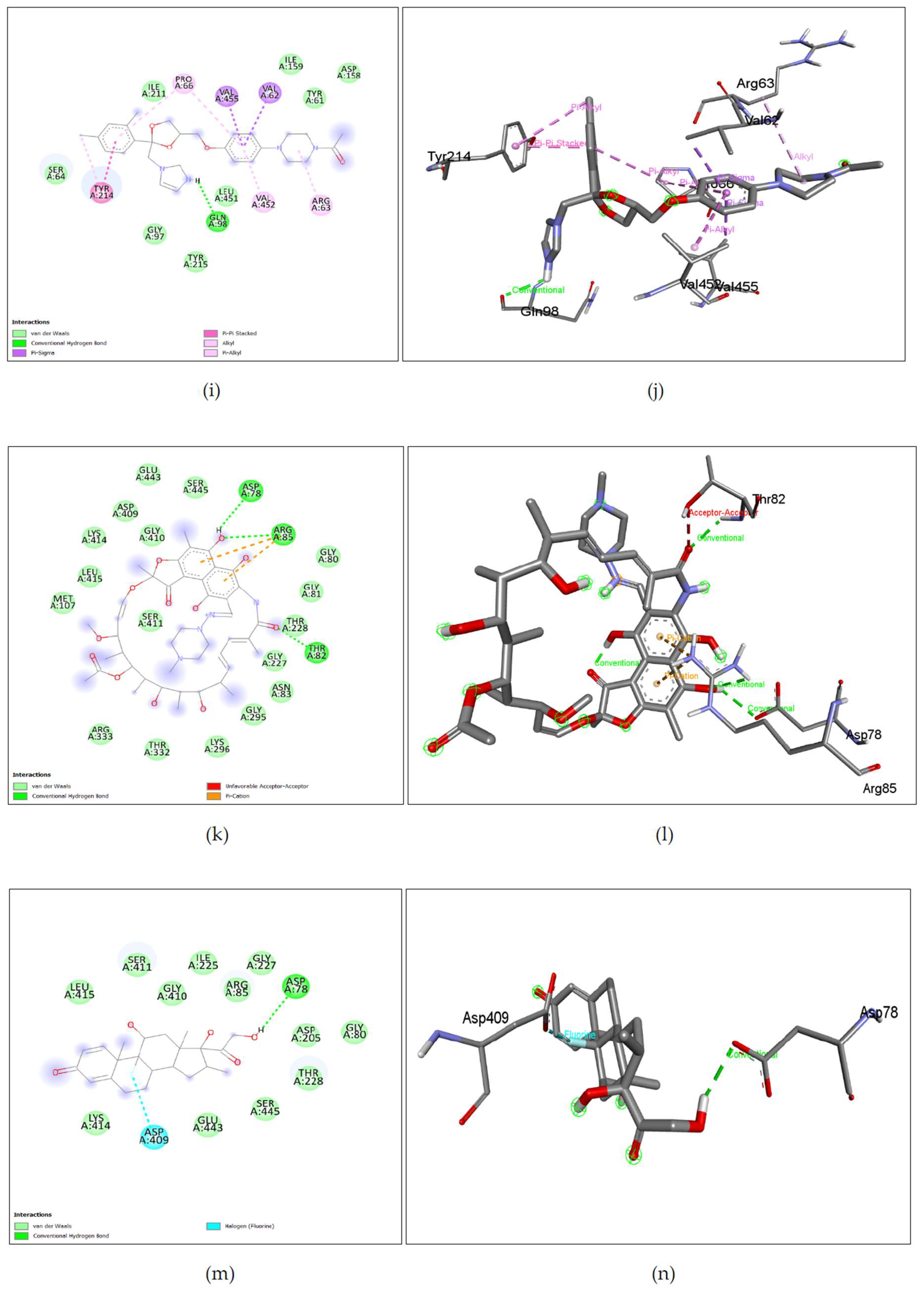
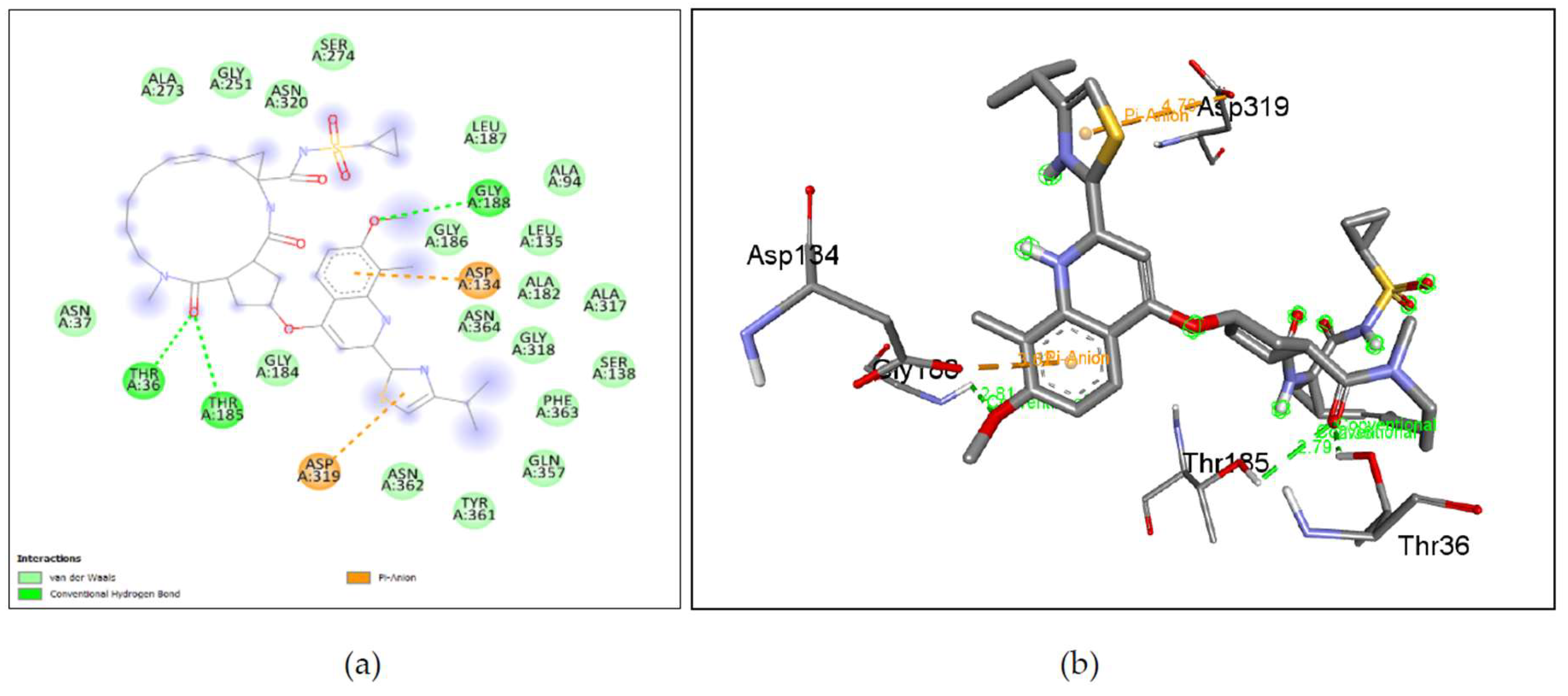
Figure 7. (i), (j), Ketoconazole shows binding with nGLK around ASN320, ARG39, THR36, ALA35, THR185, THR105, GLU246, GLY188, ASN364, ASP134, GLY318, GLY34, ASN364, ASN320 and interacts with ASP319, GLY184, GLY186 via carbon hydrogen bond, ASN106, ASN37 via conventional hydrogen bond, ALA182, ALA94 via pi-alkyl interactions, ASP319 via pi-anion interaction. Also,
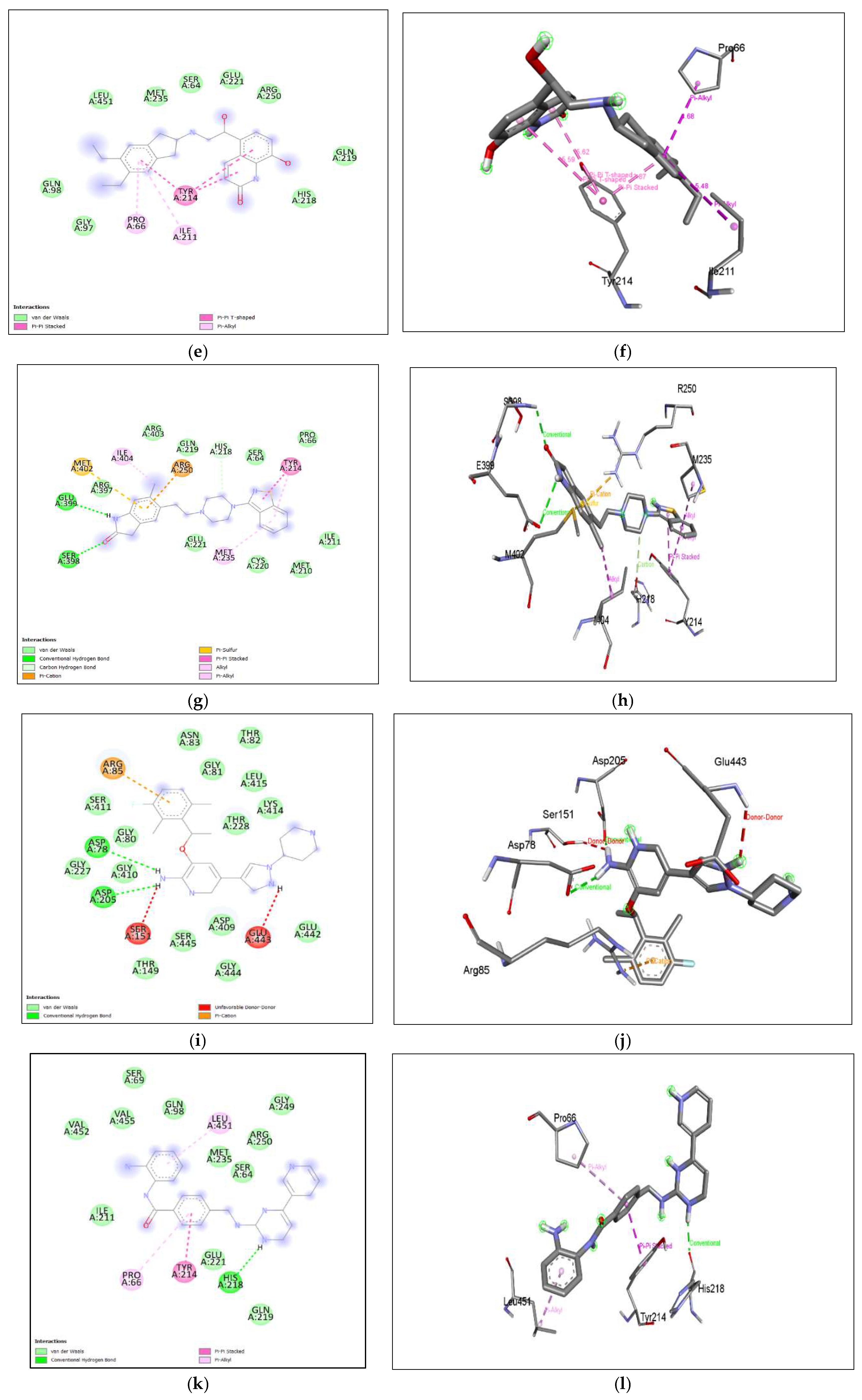
Figure 8. (i), (j), shows Ketoconazole binding with hGLK around SER64, GLY97, TYR215, LEU451, TYR61, ILE159, ASP158, ILE211 and interacts with GLN98 via conventional hydrogen bond, PRO66, VAL452, via pi-alkyl, ARG63 via alkyl interactions, TYR214 via pi-pi- stacked and pi-alkyl interactions, VAL455, VAL62 via pi-sigma interactions.
Supplementary Figure 3. (e) and
Supplementary Figure 5. (e) show amino acids of nGLK and hGLK within 5 A of ketoconazole.
2.3.6. Activity of Rifampin against nGLK and hGLK
Rifampin is also known as rifampicin is a semisynthetic antibiotic that can acts as an RNA polymerase inhibitor, DNA synthesis inhibitor, protein synthesis inhibitor, and well-known anti-tuberculosis drug[
34]. It has neuroprotective, antiangiogenic, leprostatic, antiameobic, and antineoplastic activity[
35]. Rifampin has been known to eliminate bacteria from noses and throats and inhibits tau progression in Alzheimer’s[
36]. This is important considering the route taken by
Naegleria fowleri is through the nasal pathway. Docking of rifampin against glucokinase revealed high binding affinity scores of -9.2kcal/mol. This could mean that rifampin could have been hampering carbohydrate metabolism and the pentose phosphate pathway of nucleotide synthesis by binding to glucokinase. The binding affinity of rifampin against human glucokinase is -8.3kcal/mol. As shown in
Figure 7. (k), (l), Rifampin shown binding with nGLK around ASN62, THR107, THR105, THR36, THR185, GLY251, GLU255, GLU259, ARG270, PHE256, ASP247, LYS108 and interacts with ASN106, GLU109 via a conventional hydrogen bond, ARG252, TYR237 via carbon hydrogen bond, GLU109 via attractive charge interaction. Also,
Figure 8. (k), (l), shows Rifampin binding with hGLK around MET107, LEU415, LYS414, ASP409, GLU443, GLY410, SER445, ARG333, THR332, LYS296, GLY295, ASN83, GLY227, THR228, GLY81, GLY80 and interacts with ASP78, ARG85, THR82 via conventional hydrogen bond, ARG85 via pi-cation interactions and THR82 via unfavorable interactions. Rifampin is also known to enhance the glucose-lowering effects of metformin[
37].
Supplementary Figure 3. (f) and
Supplementary Figure 5. (f) show amino acids of nGLK and hGLK within 5 A of rifampin.
2.3.7. Activity of Dexamethasone against nGLK and hGLK
Dexamethasone is a synthetic corticosteroid that has an adrenergic, antineoplastic, immunosuppressive, and anti-inflammatory effect. Dexamethasone was used for the treatment of COVID-19 and reduced the deaths of patients on ventilation by one-third and one-fifth for patients on oxygen[
38]. It is also used in gastrointestinal, dermatological, and allergic conditions. It can affect the central nervous system[
39]. It is known to increase blood glucose in humans[
40]. The docking of dexamethasone against pathogen glucokinase gave a moderate binding affinity of -7.9kcal/mol. This could indicate that it may hamper the efficiency of glucokinase in
Naegleria fowleri and affect carbohydrate catabolism negatively. It showed a binding affinity of -7.6kcal/mol when docked against hGLK. As shown in
Figure 7. (m), (n), Dexamethasone shows binding with nGLK around ASP134, ALA317, PHE363, TYR361, GLY318, ASP319, ARG39, GLY34, ALA35, GLY186, THR185, GLY184, ALA182, LEU135, SER138, ASN362 and interacts with ASN364, ASN37, THR36 via conventional hydrogen bond. Also,
Figure 8. (m), (n), shows Dexamethasone binding with hGLK around LEU415, LYS414, GLU443, SER445, THR228, ASP205, GLY80, GLY227, ARG85, ILE225, GLY410, SER411 and interacts with ASP78 via conventional hydrogen bond and ASP409 via halogen interaction.
Supplementary Figure 3. (g) and
Supplementary Figure 5. (g) show amino acids of nGLK and hGLK within 5 A of dexamethasone.
The binding affinities against human homolog of glucokinase indicated that all these FDA-approved drugs may induce inhibition of carbohydrate metabolism. This could help understand some of the side effects caused by these drugs. Though approved for medical use, these drugs cause severe negative effects. The need for drugs that specifically target the pathogen alone is necessary to decrease the side effects of Naegleria fowleri treatment.
2.3.8. Virtual Screening and Visualization of Ligands against nGLK and hGLK
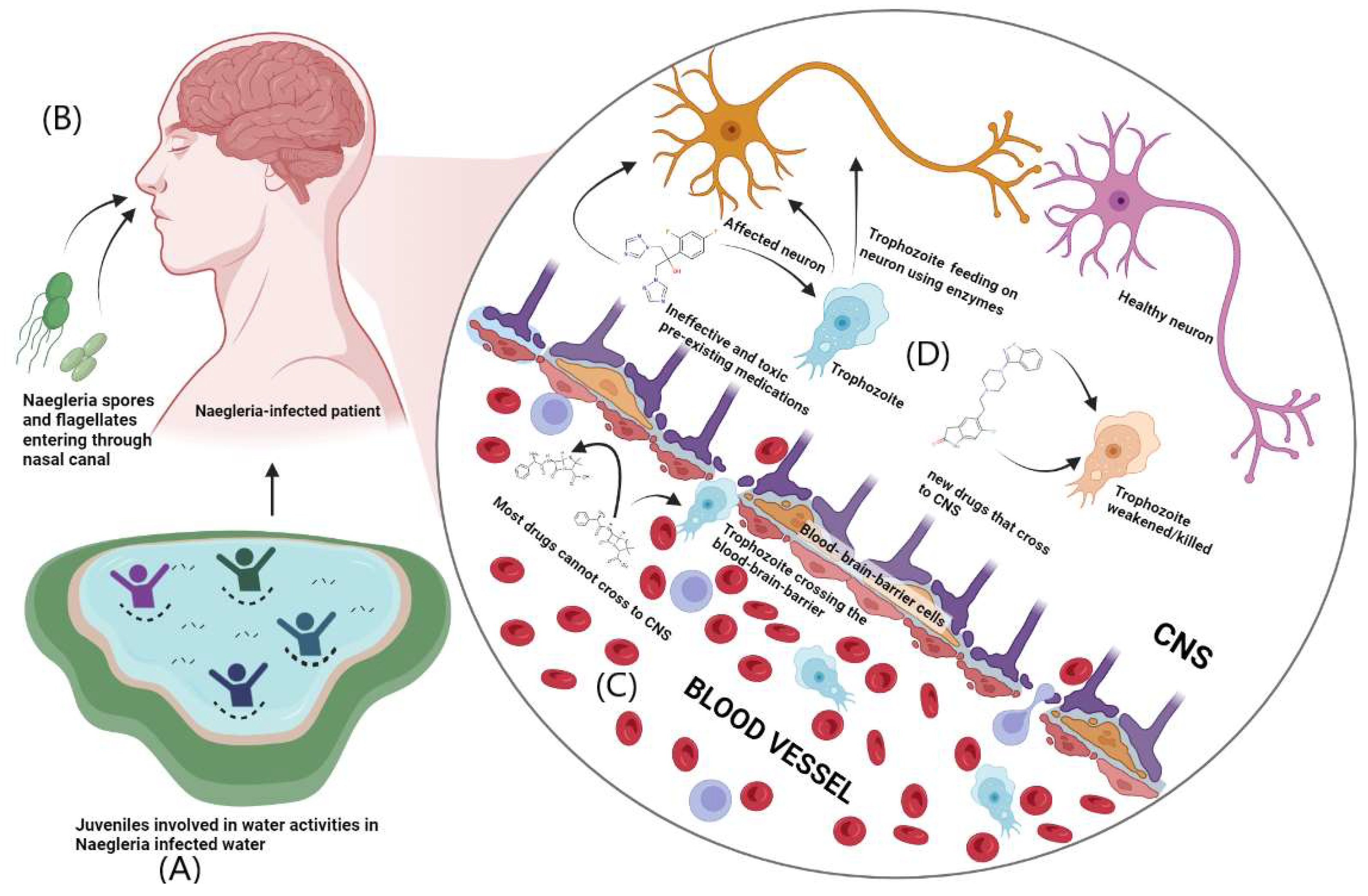
Since the FDA approved CDC recommended drugs for the treatment of PAM could bind to both glucokinases i.e., nGLK and hGLK, even though the intended targets are different partially explains why there is a drop in blood sugar levels upon treatment with these drugs. Hence, new ligands were screened against both nGLK and hGLK to identify potential drugs that could specifically bind to only the pathogenic nGLK. The compounds represented here are selected based on the specificity of binding of the compounds to the nGLK.
The top compounds selected post screening based on pharmacokinetic properties and drug-likeness. Though most compounds may have a higher binding affinity towards the pathogenic enzyme nGLK, but were disregarded due to poor ADME and pharmacokinetic properties. Most compounds could not cross the blood-brain barrier. The complexity of finding a drug that specifically binds to the target and passes all the criteria was challenging.
Most ligands showed excellent binding to the glucokinase enzymes. It was challenging to find a compound that specifically bonded with nGLK only. The difference in binding affinities between the two proteins was within the range of 3kcal/mol. The binding affinities of the compounds ranged from -4kcal/mol to -10.7kcal/mol. The ligands that had the best binding affinity and largest difference in binding between the nGLK and hGLK were chosen, along with pharmacokinetic properties in mind. Drugs that could not cross the blood-brain barrier would not be suitable for PAM treatment but were further tested to check whether they could potentially inhibit the human homolog hGLK. As mentioned in earlier sections, hGLK plays a critical role in the progression of diabetes and is a prime target for diabetes treatment[
13,
17,
33].
Table 4.
Comparative analysis of binding affinities of compounds against nGLK and hGLK.
Table 4.
Comparative analysis of binding affinities of compounds against nGLK and hGLK.
| Ligands |
Binding affinity with nGLK (Kcal/mol) |
Binding affinity with hGLK (Kcal/mol) |
| Simeprevir (PCID: 24873435) |
-10.7 |
-9 |
| Levomefolic acid (PCID: 135398561) |
-10.4 |
-9 |
| Indacaterol (PCID: 6918554) |
-10.4 |
-8.1 |
| Ziprasidone (PCID: 60854) |
-10.2 |
-8.4 |
| Crizotinib (PCID: 11626560) |
-9.8 |
-7.8 |
| Mocetinostat (PCID: 9865515) |
-10 |
-8.8 |
| Risperidone (PCID: 5073) |
-10 |
-8.9 |
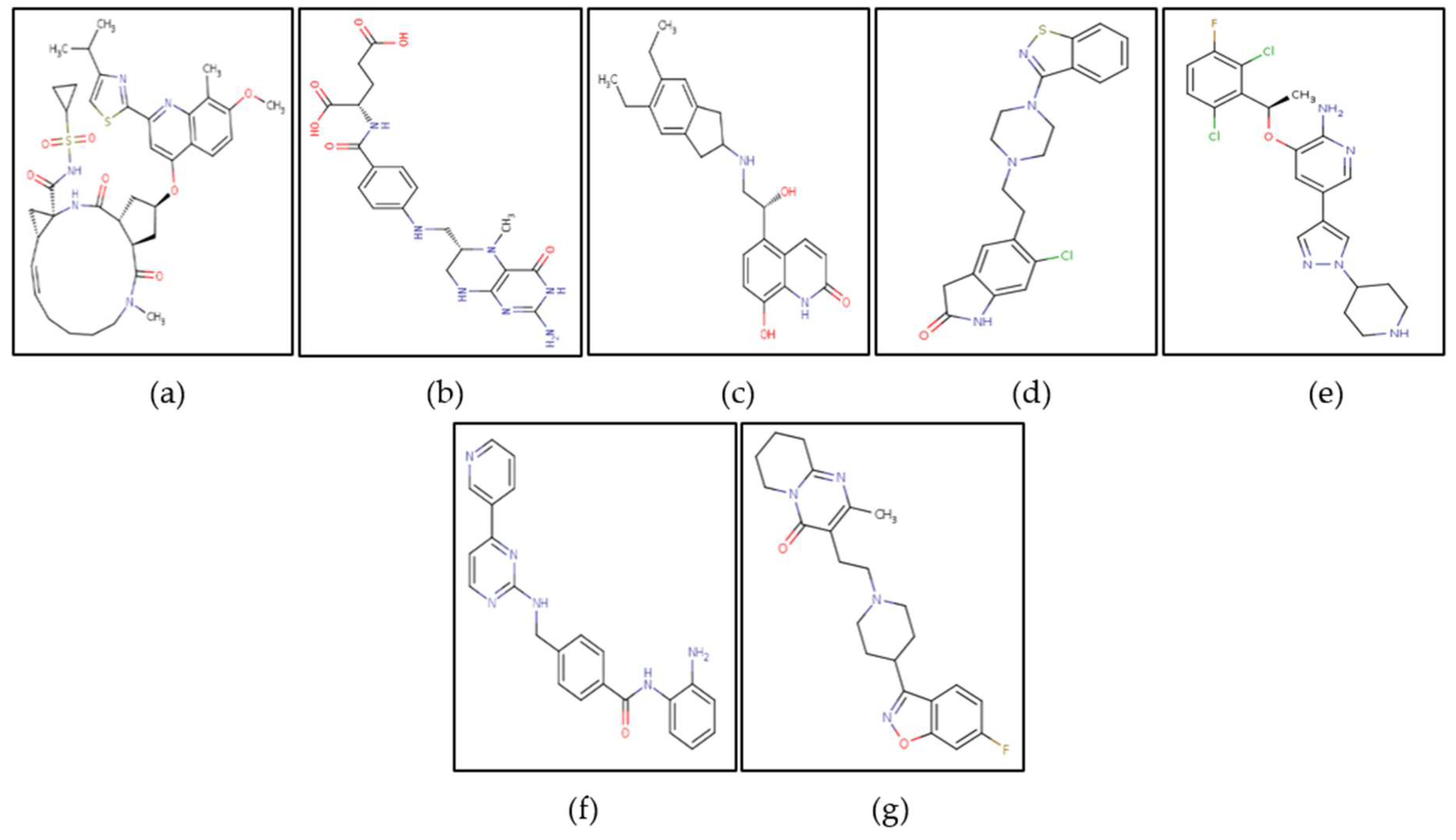
Figure 9.
Top potential drugs identified against nGLK and hGLK: (a) Simeprevir (b) Levomefolic acid (c) Indacaterol (d) Ziprasidone, and (e) Crizotinib.
Figure 9.
Top potential drugs identified against nGLK and hGLK: (a) Simeprevir (b) Levomefolic acid (c) Indacaterol (d) Ziprasidone, and (e) Crizotinib.
Though the pharmacokinetic studies suggest that Simeprevir and Levomefolic acid have violations of druglikeness and ADME properties, but they are approved by the FDA and have been used for the treatment of various diseases. The pharmacokinetic properties of the compounds shown in Figure. 14 are given in Table 5.
Table 5.
Drug-likeness and ADME properties of top ligands.
Table 5.
Drug-likeness and ADME properties of top ligands.
| Ligand |
Molecular weight (g/mol) |
Lipophilicity (Log P o/w (XLOGP3)) |
Hydrogen bond acceptors, donors |
Water solubility (Log S (ESOL)) |
| Simeprevir |
749.94 |
4.81 |
9,2 |
-7.14 Poorly soluble |
| Levomefolic |
459.46 |
-0.49 |
7,7 |
-1.99 Very soluble |
| Indacaterol |
392.49 |
3.27 |
4,4 |
-4.40 Moderately soluble |
| Ziprasidone |
412.94 |
4.02 |
3,1 |
-5.07 Moderately soluble |
| Crizotinib |
450.34 |
3.69 |
5,2 |
-5.05 Moderately soluble |
| Mocetinostat |
396.44 |
2.76 |
4,3 |
-4.17 Moderately soluble |
| Risperidone |
410.48 |
2.72 |
6,0 |
-4.20 Poorly soluble |
Table 6.
Pharmacokinetic properties of top ligands.
Table 6.
Pharmacokinetic properties of top ligands.
| Ligand |
Druglikeness violations (Lipinski, Ghose, Veber, Egan, Muegge) |
Bioavailability |
Blood-brain barrier crossing |
GI absorption |
Skin permeability (cm/s) |
| Simeprevir |
Yes (Lipinski-2, Ghose-3, Verber-1, Egan-1, Mugge-2) |
0.17 |
No |
Low |
-7.46 |
| Levomefolic acid |
Yes (Lipinski-2, Ghose-1, Verber-1, Egan-1, Mugge-2) |
0.11 |
Yes |
Low |
-9.45 |
| Indacaterol |
No |
0.55 |
No |
High |
-6.32 |
| Ziprasidone |
No |
0.55 |
Yes |
High |
-5.96 |
| Crizotinib |
No |
0.55 |
No |
High |
-6.43 |
| Mocetinostat |
No |
0.55 |
No |
High |
-6.76 |
| Risperidone |
No |
0.55 |
Yes |
High |
-6.87 |
Table 7.
Drugs against nGLK.
Table 7.
Drugs against nGLK.
| nGLK inhibitors |
Drug type |
| Levomefolic acid |
Human metabolite |
| Ziprasidone |
Antipsychotic |
| Risperidone |
Antipsychotic |
Table 8.
Drugs against hGLK.
Table 8.
Drugs against hGLK.
| hGLK inhibitors |
Drug type |
| Simeprevir |
Antiviral |
| Indacaterol |
Bronchodilator |
| Crizotinib |
Anticancer |
| Mocetinostat |
Anticancer |
Based on the blood-brain barrier crossing ability, the drugs were divided into two categories as shown in Table 7. and Table 8. The drugs that crossed the blood-brain barrier could serve as potential drugs against Naegleria fowleri while those that could not cross into the CNS could be used as hGLK inhibitors for the treatment of diabetes. Supplementary Figure 6. And Supplementary Figure 8. shows the binding of potential drugs around nGLK and hGLK respectively.
2.3.9. Repurposing of Antiviral Drug Simeprevir against nGLK and Comparing Ligand Interactions against hGLK
Simeprevir is a NS3/4A inhibitor used against the Hepatitis C virus protease complex, where it binds to the active site and disrupts viral protein processing and replication[
41,
42]. This azamacrocycle, was approved for use by the FDA in November 2014 and marketed under the name Olysio. It is known to bind albumin and alpha 1 acid glycoprotein which might assist it to cross the blood-brain barrier. It was predicted to cross the blood-brain barrier according to one in-silico study, though, in our ADME analysis and other literature, it showed to not cross the blood-brain barrier [
43,
44]. It shows binding affinity of -10.7kcl/mol and -9kacl/mol against nGLK and hGLK respectively. As shown in
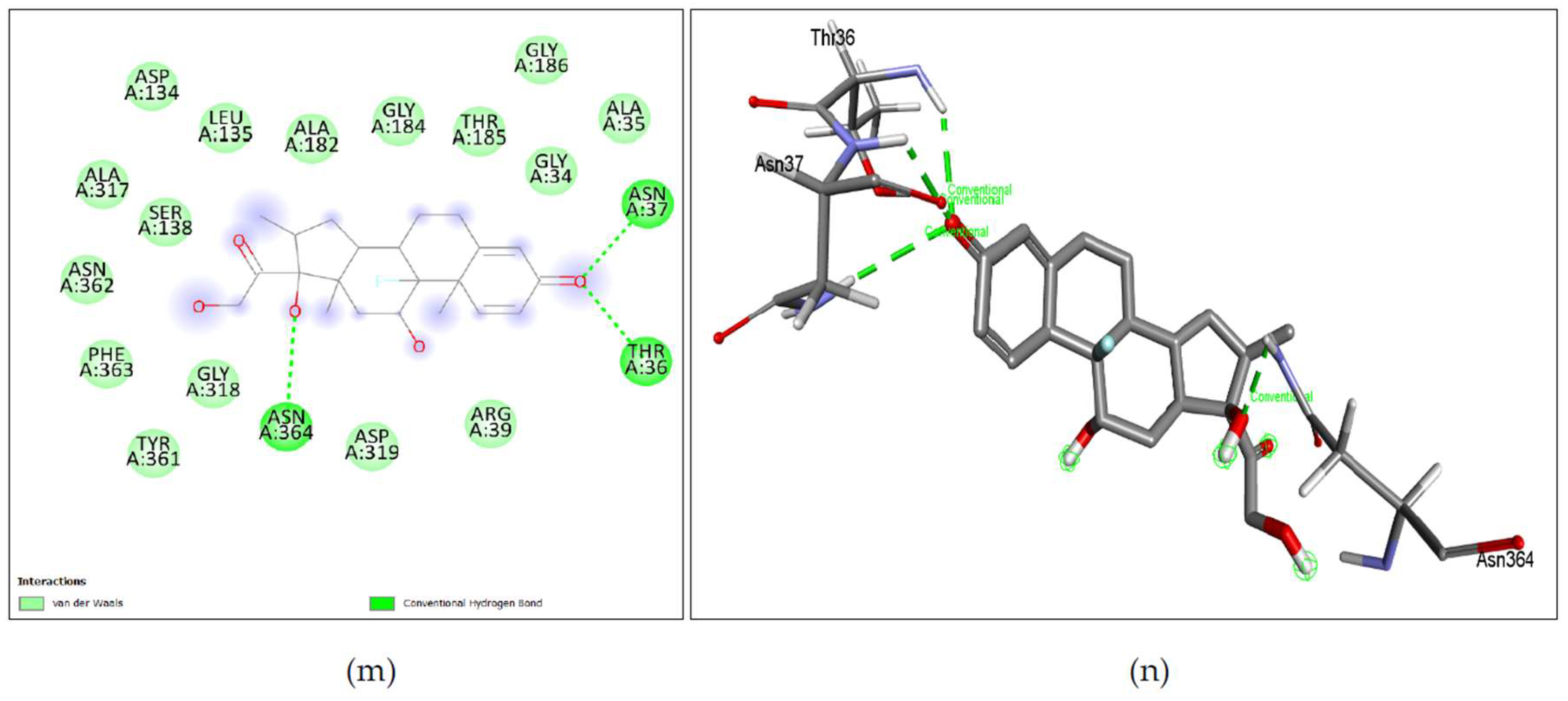
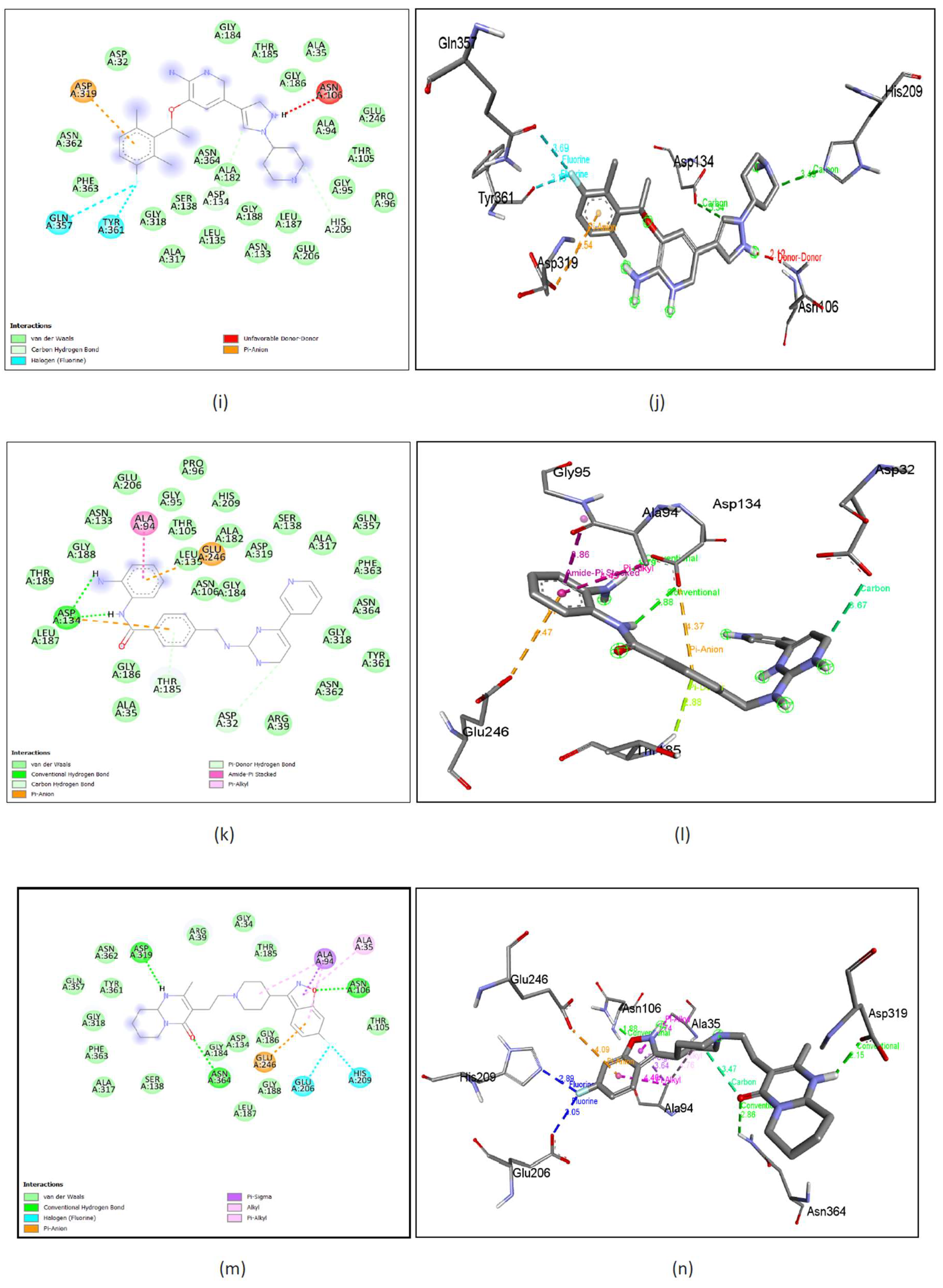
Figure 10. (a), (b) Simeprevir shows binding with nGLK around amino acids ASN37, GLY184, ASN362, TYR361, GLN357, PHE363, SER138, GLY318, ASN364, ALA182, ALA317, LEU135, ALA94, GLY186, LEU187, SER274, ASN320, GLY251, ALA273 and shows interactions with THR36, THR185, GLY188 via conventional hydrogen bond, ASP319, ASP134 via pi-anion bond. The interacting amino acids came under the glucokinase domain of the pathogen’s protein. Also, in
Figure 11. (a), (b) Simeprevir shows binding with hGLK around amino acids GLY410, GLU331, THR82, ALA282, GLY170, THR116, MET115, GLU112, ARG327, SER281, GLY328, GLU300, TYR297, GLY295, THR332, THR228, GLY227 and interacts with SER411 via a conventional hydrogen bond, SER280 via carbon hydrogen bond, ARG333, VAL227 via pi-alkyl interaction, LYS296 via alkyl and via pi-cation interaction. The interacting amino acids fall under the glucokinase domain of the human protein.
Supplementary Figure 7. (a) and
Supplementary Figure 9. (a) show amino acids of nGLK and hGLK within 5 A of Simeprevir.
Figure 10.
Lig-plots and 3D images showing interactions between nGLK and potential drugs. (a) Simeprevir interacts with target glucokinase through Van Der Waals interactions (light green), conventional hydrogen bonds (dark green), and pi-anion interactions (yellow). (b) Simeprevir interactions visualized in 3D. (c) Levomefolic acid interacts with target glucokinase through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), carbon hydrogen bond (light green), pi-donor hydrogen bond (light green), pi-alkyl interaction (light pink). (d) Levomefolic acid interactions visualized in 3D. (e) Indacaterol interacts with target glucokinase through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), carbon hydrogen bond (light green), pi-donor hydrogen bond (light green), pi-alkyl interaction (light pink), pi-anion interaction (yellow), alkyl interaction (light pink), amide-pi stacked interaction (dark pink), pi-sigma interaction (purple). (f) Indacaterol interactions visualized in 3D. (g) Ziprasidone interacts with target glucokinase through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), alkyl interaction (light pink), and pi-sigma interaction (purple). (h) Ziprasidone interactions visualized in 3D. (i) Crizotinib interacts with target glucokinase through Van Der Waals interactions (light green), carbon hydrogen bond (dark green), Halogen interactions (blue), unfavorable donor-donor (red), and pi-anion interaction (yellow). (j) Crizotinib interactions visualized in 3D. (k) Mocetinostat interacts with target glucokinase through Van Der Waals interactions (light green), carbon hydrogen bond (dark green), conventional hydrogen bond (dark green), pi-donor hydrogen bond (light green), pi-anion interaction (yellow), amide-pi stacked (dark pink), pi-alkyl interaction (light pink). (l) Mocetinostat interactions visualized in 3D. (m) Risperidone interacts with target glucokinase through Van Der Waals interactions (light green), carbon hydrogen bond (dark green), conventional hydrogen bond (dark green), pi-anion interaction (yellow), alkyl interaction (light pink), pi-alkyl interaction (dark pink), pi-sigma interaction (purple), halogen interactions (blue). (n) Mocetinostat interactions visualized in 3D.
Figure 10.
Lig-plots and 3D images showing interactions between nGLK and potential drugs. (a) Simeprevir interacts with target glucokinase through Van Der Waals interactions (light green), conventional hydrogen bonds (dark green), and pi-anion interactions (yellow). (b) Simeprevir interactions visualized in 3D. (c) Levomefolic acid interacts with target glucokinase through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), carbon hydrogen bond (light green), pi-donor hydrogen bond (light green), pi-alkyl interaction (light pink). (d) Levomefolic acid interactions visualized in 3D. (e) Indacaterol interacts with target glucokinase through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), carbon hydrogen bond (light green), pi-donor hydrogen bond (light green), pi-alkyl interaction (light pink), pi-anion interaction (yellow), alkyl interaction (light pink), amide-pi stacked interaction (dark pink), pi-sigma interaction (purple). (f) Indacaterol interactions visualized in 3D. (g) Ziprasidone interacts with target glucokinase through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), alkyl interaction (light pink), and pi-sigma interaction (purple). (h) Ziprasidone interactions visualized in 3D. (i) Crizotinib interacts with target glucokinase through Van Der Waals interactions (light green), carbon hydrogen bond (dark green), Halogen interactions (blue), unfavorable donor-donor (red), and pi-anion interaction (yellow). (j) Crizotinib interactions visualized in 3D. (k) Mocetinostat interacts with target glucokinase through Van Der Waals interactions (light green), carbon hydrogen bond (dark green), conventional hydrogen bond (dark green), pi-donor hydrogen bond (light green), pi-anion interaction (yellow), amide-pi stacked (dark pink), pi-alkyl interaction (light pink). (l) Mocetinostat interactions visualized in 3D. (m) Risperidone interacts with target glucokinase through Van Der Waals interactions (light green), carbon hydrogen bond (dark green), conventional hydrogen bond (dark green), pi-anion interaction (yellow), alkyl interaction (light pink), pi-alkyl interaction (dark pink), pi-sigma interaction (purple), halogen interactions (blue). (n) Mocetinostat interactions visualized in 3D.
Figure 11.
Lig-plots and 3D images showing interactions between hGLK and potential drugs. (a) Simeprevir interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), carbon hydrogen bond (light green) and pi-cation interactions (yellow), pi-alkyl interactions, alkyl interactions (light pink). (b) Simeprevir interactions visualized in 3D. (c) Levomefolic acid interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), pi-alkyl interactions (light pink) and amide-pi stacked interactions (dark pink). (d) Levomefolic acid interactions visualized in 3D. (e) Indacaterol interacts with hGLK through Van Der Waals interactions (light green pi-pi stacked interactions (dark pink), pi-alkyl interactions (light pink), and pi-pi-T shaped interactions (dark pink). (f) Indacaterol interactions visualized in 3D. (g) Ziprasidone interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), carbon hydrogen bond (light green) and pi-cation interactions (yellow), pi-sulfur interactions (yellow), pi-pi stacked (dark pink), pi-alkyl interactions (light pink), alkyl interactions (light pink). (h) Ziprasidone interactions visualized in 3D. (i) Crizotinib interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), pi-cation interaction (yellow) and unfavorable donor-donor interactions (red). (j) Crizotinib interactions visualized in 3D. (k) Mocetinostat interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), pi-pi stacked interactions (dark pink), and pi-alkyl interactions (light pink). (l) Mocetinostat interactions visualized in 3D. (m) Risperidone interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), carbon-hydrogen bond (light green), and halogen interactions (light and dark blue). (n) Risperidone interactions visualized in 3D.
Figure 11.
Lig-plots and 3D images showing interactions between hGLK and potential drugs. (a) Simeprevir interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), carbon hydrogen bond (light green) and pi-cation interactions (yellow), pi-alkyl interactions, alkyl interactions (light pink). (b) Simeprevir interactions visualized in 3D. (c) Levomefolic acid interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), pi-alkyl interactions (light pink) and amide-pi stacked interactions (dark pink). (d) Levomefolic acid interactions visualized in 3D. (e) Indacaterol interacts with hGLK through Van Der Waals interactions (light green pi-pi stacked interactions (dark pink), pi-alkyl interactions (light pink), and pi-pi-T shaped interactions (dark pink). (f) Indacaterol interactions visualized in 3D. (g) Ziprasidone interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), carbon hydrogen bond (light green) and pi-cation interactions (yellow), pi-sulfur interactions (yellow), pi-pi stacked (dark pink), pi-alkyl interactions (light pink), alkyl interactions (light pink). (h) Ziprasidone interactions visualized in 3D. (i) Crizotinib interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), pi-cation interaction (yellow) and unfavorable donor-donor interactions (red). (j) Crizotinib interactions visualized in 3D. (k) Mocetinostat interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), pi-pi stacked interactions (dark pink), and pi-alkyl interactions (light pink). (l) Mocetinostat interactions visualized in 3D. (m) Risperidone interacts with hGLK through Van Der Waals interactions (light green), conventional hydrogen bond (dark green), carbon-hydrogen bond (light green), and halogen interactions (light and dark blue). (n) Risperidone interactions visualized in 3D.
2.3.10. Repurposing of Human Metabolite/Supplement Levomefolic Acid against nGLK and Comparing Ligand Interactions against hGLK
Levomefolic acid, an active form of folate is a naturally found metabolite in
Homo sapiens. Though the SwissADME results indicate that the compound cannot cross the blood-brain barrier, research confirms that it can transfer across all membranes even the blood-brain-barrier[
45]. It plays a vital role in DNA synthesis and the cysteine cycle. It is being studied for use as a psychedelic drug[
45,
46]. It is a food additive and a supplement and is considered an excellent drug due to its ability to cross blood-brain-barrier. The possibility of side effects due to a naturally found metabolite seems unlikely. It showed a binding affinity of -10.4kcal/mol and -9kcal/mol with nGLK and hGLK, respectively. The possibility of toxicity against humans seems minimal as it has a half-life of 3 hours and is eliminated via the renal or fecal route (
https://www.caymanchem.com/msdss/21616m.pdf). Levomefolic acid is metabolized into tetrahydrofolate and then converted to polyglutamate form. It has been used to prevent the risk of neural tube defects in pregnant women as a folate supplement. As shown in
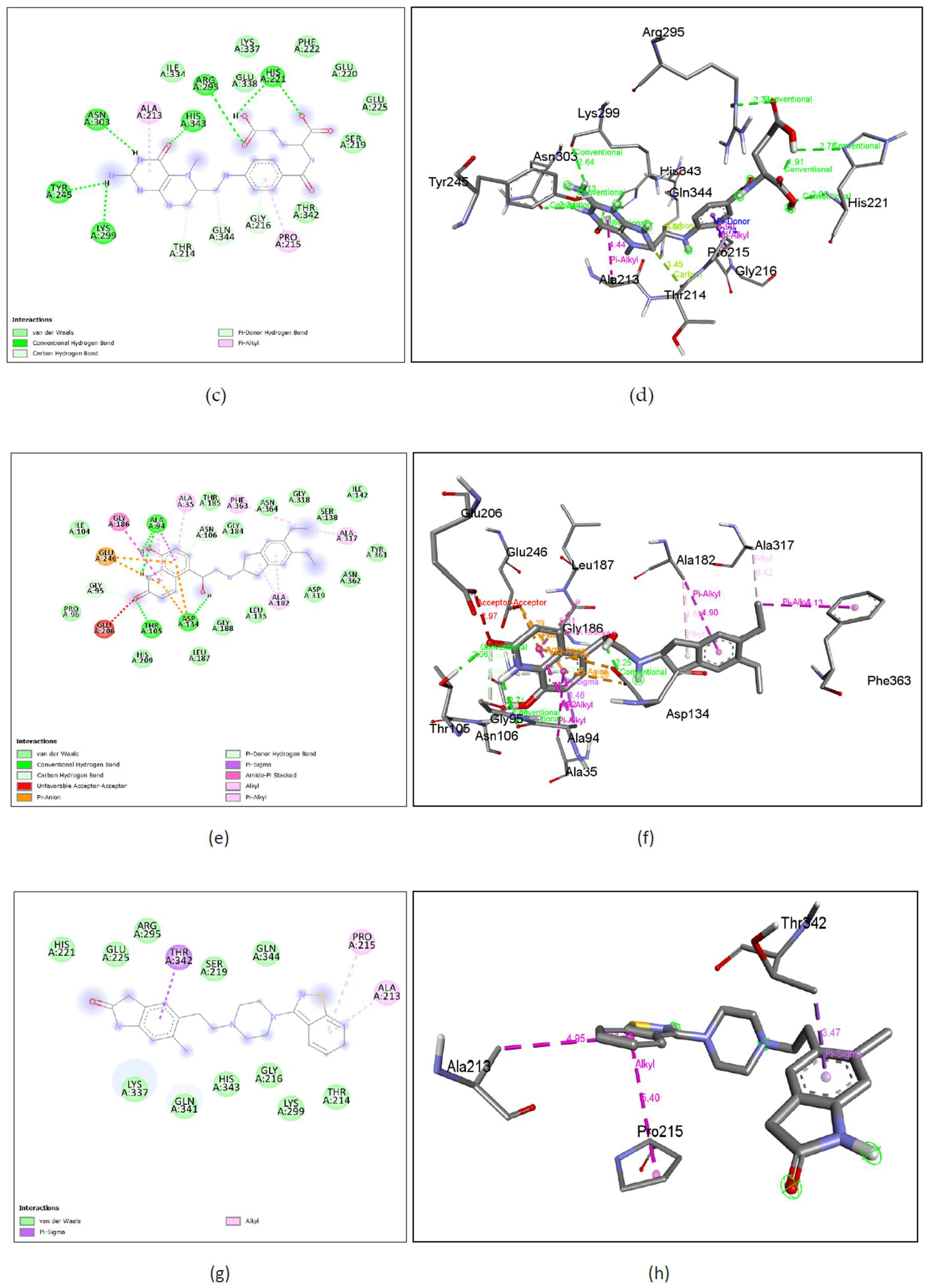
Figure 10. (c), (d) Levomefolic acid shows binding with nGLK around amino acids ILE334, LYS337, GLU338, PHE222, GLU220, GLU225, SER219, THR342 and shows interactions with ASN303, HIS343, TYR245, LYS299, ARG295, HIS221 via conventional hydrogen bond, THR214, GLN344, GLY216 via carbon hydrogen bond and PRO215, ALA213 via pi-alkyl bond. The interacting amino acids come under the glucokinase domain of the pathogen’s protein. Also,
Figure 11. (c), (d) Levomefolic acid shows binding with hGLK around amino acid THR149, ASP409, THR209, ILE225, GLY410, GLY295, VAL412, HIS416, SER411, THR228, GLY227, LYS169, GLY80, ASP205, ASP78, GLU443, GLY444 and interacts with SER445, SER151, ARG85, LEU415 via conventional hydrogen bond, THR332 via amide pi-stacked interactions, ARG333 via pi-alkyl interactions and SER336 via unfavorable interactions. The interacting amino acids come under the glucokinase domain of the human protein. As the only form of folate that can cross the blood-brain barrier, it acts as a cofactor in the production of monoamine neurotransmitters such as dopamine, serotonin, and norepinephrine.
Supplementary Figure 7. (b) and
Supplementary Figure 9. (b) show amino acids of nGLK and hGLK within 5 A of levomefolic acid.
2.3.11. Repurposing of Bronchodilator Indacaterol against hGLK and Comparing Ligand Interactions against nGLK
Indacaterol is a monohydroxyquinoline that is a potent inhibitor of β (2)-adrenoceptor agonist used for the treatment of asthma and pulmonary disease[
47]. It is a bronchodilator and is an approved drug by the European Medicines Agency and FDA. It showed no druglikeness violations, excellent bioavailability, and high gastrointestinal absorption. The binding affinity of indacaterol to the nGLK is -10.4kcal/mol and hGLK is -8.1kcal/mol. As shown in
Figure 10. (e), (f) Indacaterol shows binding with nGLK around amino acids ILE104, PRO96, HIS209, LEU187, GLY188, LEU135, ASP319, ASN362, TYR361, SER138, ILE142, GLY318, ASN364, GLY184, THR185 and interacts with ALA94, ASP134, THR105, via conventional hydrogen bond, ASN106 via Pi-donor hydrogen bond, GLY95 via a carbon hydrogen bond, ALA182, PHE363, ALA317, ALA94, ALA35 via pi-alkyl bond, ALA94 via pi-sigma bond, ALA182, ALA94 via alkyl interactions, GLU246, ASP134 via pi-anion bond, GLU206 via unfavorable acceptor-acceptor interactions. The interacting amino acids come under the glucokinase domain of the pathogen’s protein. Also,
Figure 11. (e), (f) Indacaterol shows binding with hGLK around amino acids GLY97, GLN98, LEU451, MET235, SER64, GLU221, ARG250, GLN219, HIS218, and interacts with TYR214 via pi-pi-T stacked interactions, PRO66, ILE211 via pi-alkyl interactions. The interacting amino acids come under the glucokinase domain of the human protein.
Supplementary Figure 7. (c) and
Supplementary Figure 9. (c) show amino acids of nGLK and hGLK within 5 A of indacaterol.
2.3.12. Repurposing of Antipsychotic Ziprasidone against hGLK and Comparing Ligand Interactions against nGLK
Ziprasidone, a piperazine derivative which is a second-generation antipsychotic drug used to treat schizophrenia and bipolar disorder[
48]. It functions as an antagonist to dopamine, histamine, muscarinic, and serotonin receptor, and is an inhibitor of synaptic reuptake of serotonin, and norepinephrine. It is used to treat schizophrenia and bipolar disorder[
49]. This compound has the best drug-likeness and pharmacokinetic properties to be considered a promising lead drug for inhibiting nGLK. The binding affinity of the ligand against nGLK is -10.2kcal/mol and hGLK is -8.1kcal/mol. The potential of ziprasidone to inhibit hGLK is yet to be reported. A previous study has shown that it has no effect on the blood glucose levels while an epidemiological study found no link between ziprasidone and increased risk of hyperglycemia unlike olanzapine and clozapine [
50]. It has a half-life of 6-7 hours and is heavily metabolized in the liver into 12 different metabolites by CYP3A4 and aldehyde oxidase enzymes. As shown in
Figure 10. (g), (h), Ziprasidone shows binding to nGLK around amino acids LYS337, GLN341, HIS343, GLY216, LYS299, THR214, GLN344, SER219, ARG295, GLU225, HIS221 and interacts with THR342 via pi-sigma bond, PRO215, ALA213 via alkyl interaction. The interacting amino acids come under the glucokinase domain of the pathogen’s protein. Also, in
Figure 11. (g), (h), Ziprasidone shows binding with hGLK around amino acids ARG397, ARG403, GLN219, SER64, PRO66, CYS220, MET210, GLU221 and ILE211 and interacts with SER398, GLU399 via conventional hydrogen bonds, HIS218 via carbon-hydrogen bond, TYR214, ILE404, MET235 via pi-alkyl and alkyl interactions, TYR214 via pi-pi stacked interaction, ARG250 via pi-cation interaction and MET402 via pi-sulfur interaction. The interacting amino acids come under the glucokinase domain of the human protein. Also, multiple antipsychotic drugs are known to influence glucose metabolism[
51]. In one study, it was found that olanzapine and clozapine bind strongly to muscarinic M
3 and inhibit insulin release[
52]. Ziprasidone is known to reverse olanzapine and clozapine-induced hyperglycemia[
52,
53]. Supplementary
Figure 7. (d) and
Figure 9. (d) show amino acids of nGLK and hGLK within 5 A of ziprasidone.
2.3.13. Repurposing of Anticancer Crizotinib against hGLK and Comparing Ligand Interactions against nGLK:
Crizotinib is a specific aminopyridine inhibitor of tyrosine kinase anaplastic lymphoma kinase receptor and c-Met/hepatocyte growth factor receptor. It binds to tyrosine kinase anaplastic lymphoma kinase by competing with ATP[
54]. Tyrosine kinase anaplastic lymphoma kinase plays a significant role in neuronal development. Crizotinib is known to be used in certain cases against non-small cell lung cancer (PubChem). It is an oral drug and has side effects[
55]. The binding affinity of Crizotinib to nGLK is -9.8kcal/mol and hGLK is -7.8kcal/mol. The drug has no drug-likeness violations but must be considered before use as a drug against N.
fowleri due to its inability to cross-blood the brain barrier. Though the binding to hGLK is moderately strong, yet, it is unclear whether the side effects are due to the inhibition of hGLK. As shown in
Figure 10. (i), (j), Crizotinib shows binding with nGLK around amino acids ASP32, GLY184, THR185, GLY186, ALA35, ALA94, GLU246, THR105, GLY95, PRO96, GLU206, LEU187, GLY188, ASN133, LEU135, ALA182, ASN364, SER138, GLY318, PHE363, ASN362 and interacts with amino acids ASP319 via pi-anion bond, HIS209, ASP134 via carbon hydrogen bond, GLN357, TYR361 via halogen interactions and ASN106 via unfavorable interactions. Also, in
Figure 11. (i), (j), Crizotinib shows binding with hGLK around amino acids GLY80, GLY227, GLY410, THR149, SER445, ASP409, GLY444, GLU442, THR228, LYS414, LEU415, GLY81, THR82, ASN83, SER411 and interacts with ASP78, ASP205 via conventional hydrogen bond, ARG85 via pi-cation interaction, SER151, GLU443 via unfavorable interaction. Crizotinib has been shown to moderately decrease blood glucose levels in rats[
56].
Supplementary Figure 7. (e) and
Supplementary Figure 9. (e) show amino acids of nGLK and hGLK within 5 A of crizotinib.
2.3.14. Repurposing of Anticancer Mocetinostat against hGLK and Comparing Ligand Interactions against nGLK:
Mocetinostat is a. novel, oral drug used to treat tumor cells, causing apoptosis of cells[
57,
58]. This benzanilide derivative binds to Histone deacetylase 1, 2, 3 and inhibits its activity. It leads to the upregulation of tumor-suppressive genes, and cell cycle arrest inhibition of DNA repairs. Though the exact mechanism is not defined, it is being studied to be used in trials for the treatment of urothelial carcinoma, lymphoma, and metastatic leiomyosarcoma[
59]. The binding of mocetinostat is -10kcal/mol and -8.8kcal/mol for nGLK and hGLK, respectively. As shown in
Figure 10. (k), (l), Mocetinostat interacts with THR189, GLY188, ASN133, GLU206, GLY95, PRO96, THR105, LEU135, ALA182, HIS209, GLY184, ASP319, ALA317, GLN357, PHE363, ASN364, GLY318, ASN362, ARG39, ALA35, GLY186, LEU187, SER138 and interact with ASP134 via conventional hydrogen bond, GLU246, ASP134 via pi-anion bond, ALA94 via amide-pi stacked, THR185, ASP32 via pi-donor hydrogen bond. Also, in
Figure 11. (k), (l), Mocetinostat shows binding with hGLK around amino acids VAL452, VAL455, ILE211, GLU221, GLN219, SER64, MET235, ARG250, GLY249, GLN98, SER69 and interacts with HIS218 via conventional hydrogen bond, TYR214 via pi-pi stacked interaction and PRO66, LEU415 via pi-alkyl interaction.
Supplementary Figure 7. (f) and
Supplementary Figure 9. (f) show amino acids of nGLK and hGLK within 5 A of mocetinostat.
2.3.15. Repurposing of Antipsychotic Risperidone against nGLK and Comparing Ligand Interactions against hGLK
Risperidone is a second-generation antipsychotic drug used in the treatment of hallucinations, delusions, and other psychotic effects associated with schizophrenia and bipolar disorder. It is derived from benzisoxazole and is a pyridopyrimidine. It has an inhibitory action on both dopaminergic D2 receptors and serotonergic 5-HT2A receptors but has a 10-fold higher activity against the latter as compared to D2 receptors in the central nervous system. It is mainly metabolized in the liver with a half-life of 3 hours and is cleared via the kidneys. This drug has also been tested for the treatment of irritability in autistic children[
60]. The binding of Risperidone is -10kcal/mol and -8.9kcal/mol for nGLK and hGLK, respectively. As shown in
Figure 10. (m), (n), Risperidone shows binding with nGLK around amino acids ALA317, PHE363, GLY318, TYR361, GLN357, ASN362, ARG39, GLY34, THR185, THR105, GLY188, LEU187, ASP134, GLY184, GLY186 and interacts with ASP319, ASN364, ASN106 via conventional hydrogen bond, ALA94, ALA35 via pi-alkyl interactions, ALA94 via alkyl and pi-sigma interactions, GLU246 via pi-anion interactions, GLU206, HIS209 via halogen interactions. Also, in
Figure 11. (m), (n), Risperidone shows binding with hGLK around amino acids ASP78, ARG85, ASP205, SER151, GLY81, GLY80, GLY295, LYS296, GLU331, ARG333, SER411, GLY410, ASP409, SER445 and interacts with THR228, THR332 via conventional hydrogen bond, GLY227 via carbon hydrogen bond and GLY328 via halogen interactions. Research showed that mice that were treated with risperidone had elevated fasting blood glucose levels and lower serum insulin, indicating that these drugs do influence glucose metabolism through various mechanisms [
61]. Thus, our computational research data is in line with previously published research and adds a new perspective to it by showing that it binds strongly to the glucokinase of both nGLK and hGLK.
Supplementary Figure 7. (g) and
Supplementary Figure 9. (g) show amino acids of nGLK and hGLK within 5 A of Risperidone.
Though the binding of these drugs is strong, a standard is required to ensure that the numbers are relevant. Another set of docking was done to identify the binding affinity of known inhibitors of hGLK to be used as a standard for comparison. Three known inhibitors of the hGLK enzyme, namely 2-Deoxy-D-glucose, lonidamine, and 3-bromopyruvate were docked against the target. The binding affinities were -5.3kcal/mol, -8.3kcal/mol and -4.5kcal/mol, respectively (data not shown). This indicates that even well-known enzyme inhibitors may have a binding affinity ranging from as low as -4.5kcal/mol.
The binding affinities of these drugs indicate that there is a strong interaction between them and the targets. Several research articles have also attributed some of these drugs to influence glucose metabolism through various mechanisms. According to our results, one mechanism by which these drugs influence our metabolism could be by directly inhibiting the glucokinase enzyme.