1. Introduction
A large majority of proteins from superfamilies of trypsin-like serine proteases and alpha/beta-hydrolases are enzymes that function through the use of a catalytic triad [1, 2]. According to the MEROPS database, enzymes of these two superfamilies belong to clan PA (mixed C/S/T catalytic type) serine peptidases and clan SC serine peptidases, respectively [
3]. Despite difference in protein class and fold (SCOP database [
4]), proteins of both superfamilies show structural similarity in the arrangement of their active sites [5, 6]. For example, both have an additional “fourth” conserved residue interacting with the catalytic triads and bound to the respective catalytic bases by weak hydrogen bonds. Derewenda (2023) recently wrote a comprehensive review on the role of weak hydrogen bonds in structures of proteins and nucleic acids [
7]. Independently, it was also reported that subtilisin-like superfamily proteins [8, 9], which belong to clan SB serine peptidases [
3], also have a similar 3D arrangement of four catalytic residues mentioned above [5, 6], which we will refer to as the “catalytic tetrad” of serine proteases when specifically speaking about the three superfamilies above. As opposed to trypsin-like serine proteases, the alpha/beta-hydrolases have five key residues (the catalytic triad plus two) to carry out protein function, which therefore we call a “catalytic pentad” [
10].
Earlier, we had described the structural catalytic core (SCC) in trypsin-like serine proteases and in alpha/beta-hydrolases [11, 12]. The core description method was based on the assumption that because there are similar arrangements of key amino acids, such as the catalytic triads – which can be found in the active sites of unrelated proteins where these key amino acids (acid, base and nucleophile in a catalytic triad) are positioned in specific places with respect to each other – then it should not be surprising to find other supporting/interacting amino acids that could also be placed in the equivalent positions in space (structurally conserved), simply because they are interacting with the same groups in a similar way and which altogether create similar local structural environments that we call “Structural Catalytic Cores” (SCCs) in the functionally unrelated proteins.
Furthermore, these overall similar structural environments, SCCs, in the active sites were divided into “bricks”, i.e. smaller structural units of several amino acids size, which usually contain one or several key amino acids, their supporting amino acids, and importantly, they are intra-locked by bonds, and thus can be considered as pseudo-independent, closed, small structural units that can be repeatedly found in the active sites of different proteins. These small conserved and closed structural brick units, which usually reflect environments around one or several key functional groups, we call “zones”, giving rise to names like the “catalytic acid zone” or the “acid-base zone”, and so on. In order for a structural arrangement to be a zone, it should be: (1) a small unit of several amino acid size; (2) contain a functional element; (3) be considered as a structurally independent, closed or sometimes circular substructure, i.e. it is intra-locked by bonds and hydrophobic interactions; and (4) it could be found in several different proteins. Each zone incorporates a segment of SCC and governs its respective element of protein functional machinery through a network of conserved hydrogen bonds and other interactions.
Here, we aim to describe the SCC in subtilisin-like superfamily proteins and compare it to SCCs of trypsin-like serine proteases and alpha/beta-hydrolases, who’s SCCs were found to be different from the subtilisin-like superfamily proteins despite the similarity in the 3D arrangement of their catalytic triads.
2. Results and Discussion
As described in the Introduction, various clans of serine proteases and alpha/beta-hydrolases have sets of four or five key residues called catalytic tetrads or catalytic pentads to carry out their function. These sets of key residues are usually incorporated into their respective zones, which taken together constitute the Structural Catalytic Core (SCC). The zones can be used as a convenient tool to compare active sites of different “catalytic triad” enzymes with other proteins of the same fold, but different function. Here, we start with the previously observed fact that subtilisin-like proteins have a catalytic tetrad as seen in trypsin-like serine proteases and alpha/beta-hydrolases, and proceed with identification of the subtilisin SCC, making an inventory of key catalytic residues and the other elements of the SCC, and further comparing the respective SCCs in subtilisin-like proteins, trypsin-like serine proteases and alpha/beta-hydrolases to determine how similar or different catalytic cores are in these three superfamilies of enzymes.
2.1. Creating a Dataset of the Subtilisin-Like Superfamily Proteins
In the SCOP database, the subtilisin-like superfamily incorporates two families of subtilases and serine-carboxyl proteinases (SCP) with 3D structures of 42 and 3 different proteins, respectively [
4]. For each of the 45 proteins from SCOP, one representative Protein Data Bank (PDB [
13]) ID 3D structure with the highest resolution has been chosen (
Table S1). Additionally, outside of the SCOP database, eight different structures, seven subtilases and one SCP, were identified to be included within the subtilisin-like superfamily, thus totaling 53 representative PDB ID entries (
Table S1).
2.2. SCC in Subtilisin Savinase (Representative Structure of the Subtilases Family; Subtilisin-Like Superfamily)
2.2.1. Five Key Functional Amino Acids in Subtilisin Savinase
Based on the criteria above, the structure of the subtilisin savinase (PDB ID: 1GCI; R = 0.78 Å) [
14] can be accepted as the representative structure of the overall subtilisin-like superfamily. Unlike trypsin-like serine proteases but similar to alpha/beta-hydrolases, subtilisin savinase displays a catalytic pentad of key functional residues. Three of the five residues, Asp32 (Acid), His64 (Base) and Ser221 (Nucleophile (Nuc)), are the actual catalytic triad (
Table 1 and
Table S1). The fourth residue, Ser125, forms a weak CH-O hydrogen bond between its carbonyl oxygen and the side-chain group of the catalytic base. We will refer to this amino acid as the “CHO”. And finally, the last amino acid of the catalytic pentad is Asn155 (Oxy). The backbone amide of Ser221 forms the oxyanion hole, which does not require introduction [
15].
2.2.2. AcidBaseCHO Zone
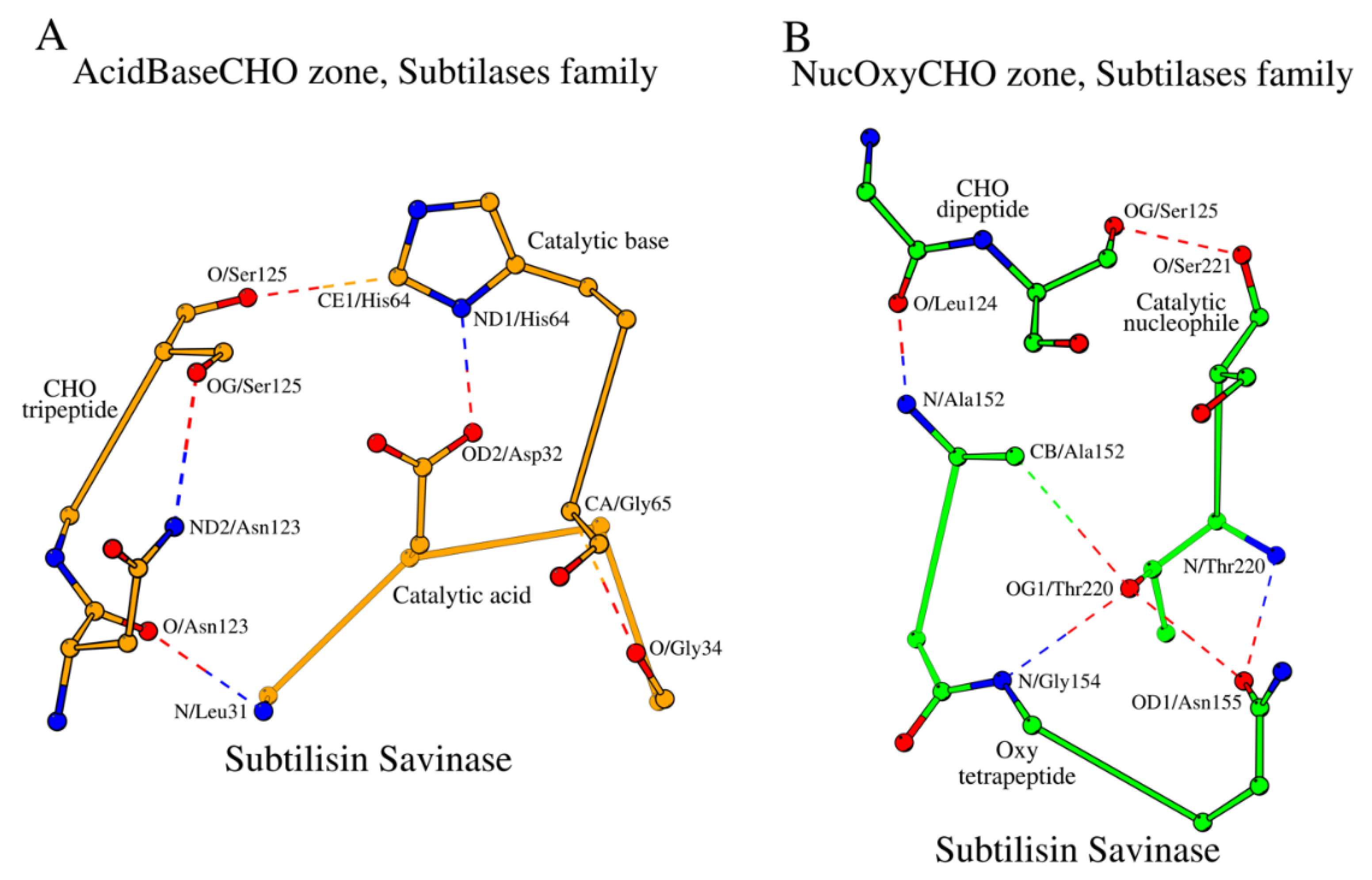
Let us consider local sub-structures, which govern the five key amino acids described above. Three continuous fragments of the subtilisin savinase – (1) the tetrapeptide Leu31-Gly34, which includes the catalytic acid Asp32, (2) the dipeptide His64-Gly65, which includes the catalytic base His64, and (3) the tripeptide Asn123-Ser125, which includes the CHO residue Ser125 – form a closed structural formation locked by hydrogen bonds, which we will refer to as the “AcidBaseCHO zone” (
Figure 1A;
Table 2). For convenience purposes, we will refer to amino acids of the zone as, for example, the first amino acid from the segment containing the catalytic acid is referenced as “Acid1”, and so on. In addition to the contacts that lock the AcidBaseCHO zone,
Figure 1A and
Table 2 also show contacts between catalytic residues, as well as the other internal stabilizing contacts of the segments of the zone.
2.2.3. NucOxyCHO Zone
Two of the five key functional amino acids of subtilisin, the catalytic nucleophile (Nuc; Ser221 in 1GCI) and the oxyanion hole (Oxy; Asn155 in 1GCI), fall out of the AcidBaseCHO zone. They form their own closed substructure, the NucOxyCHO zone (
Figure 1B;
Table 3). The NucOxyCHO zone is formed through interlocking of the ends of the Nuc dipeptide (Thr220 – Ser221), the Oxy tetrapeptide (Ala152 - Asn155) and the Leu124 - Ser125 segment of the CHO tripeptide (
Figure 1B). The OG1 atom of Thr220 seems to be the center of coordination of the “NucOxy” sub-zone due to its contacts with atom CB/Ala152, N/Gly154 and OD1/Asn155 (
Figure 1B;
Table 3).
2.2.4. SCC as a Structural Association of AcidBaseCHO and NucOxyCHO Zones
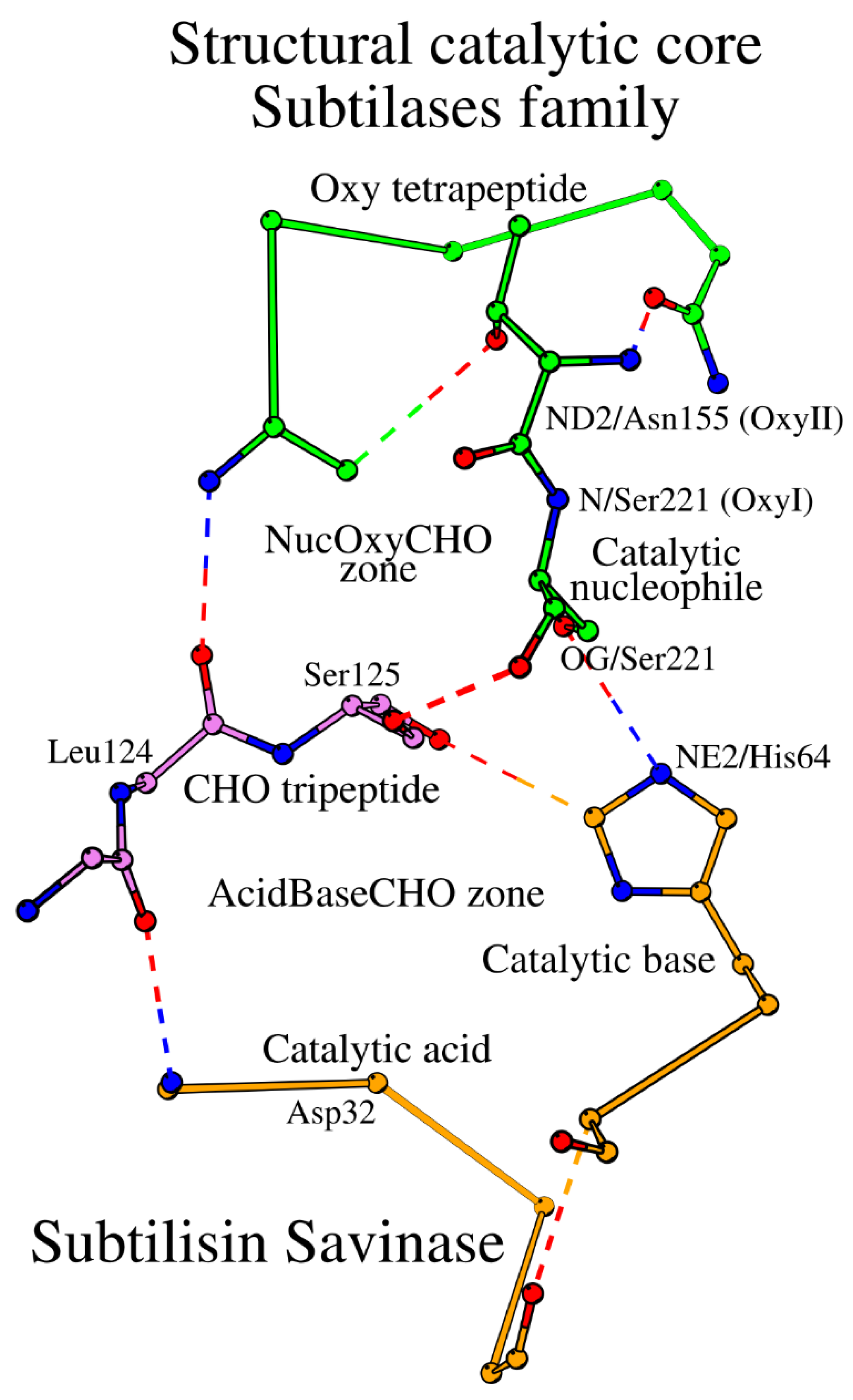
As it is demonstrated above, the catalytic acid / catalytic base and the catalytic nucleophile / oxyanion hole environments are two different localized environments, referred to as the AcidBaseCHO and the NucOxyCHO zones, respectively. The two zones are linked by their common element, the Leu124-Ser125 dipeptide of the CHO tripeptide (
Figure 2). Taken together, the two zones constitute the SCC of the subtilisin savinase, which includes 15 amino acids from 5 different peptides (
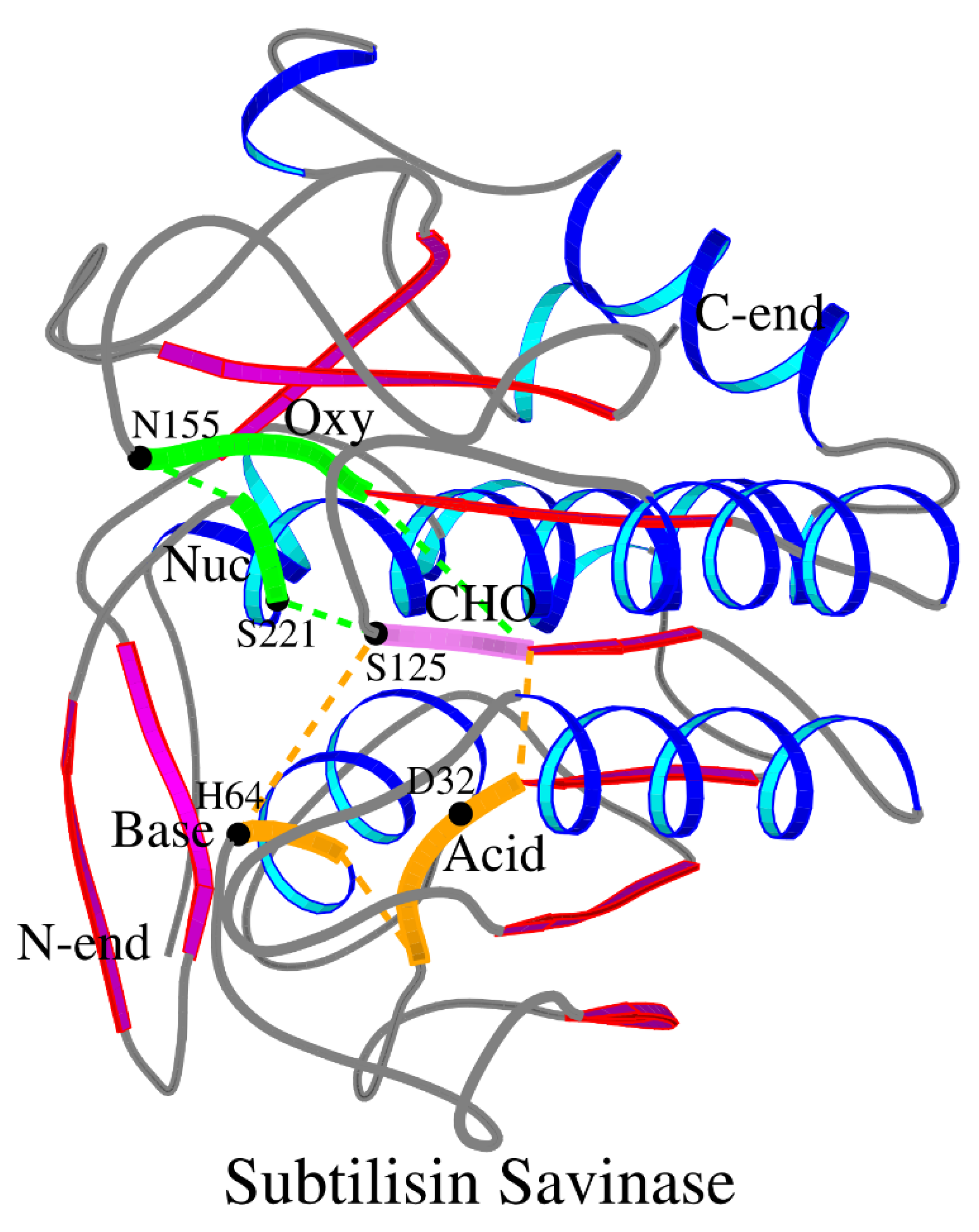
Table 1). The location of the SCC within the 3D structure of savinase is shown in
Figure 3.
2.3. SCC of the Other Subtilases (Subtilisin-Like Superfamily). Variations in the CHO Peptide
After examining the subtilisin savinase, the remaining representative structures from the subtilase family of subtilisin-like proteins were similarly analyzed for the SCCs formed from fifteen residues in five peptides and incorporating five key functional amino acids. The results are summarized in
Table S1 in the form of a structural alignment. All structural superpositions were done using the Dali server [
21]. As shown above, the CHO peptide belongs to both the AcidBaseCHO and NucOxyCHO zones and joins them together into the SCC. All subtilases have one of three types of the CHO peptide: (1) where CHO1=Asn (the Asn group; 33 structures in
Table S1); (2) where CHO1=Ser/Thr (the Ser/Thr group; 14 structures in
Table S1); and (3) two exceptions in the proprotein convertase subtilisin/kexin type 9 and thiazoline oxidase/subtilisin-like protease, where CHO1 is not Asn, Ser or Thr, and the polar CHO1-CHO3 contact is missing (the Xaa group in
Table S1). In proteins of the Asn group and the Ser/Thr group CHO3=Ser. The change from Asn to Ser at the CHO1 position in group (2) results in inclusion of the water molecule, HOHI, as an intermediate link between CHO1 and CHO3 (
Table 2 and
Table S1).
Due to the important role of the CHO peptide in the formation of the native functional contact between the catalytic nucleophile and the catalytic base, it can be speculated that there should be three structurally different active sites in the subtilase family.
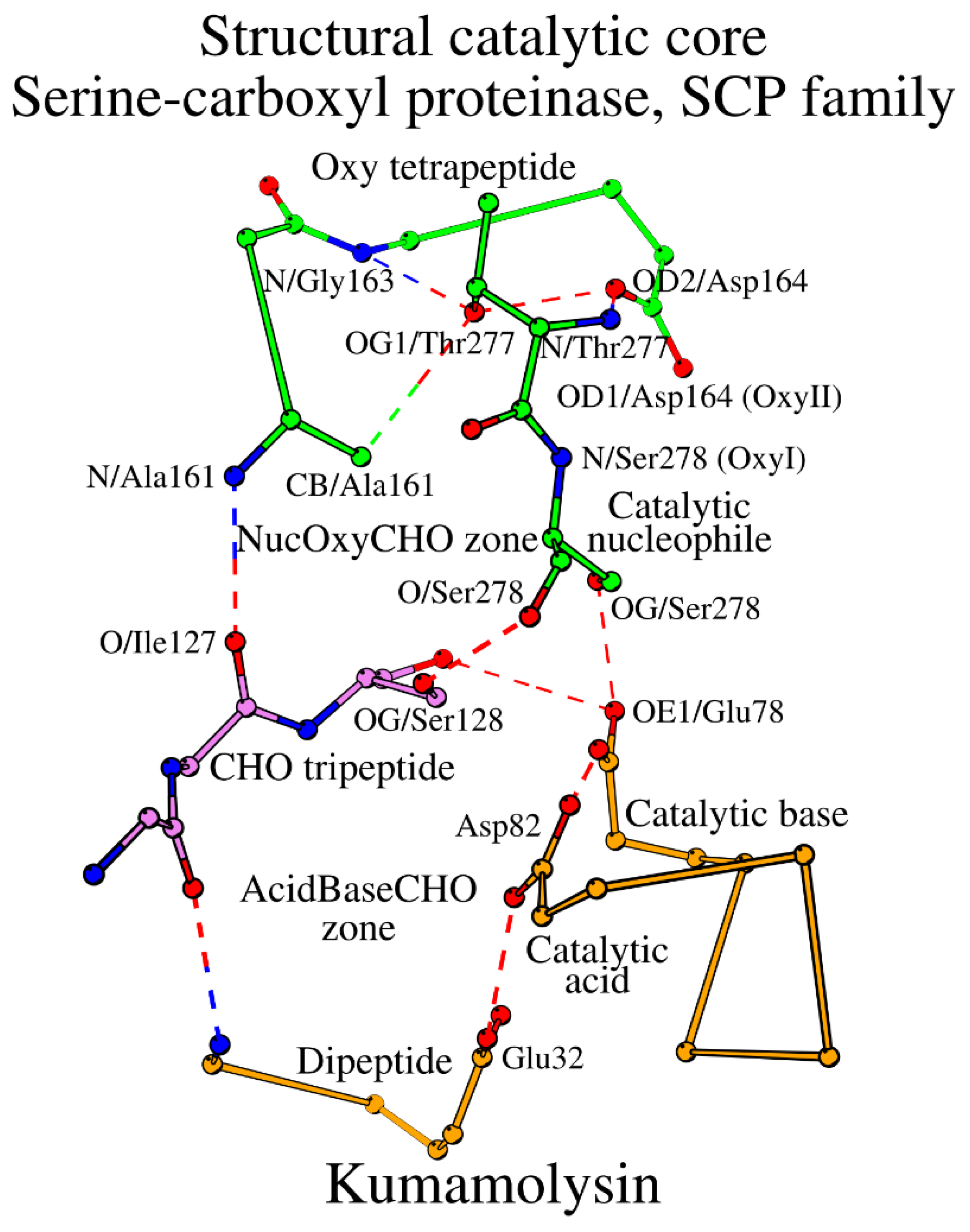
2.4. SCC in Serine-Carboxyl Proteinases (SCP Family; Subtilisin-Like Superfamily)
The SCP family of subtilisin-like proteins includes only a few known representative structures (
Table S1). Similar to the subtilases, the SCP enzymes can be divided into Asn and Ser/Thr groups according to the structure of the CHO peptide. Kumamolysin, a member of the Ser/Thr group, can be selected as the representative structure of the SCP family (
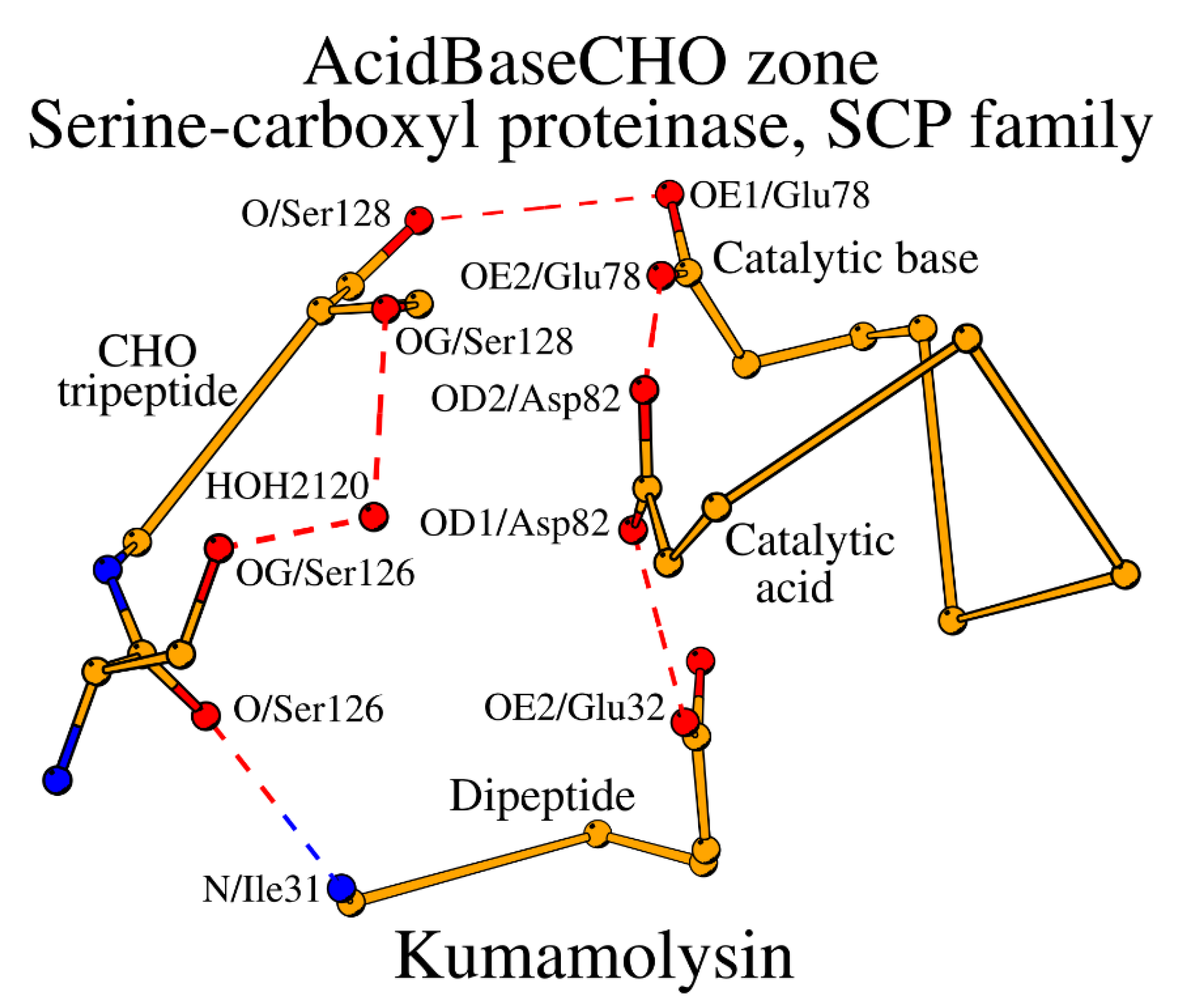
Table 1). Both subtilases and the SCP enzymes have five similarly placed key catalytic residues: the catalytic triad plus CHO plus Oxy. However, the main difference between subtilases and the SCP family is in the construction of their SCCs. The key catalytic acid in SCP occupies the typical location in the structure, but in sequence it moves to the C-terminal end of the peptide, which contains the catalytic base, i.e. changing from the peptide “Acid” location in subtilases to the peptide “Base” location (
Table 1;
Figure 4). Additionally, the catalytic base in SCP is always Glu instead of His. As the result, the “Acid” tetrapeptide turns into a dipeptide in SCP, and the “Base” dipeptide becomes chimeric “BaseAcid” pentapeptide, which for transparency reasons we will label “Base” in
Table 1, having the following structure: “catalytic base (BaseAcid1) – X2 – X3 – X4 – catalytic acid (BaseAcid5)”. Incorporation of both the catalytic base and acid in the same peptide while maintaining their functional relation is carried out by the α-helical conformation of the BaseAcid peptide. In a canonical α-helix, the first residue (BaseAcid1; catalytic base) and the fifth residue (BaseAcid5; catalytic acid) are usually connected by a canonical helix-forming hydrogen bond; at the same time they serve the enzymatic function as part of the catalytic triad. Thus, taken together, the subtilisin-like proteins can be divided into two major groups, where (1) the catalytic acid occupies its own structural segment within the SCC (“Acid” in
Table 1) or (2) the catalytic acid is placed within the same structural segment as the catalytic base (“Base” in
Table 1). A similar division had been also observed in alpha/beta-hydrolases [
22].
Due to the structural rearrangement, the number of amino acids forming SCC in the SCP family is 16. Additional changes include the Oxy4 residue transition Asn → Asp, and the presence of the water-mediator at the CHO1-CHO3 contact (
Figure 4,
Table 2). The entire SCC of the SCP family of subtilisin-like proteins is shown in
Figure 5 and described in
Table 3.
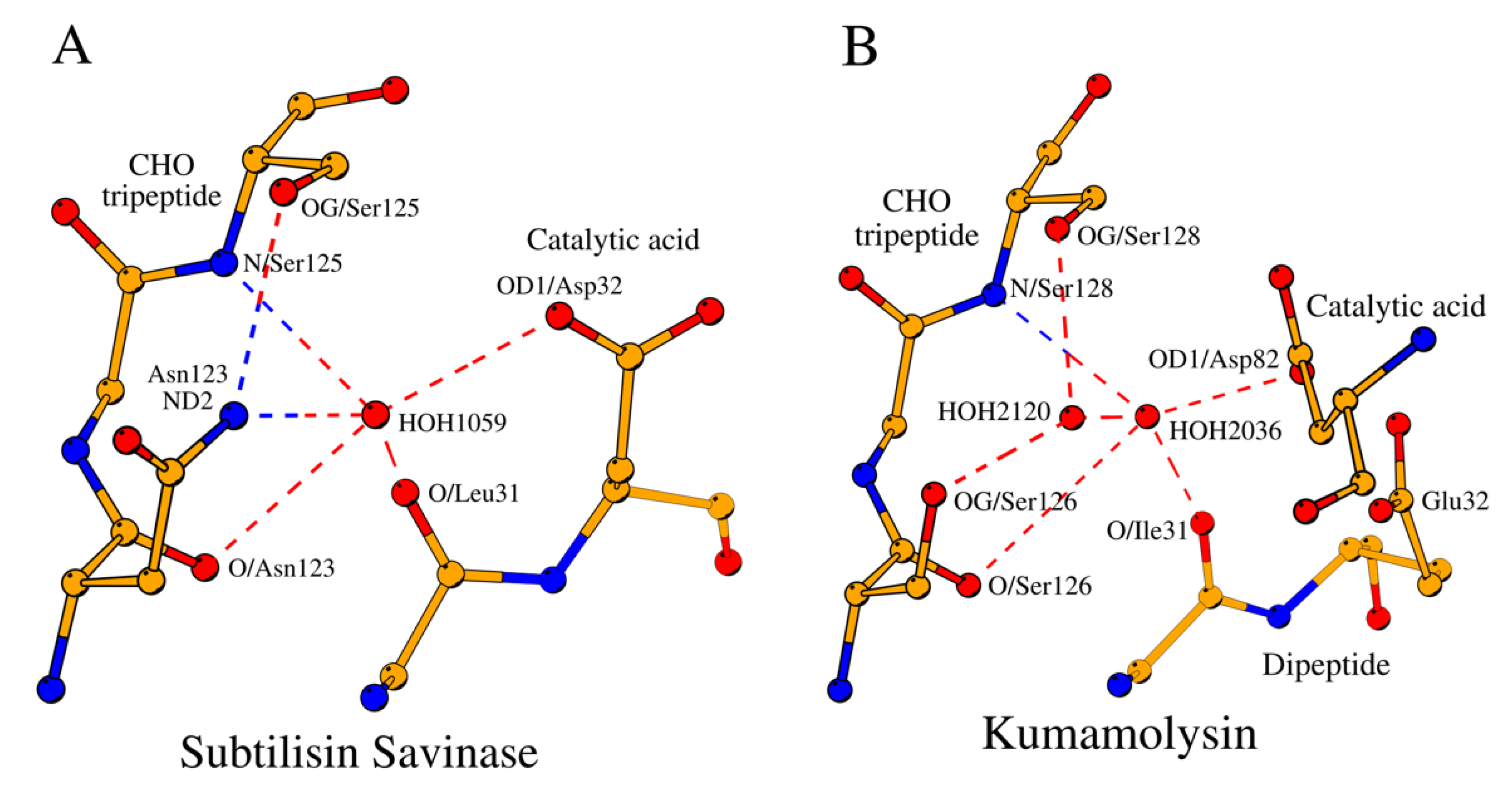
2.5. Invariant Water Molecule of the AcidBaseCHO Zone in the Subtilisin-Like Superfamily Proteases
As it is shown in
Section 2.2.3, in subtilases the OG1 atom of Thr220 seem to be the center of coordination of the NucOxyCHO zone due to its contacts with CB/Ala152, N/Gly154 and OD1/Asn155 (
Figure 1B). A somewhat similar coordinating role in the AcidBaseCHO zones in both the subtilases and SCP is played by the conserved structural water molecules (
Figure 6A and
Figure 6B;
Table 1,
Table 4 and
Table S1).
In subtilases, a water molecule (HOHII in
Table 4) coordinates the location of the catalytic acid with respect to the functionally important CHO3 residue (
Figure 6A). Of the 53 analyzed structures, HOHII water was not identified only in two cases: in the intracellular serine protease (PDB ID: 7Y6M;
Table S1) likely due to low resolution of the structure, and in the proprotein convertase subtilisin/kexin type 9 (PDB ID: 6U26;
Table 4 and
Table S1) due to proline at the CHO3 position.
Above we described that based on the composition of the CHO peptide the subtilisin-like enzymes belong to either the Asn group and or the Ser/Thr group. The two groups differ by the presence or absence of the second coordinating water molecule, HOHI, in the structure of the AcidBaseCHO zone that separates the two groups (
Table S1).
2.6. Comparison of Subtilisin-Like Enzymes and Alpha/Beta-Hydrolases. Catalytic Pentads
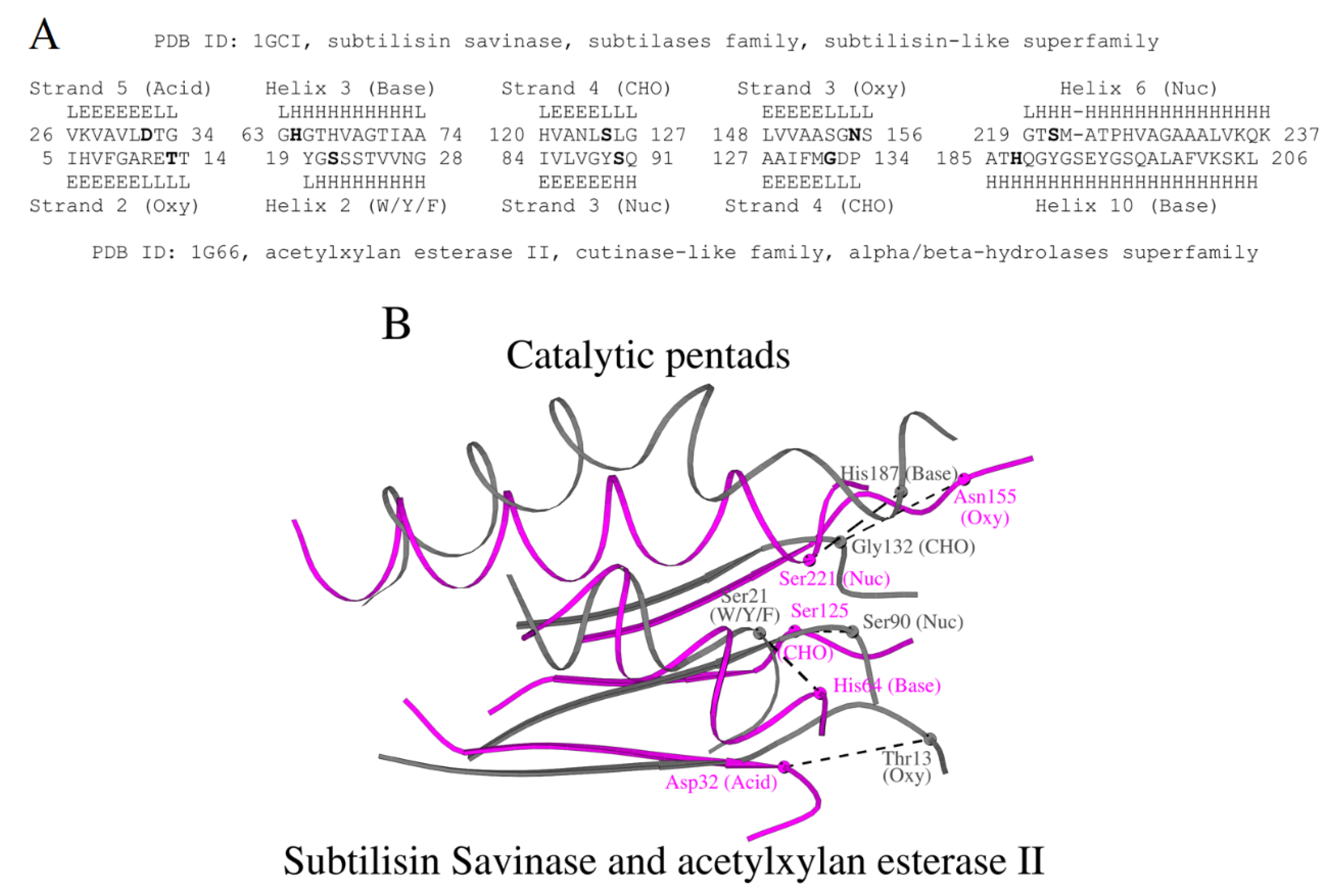
Both subtilisin-like superfamily enzymes and alpha/beta-hydrolases share a central parallel β-sheet of 7 strands [
4], and the pairwise superposition of the subtilisin savinase (PDB ID: 1GCI) and acetylxylan esterase II (PDB ID 1G66, [
23]) using the Dali server [
21] shows a z-score of 2.9 and RMSD of 3.7 Å over 108 residues. The resulting structural alignment between subtilisin and acetylxylan esterase II, aligned segments in
Figure 7A, includes five functionally important residues together with the adjacent secondary structure fragments: two α-helixes and three β-strands. From the alignment it is seen that few identical amino acids are aligned; nevertheless, the five catalytic residues are located at the same termini of the same fragments of the secondary structure, albeit not in identical positions, but forming functional pentads at the same locations of the overall fold (
Figure 7B). Note that in
Figure 7B, the catalytic acid-containing fragment of acetylxylan esterase II is shown as W/Y/F as described earlier in [
24].
3. Materials and Methods
The SCOP classification database [
4] and Protein Data Bank (PDB,
http://www.rcsb.org/; August 2024 [
13]) were used to retrieve 53 representative structures of proteins from the subtilisin-like superfamily (SCOP ID: 3000226). Detailed structural information from the above set of PDB files is given in the
Section 2.1.
Structure visualization and structural analysis of interactions between amino acids in proteins (hydrogen bonds, hydrophobic, other types of weak interactions) was carried out using Maestro (Schrödinger Release 2023-1: Schrödinger, LLC, New York, NY, 2021;
http://www.schrodinger.com/ release August 2024) and the software to determine ligand-protein contacts (LPCs) and between structural units (CSUs) [
25].
Pairwise structural superpositions were done using the Dali server (
http://ekhidna2.biocenter.helsinki.fi/dali/; August 2024) [
21]. Weak hydrogen bonds were identified based on geometrical criteria [
7]. The π-π stacking interactions and other interactions were analyzed using the Residue Interaction Network Generator (RING,
https://ring.biocomputingup.it/; August 2024) [
20]. Figures were drawn with MOLSCRIPT [
26].
4. Conclusions
Here, we have described a structural scaffold incorporating catalytic residues – the Structural Catalytic Core (SCC) – in the subtilisin-like superfamily of enzymes. We showed that the SCC is roughly divided into two halves, which are two structurally conserved and locally interconnected structural organizations or zones, the AcidBaseCHO and the NucOxyCHO zone. The AcidBaseCHO zone governs positioning of the catalytic acid and base, and the NucOxyCHO zone governs positioning of the catalytic nucleophile and the oxyanion hole. The two zones are connected by the CHO peptide, which can only be of two types, dividing subtilisin-like enzymes into two groups, the Asn group and the Ser/Thr group. The AcidBaseCHO zone incorporates structurally conserved water molecules for coordination of the catalytic acid, which are the key elements separating the Asn (one water) and Ser/Thr (two water molecules) groups.
Although the two known families within the subtilisin-like superfamily, the family of subtilases and the family of serine-carboxyl proteinases (SCP), have a similar arrangement of the catalytic residues and the catalytic cores, they differ in the positioning of the catalytic acid along the sequence. In SCP, the catalytic acid does not reside in its own segment of the SCC as seen in subtilases, but “moves” to the same segment of SCC containing the catalytic base.
The comparisons reveal that the subtilisin-like proteins mimic the alpha/beta-hydrolases, but not the trypsin-like serine proteases. Similarly to the alpha/beta-hydrolases, subtilisin-like proteins have five key functional amino acids, which are positioned at the same general location of the supersecondary structure; most importantly, in the alpha/beta-hydrolases, we see the same division into Asn and Ser/Thr groups together with the “walk” of the catalytic acid to the structural segment of the catalytic base [
22].
In short, the structural catalytic core (SCC) of subtilisin-like proteins consists of five key catalytic residues in five respective structural segments, and can be separated into two closed zones, AcidBaseCHO and NucOxyCHO, which are interlocked by a network of hydrogen bonds and non-polar interactions. The AcidBaseCHO zone governs positioning of the catalytic acid and base, and the NucOxyCHO zone governs positioning of the catalytic nucleophile and the oxyanion hole. The two zones are connected by the CHO peptide, which can only be of two types, dividing subtilisin-like enzymes into two groups, the Asn group and the Ser/Thr group. The two families of the subtilisin-like superfamily, subtilases and serine-carboxyl proteinases (SCP), have a similar arrangement of the catalytic residues and catalytic cores, but they differ in the positioning of the catalytic acid. Subtilisin-like proteins mimic the alpha/beta-hydrolases but not the trypsin-like serine proteases.
Supplementary Materials
The following supporting information can be downloaded at:
www.mdpi.com/xxx/s1, Table S1. Structural alignment of five peptides and water molecules forming the SCC in 53 representative structures of the subtilisin-like superfamily proteases.
Author Contributions
A.I.D.: Study design, Formal analysis, Methodology, Visualization, Writing – Original Draft, Writing – Review & Editing; K.D.: Formal analysis, Methodology, Visualization, Writing – Original Draft, Writing – Review & Editing; M.S.J.: Formal analysis, Methodology, Writing – Original Draft; V.N.U.: Study design, Formal analysis, Methodology, Visualization, Investigation, Writing – Original Draft, Writing – Review & Editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Acknowledgments
We thank the Biocenter Finland Bioinformatics Network (Jukka Lehtonen) and CSC IT Center for Science for computational support for the project. The Structural Bioinformatics Laboratory is part of the Solution for Health strategic area of Åbo Akademi University and within the InFLAMES Flagship program on inflammation and infection, Åbo Akademi University and the University of Turku, funded by the Academy of Finland.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Dodson G, Wlodawer A. Catalytic triads and their relatives. Trends Biochem Sci. 1998; 23(9):347-352. [CrossRef]
- Polgár, L. The catalytic triad of serine peptidases. Cell Mol Life Sci. 2005; 62(19-20):2161-2172. [CrossRef]
- Rawlings ND, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2000; 28(1):323-325. [CrossRef]
- Andreeva A, Kulesha E, Gough J, Murzin AG. The SCOP database in 2020: expanded classification of representative family and superfamily domains of known protein structures. Nucleic Acids Res. 2020; 48(D1):D376-D382. [CrossRef]
- Wallace AC, Laskowski RA, Thornton JM. Derivation of 3D coordinate templates for searching structural databases: application to Ser-His-Asp catalytic triads in the serine proteinases and lipases. Protein Sci. 1996; 5(6):1001-1013. [CrossRef]
- Krem MM, Di Cera E. Molecular markers of serine protease evolution. EMBO J. 2001; 20(12):3036-3045. [CrossRef]
- Derewenda, ZS. C-H Groups as Donors in Hydrogen Bonds: A Historical Overview and Occurrence in Proteins and Nucleic Acids. Int J Mol Sci. 2023; 24(17):13165. [CrossRef]
- Siezen RJ, Leunissen JA. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 1997; 6(3):501-523. [CrossRef]
- Oda K, Dunn BM, Wlodawer A. Serine-Carboxyl Peptidases, Sedolisins: From Discovery to Evolution. Biochemistry. 2022; 61(16):1643-1664. [CrossRef]
- Janssen, DB. Evolving haloalkane dehalogenases. Curr Opin Chem Biol. 2004; 8(2):150-159. [CrossRef]
- Denesyuk AI, Johnson MS, Salo-Ahen OMH, Uversky VN, Denessiouk K. NBCZone: Universal three-dimensional construction of eleven amino acids near the catalytic nucleophile and base in the superfamily of (chymo)trypsin-like serine fold proteases. Int J Biol Macromol. 2020; 153:399-411. [CrossRef]
- Denesyuk A, Dimitriou PS, Johnson MS, Nakayama T, Denessiouk K. The acid-base-nucleophile catalytic triad in ABH-fold enzymes is coordinated by a set of structural elements. PLoS One. 2020; 15(2):e0229376. [CrossRef]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000; 28(1):235-242. [CrossRef]
- Kuhn P, Knapp M, Soltis SM, Ganshaw G, Thoene M, Bott R. The 0.78 Å structure of a serine protease: Bacillus lentus subtilisin. Biochemistry. 1998; 37(39):13446-13452. [CrossRef]
- Graycar TP, Bott RR, Power SD, Estell DA. 693. Subtilisins, in: Handbook of Proteolytic Enzymes, 3rd Edn, Elsevier Ltd., 2013; 3148–3155. [CrossRef]
- Almog O, González A, Godin N, de Leeuw M, Mekel MJ, Klein D, Braun S, Shoham G, Walter RL. The crystal structures of the psychrophilic subtilisin S41 and the mesophilic subtilisin Sph reveal the same calcium-loaded state. Proteins. 2009; 74(2):489-496. [CrossRef]
- Petrilli WL, Adam GC, Erdmann RS, Abeywickrema P, Agnani V, Ai X, Baysarowich J, Byrne N, Caldwell JP, Chang W, DiNunzio E, Feng Z, Ford R, Ha S, Huang Y, Hubbard B, Johnston JM, Kavana M, Lisnock JM, Liang R, Lu J, Lu Z, Meng J, Orth P, Palyha O, Parthasarathy G, Salowe SP, Sharma S, Shipman J, Soisson SM, Strack AM, Youm H, Zhao K, Zink DL, Zokian H, Addona GH, Akinsanya K, Tata JR, Xiong Y, Imbriglio JE. From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9. Cell Chem Biol. 2020; 27(1):32-40.e3. [CrossRef]
- Wlodawer A, Li M, Dauter Z, Gustchina A, Uchida K, Oyama H, Dunn BM, Oda K. Carboxyl proteinase from Pseudomonas defines a novel family of subtilisin-like enzymes. Nat Struct Biol. 2001; 8(5):442-446. [CrossRef]
- Comellas-Bigler M, Fuentes-Prior P, Maskos K, Huber R, Oyama H, Uchida K, Dunn BM, Oda K, Bode W. The 1.4 Å crystal structure of kumamolysin: a thermostable serine-carboxyl-type proteinase. Structure. 2002; 10(6):865-876. [CrossRef]
- Clementel D, Del Conte A, Monzon AM, Camagni GF, Minervini G, Piovesan D, Tosatto SCE. RING 3.0: fast generation of probabilistic residue interaction networks from structural ensembles. Nucleic Acids Res. 2022; 50(W1):W651-W656. [CrossRef]
- Holm L, Laiho A, Törönen P, Salgado M. DALI shines a light on remote homologs: One hundred discoveries. Protein Sci. 2023; 32(1):e4519. [CrossRef]
- Dimitriou PS, Denesyuk A, Takahashi S, Yamashita S, Johnson MS, Nakayama T, Denessiouk K. Alpha/beta-hydrolases: A unique structural motif coordinates catalytic acid residue in 40 protein fold families. Proteins. 2017; 85(10):1845-1855. [CrossRef]
- Ghosh D, Sawicki M, Lala P, Erman M, Pangborn W, Eyzaguirre J, Gutierrez R, Jornvall H, Thiel DJ. Multiple conformations of catalytic serine and histidine in acetylxylan esterase at 0.90 Å. J Biol Chem. 2001; 276(14):11159-11166. [CrossRef]
- Dimitriou PS, Denesyuk AI, Nakayama T, Johnson MS, Denessiouk K. Distinctive structural motifs co-ordinate the catalytic nucleophile and the residues of the oxyanion hole in the alpha/beta-hydrolase fold enzymes. Protein Sci. 2019; 28(2):344-364. [CrossRef]
- Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999; 15(4):327-332. [CrossRef]
- Kraulis, PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Cryst. 1991; 24:946-950. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).