Submitted:
16 October 2024
Posted:
17 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
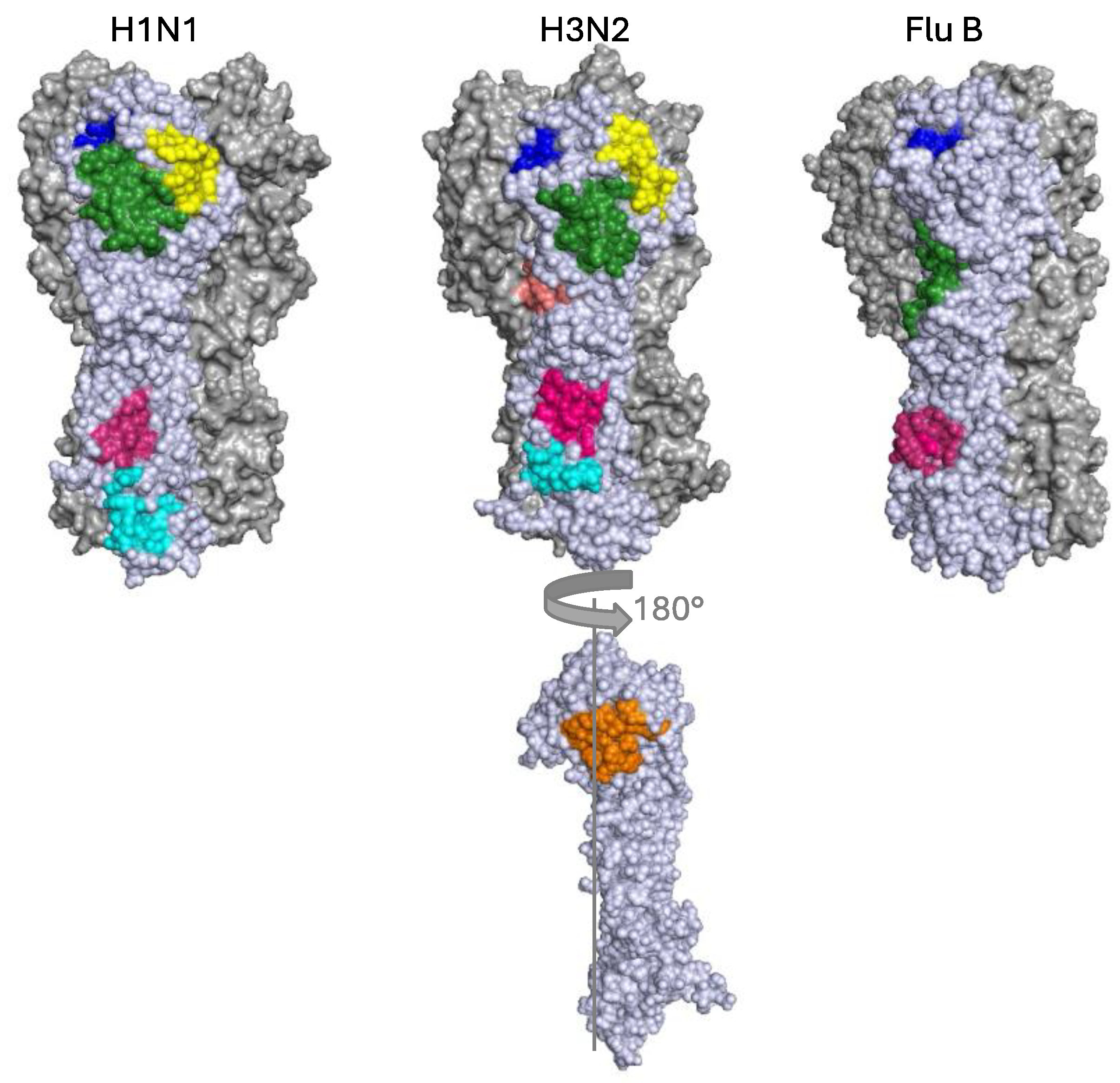
1.1. Hemagglutinin
2. bnAbs against the HA Stem
2.1. The Central Stem Epitope
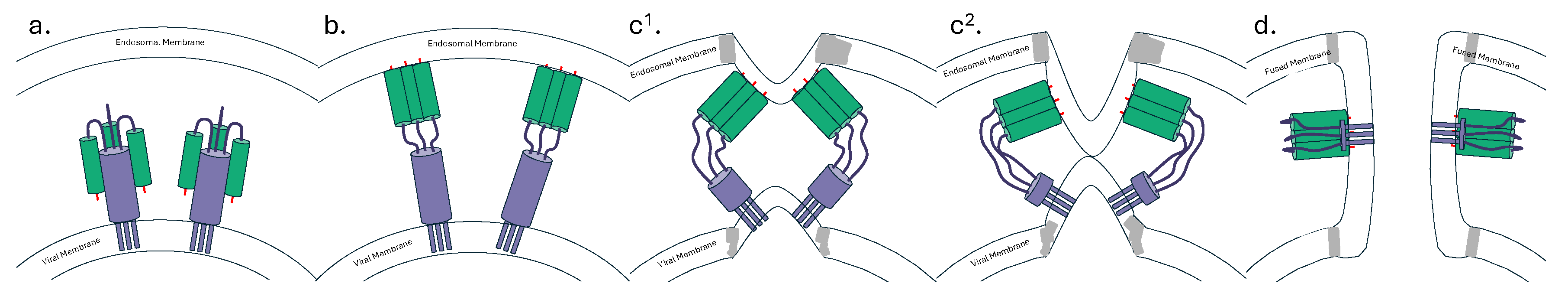
2.2. The Fusion Peptide and Anchor Epitope
3. bnAbs against the HA Head Domain
3.1. Receptor Binding Site
3.2. Lateral Patch
3.3. Vestigial Esterase
3.4. Interface and Occluded Epitope
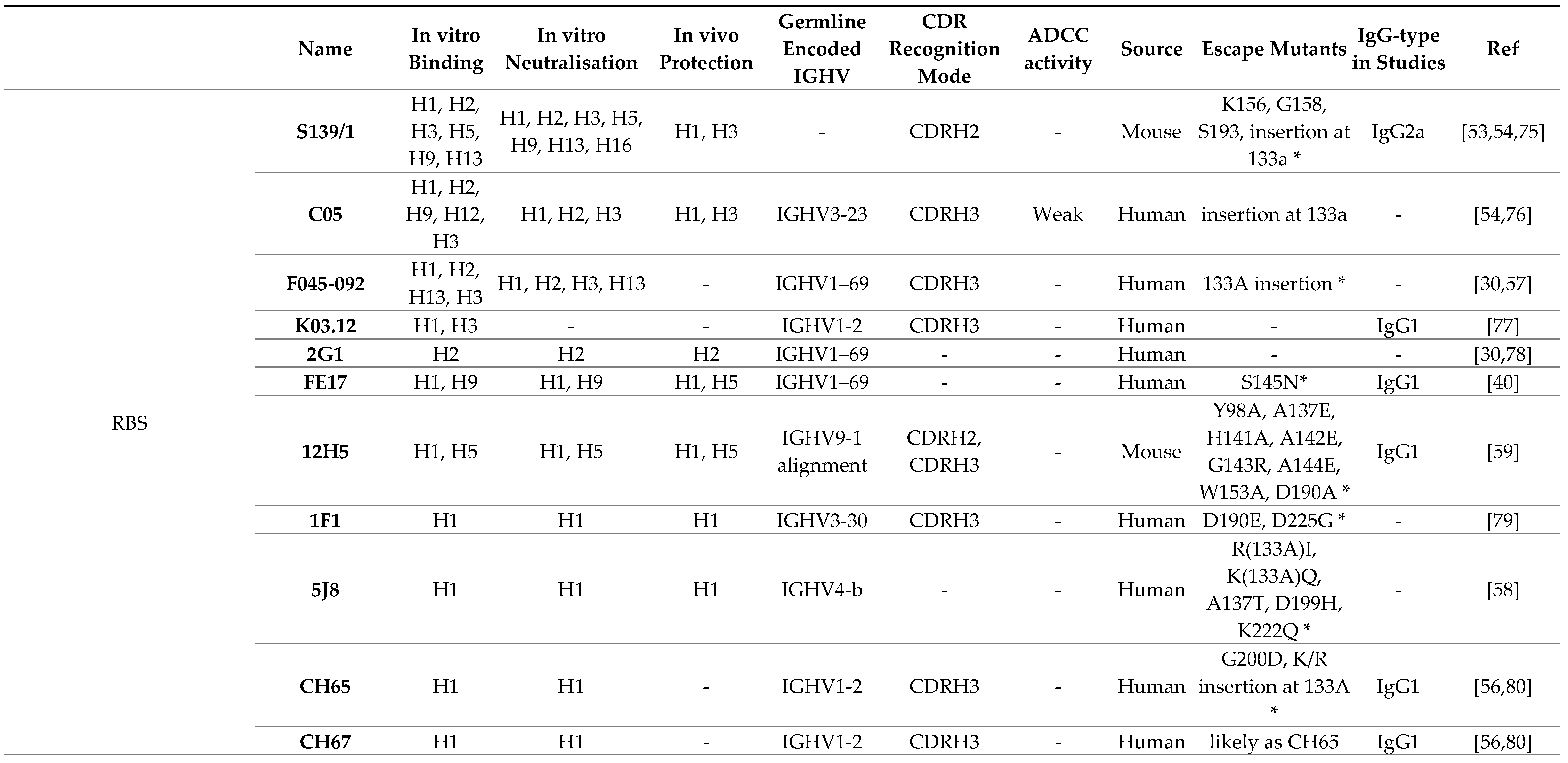 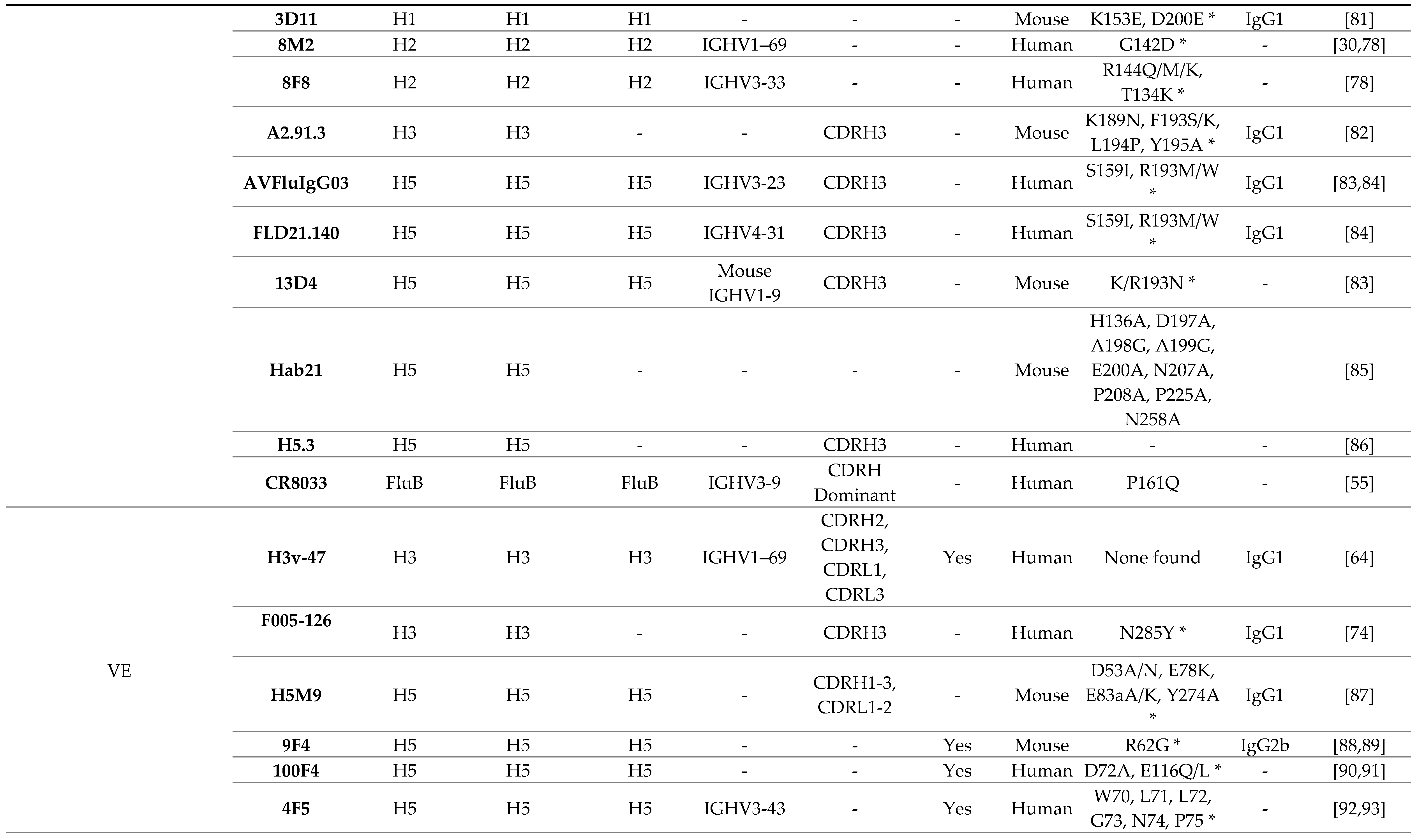 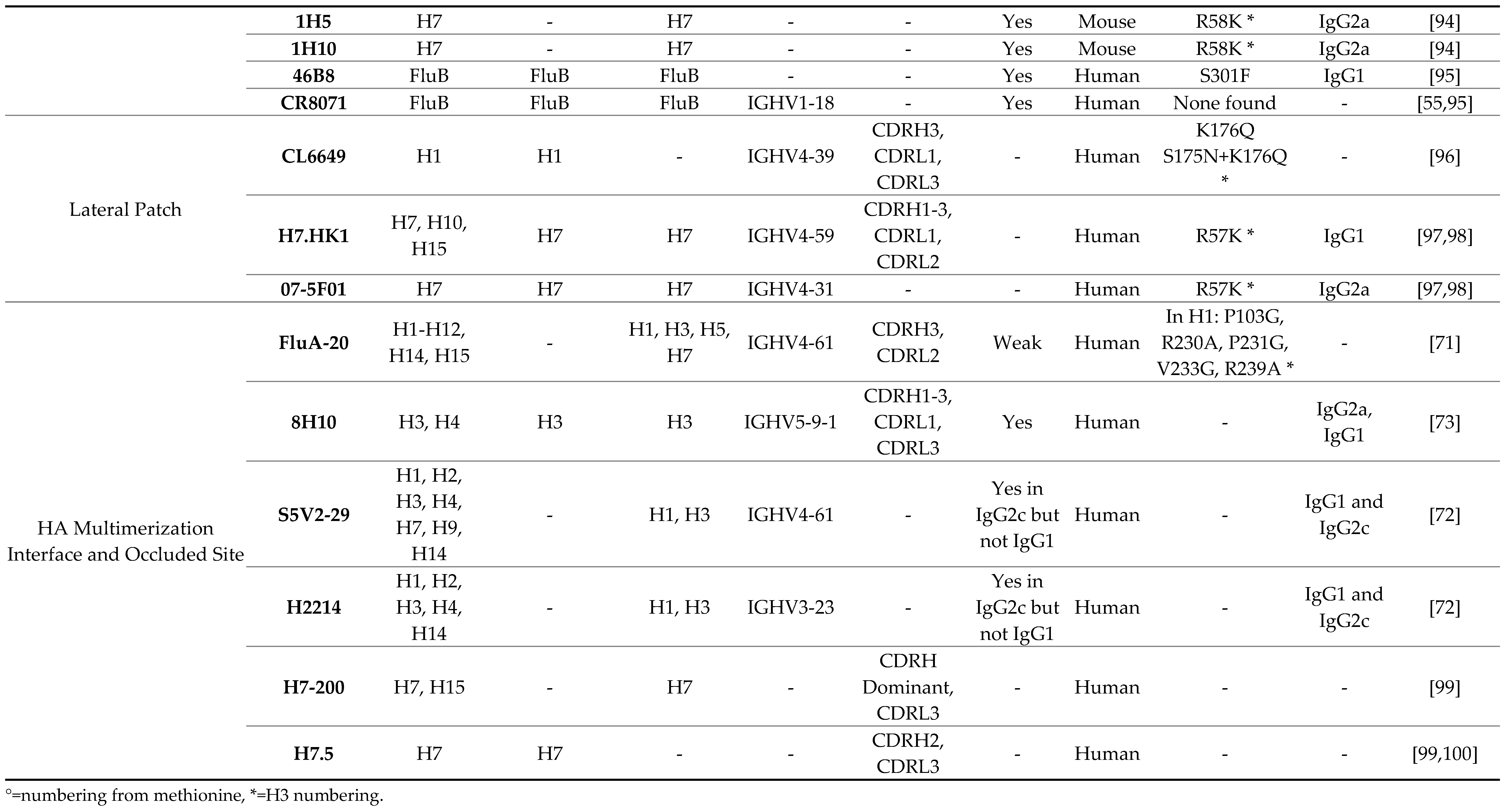
|
4. bnAbs in Clinical Trials
5. Broadly Protective Vaccines in Clinical Trials
6. bnAbs in Current and Future Directions
6.1. Escape Mutations
6.2. Immunogenicity of bnAbs
6.3. Antibody-Dependent Enhancement
References
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K; Doherty, P.C.; Palese, P; Shaw, M.L; Treanor, J.; Wesbter, R.G.; et al. Influenza. Nature Reviews Disease Primers, 2018, 4, 3. [Google Scholar] [PubMed]
- Collins, J.P., Campbell; Openo, K.; Farley, M.; Cummings, C.N.; Hill, M.; Schaffner, W.; Lindegren, *!!! REPLACE !!!*; M.L., *!!! REPLACE !!!*; Thomas, A.; Billing, L.; et al. Outcomes of Immunocompromised Adults Hospitalized With Laboratory-confirmed Influenza in the United States, 2011-2015. Clin Infect Dis, 2020, 70, 2121–2130. [Google Scholar] [CrossRef] [PubMed]
- WHO. Influenza seasonal. 2024 [cited 2024 28.08.2024]; Available from: https://www.who.int/health-topics/influenza-seasonal#tab=tab_1.
- Knobler, S.L.; Mack, A.; Mahmoud, A.; Lemon, S.M. Story of Influenza. 2005, Institute of Medicine (US): Forum on Microbial Threats.
- Fierer, J. Looney, and J.-C. Pechère, 2 - Nature and Pathogenicity of Micro-organisms, in Infectious Diseases (Fourth Edition), J. Cohen, W.G. Powderly, and S.M. Opal, Editors. 2017, Elsevier. 4-25.e1.
- Ryu, W.-S. , Chapter 15 - Influenza Viruses, in Molecular Virology of Human Pathogenic Viruses, W.-S. Ryu, Editor. 2017, Academic Press: Boston. 195-211.
- Fujimura, S.F. , Purple Death: The Great Flu of 1918, in Perspectives in Health. 2003: Pan American Health Organization.
- Gerhard, W.; Yedwell, J.; Frankel, M.E.; Webster, R. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies . Nature, 1981, 290, 713–717. [Google Scholar] [CrossRef]
- Okuno, Y.; Isegawa, F.; Sasao, F.; Ueda, S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol, 1993, 67, 2552–8. [Google Scholar] [CrossRef]
- Lousa, D.; Soares, C.M. Molecular mechanisms of the influenza fusion peptide: insights from experimental and simulation studies. FEBS Open Bio, 2021, 11, 3253–3261. [Google Scholar] [CrossRef]
- Cheung, C.S.-F.; Gorman, J.; Andrews, S.F.; Rawi, R.; Reveiz, M.; Shen, C.-H.; Wang, Y.; Harris, D.R.; Nazzari, A.F.; Olia, A.S.; et al. , Structure of an influenza group 2-neutralizing antibody targeting the hemagglutinin stem supersite. Structure, 2022, 30, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Throsby, M., van den Brink; Jongeneelen, M.; Poon, L.L.M; Alard, Ph; Cornelissen, L.; Bakker, A.; Cox, F.; van Deventer, E.; Guan, Y.; et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One, 2008, 3, e3942. [Google Scholar] [CrossRef]
- Nath Neerukonda, S. Vassell, C.D. Weiss, Neutralizing Antibodies Targeting the Conserved Stem Region of Influenza Hemagglutinin. Vaccines (Basel), 2020, 8(3).
- Sun, X., Ling, Z.; Yang, Z.; Sun, B. Broad neutralizing antibody-based strategies to tackle influenza. Curr. Opi. Virol. 2022, 53: 101207.
- Tan, H.X., Jegaskanda, S.; Juno, J.A.; Esterbauer, R.; Wong, J.; Kelly, H.G.; Liu, Y.; Tilmanis, D.; Hurt, A.C.; Yedwell, J.W.; e al. Subdominance and poor intrinsic immunogenicity limit humoral immunity targeting influenza HA stem. J Clin Invest. 2019, 129, 850–862.
- Eggink, D. Eggink, D., P.H. Goff, Palese, Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J Virol. 2014, 88, 699–704. [Google Scholar]
- Jiao, C.; Chen, P.; Jian, Y. ; Liu; J. Analysis of the conserved protective epitopes of hemagglutinin on influenza A viruses. Frontiers in Immunology, 2023, 14.
- Ekiert, D.C., Bhabha, G.; Elsliger, M.-A.; Friesen, R.H.; Jongeneelen, M.; Throsby, M.; Goudsmit, J.; Wilson, I.A. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009, 324, 246–51.
- Sui, J., Hwang, W.C.; Pérez, S .; Wei, G.; Aird, D.; Chen, L.-.m; Santelli, E.; Stec, B.; Cadwell, G.; Ali, M.; et al., Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009, 16, 265–273.
- Tharakaraman, K.; Subramanian, V.; Cain, D.; Sasisekharan, V.; Sasisekharan, R. Broadly neutralizing influenza hemagglutinin stem-specific antibody CR8020 targets residues that are prone to escape due to host selection pressure. Cell Host Microbe, 2014, 15, 644–51. [Google Scholar] [CrossRef]
- Corti, D., Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; et al., A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011, 333, 850–6.
- Limberis, M.P., Adam, V.S.; Wong, G.; Gren, J.; Kobasa, D.; Ross, T.M.; Kobinger, G.P.; Tretiakova, A.; Wilson, J.M. Intranasal antibody gene transfer in mice and ferrets elicits broad protection against pandemic influenza. Sci Transl Med. 2013, 5, 187ra72.
- Morgan, S.B., Holzer, B.; Hemmink, J.D.; Salguero, F.J.; Schwartz, J.C.; Agatic, G.; Cameroni, E.; Guarino, B.; Porter, E.; Rijal, P.; et al., Therapeutic Administration of Broadly Neutralizing FI6 Antibody Reveals Lack of Interaction Between Human IgG1 and Pig Fc Receptors. Front Immunol, 2018, 9: 865. 8: 9.
- Ekiert, D.C., Friesen; Bhabha, G.; Kwaks, T.; Jongeneelen, M.; Yu, W.; Ophorst, C.; Cox, F.; Korse, H.J.W.M.; Brandenburg, B.; et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science, 2011, 333, 843–50. [Google Scholar] [CrossRef]
- Friesen, R.H., Lee; Stoop, E.J.M.; Hoffman, R.M.B.; Ekiert, D.C.; Bhabha, G.; Yu, W.; Juraszek, J.; Koudstaal, W.; Jongeneelen, M.; et al. A common solution to group 2 influenza virus neutralization. Proc Natl Acad Sci U S A, 2014, 111, 445–50. [Google Scholar] [CrossRef]
- Guthmiller, J.J., Han; Utset, H.A.; Li, L.; Lan, L.Y.-L.; Henry, C.; Stamper, C.T.; McMahon, M.; O'Dell, G.; Fernández-Quintero, M.L.; et al. Broadly neutralizing antibodies target a haemagglutinin anchor epitope. Nature, 2022, 602, 314–320. [Google Scholar] [CrossRef] [PubMed]
- McCraw, D.M., Myers; Gulati, N.M.; Prabhakaran, M.; Brand, J.; Andrews, S.; Gallagher, J.R.; Maldonado-Puga, S.; Kim, A.J.; Torian, U.; et al. Designed nanoparticles elicit cross-reactive antibody responses to conserved influenza virus hemagglutinin stem epitopes. PLoS Pathog, 2023, 19, e1011514. [Google Scholar] [CrossRef] [PubMed]
- Sakabe, S., Iwatsuki-Horimoto; Horimoto, T.; Nidom, .C.A; Le, M.t.Q.; Takano, R.; Kubota-Koketsu, R.; Okuno, Y.; Ozawa, M.; Kawaoka, Y. A cross-reactive neutralizing monoclonal antibody protects mice from H5N1 and pandemic (H1N1) 2009 virus infection. Antiviral Res, 2010, 88, 249–55. [Google Scholar] [CrossRef]
- Lang, S., Xie; Zhu, X.; Wu, N.C., Lerner; Wilson, I.A. Antibody 27F3 Broadly Targets Influenza A Group 1 and 2 Hemagglutinins through a Further Variation in V(H)1-69 Antibody Orientation on the HA Stem. Cell Rep, 2017, 20, 2935–2943. [Google Scholar] [CrossRef]
- Chen, F., Tzarum, N.; Wilson, I.A.; Law, M. V(H)1-69 antiviral broadly neutralizing antibodies: genetics, structures, and relevance to rational vaccine design. Curr Opin Virol, 2019, 34: 149-159.
- Roubidoux, E.K. , Carreño, J.M; McMahon, M.; Jiang, K.; van Bakel, H.; Wilson, P.; Krammer, F. Mutations in the Hemagglutinin Stalk Domain Do Not Permit Escape from a Protective, Stalk-Based Vaccine-Induced Immune Response in the Mouse Model. mBio, 2021, 12(1).
- Muralidharan, A. Muralidharan, A., Gravel, C.; Harris, G.; Hashem, A.M.; Zhang, W.; Safronetz, D.; Van Domselaar, G.; Krammer,, F.; Sauve, S.; Rosu-Myles, M.; et al., Universal antibody targeting the highly conserved fusion peptide provides cross-protection in mice. Hum Vaccin Immunother. 2022, 18, 2083428. [Google Scholar]
- Sutton, T.C., Lamirande, E.W.; Bock, K.W.; Moore, I.N.; Koudstaal, W.; Rehman, M.; Weverling, G.J.; Goudsmit, J.; Subbarao, K. In Vitro Neutralization Is Not Predictive of Prophylactic Efficacy of Broadly Neutralizing Monoclonal Antibodies CR6261 and CR9114 against Lethal H2 Influenza Virus Challenge in Mice. J Virol, 2017, 91(24).
- Throsby, M., van den Brink, E.; Jongeneelen, M.; Poon, L.L.M; Alard, Ph; Cornelissen, L.; Bakker, A.; Cox, F.; van Deventer, E.; Guan, Y.; et al., Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008, 3, e3942.
- Park, J.K. , Xiao, Y.; Ramuta, M.D.; Rosas, L.A.; Fong, S.; Matthews, A.M.; Freeman, A.D.; Gouzoulis, M.A.; Batchenkova, N.A.; Yang, X.; Pre-existing immunity to influenza virus hemagglutinin stalk might drive selection for antibody-escape mutant viruses in a human challenge model. Nat Med, 2020, 26, 1240–1246. [Google Scholar] [PubMed]
- Li, G.M. , Chiu, C.; Wrammert, J.; McCausland, M.; Andrews, S.F.; Zheng, N.-Y.; Lee, J.-H.; Huang, M.; Qu, X.; Edupuganti, S.; et al., Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci U S A, 2012, 109, 9047–52. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, A.K., Steel, J.; Rubrum, A.; Estelles, A.; Briante, R.; Ilyushina, N.A.; Xu, L.; Swale, R.E.; Faynboym, A.M.; Foreman, P.K.; et al., Protection from the 2009 H1N1 pandemic influenza by an antibody from combinatorial survivor-based libraries. PLoS Pathog. 2010, 6, e1000990.
- Nakamura, G., Chai, N.; Park, S.; Chiang, N.; Lin, Z.; Chiu, H.; Fong, R.; Yan, D.; Kim, J.; Zhang, J.; et al., An In Vivo Human-Plasmablast Enrichment Technique Allows Rapid Identification of Therapeutic Influenza A Antibodies. Cell Host Microbe. 2013, 14, 93–103.
- Chai, N., Swem, L.R.; Reichelt, M.; Chen-Harris, H.; Luis, E.; Park, S.; Fouts, A.; Lupardus, P.; Wu, T.D.; Li, O.; et al., Two Escape Mechanisms of Influenza A Virus to a Broadly Neutralizing Stalk-Binding Antibody. PLoS Pathog. 2016, 12, e1005702.
- Corti, D., Suguitan Jr.; A.L.; Pinna, D.; Silacci, C.; Fernandez-Rodriguez, B.M.; Vanzetta, F.; Santos, C.; Luke, C.J.; Torres-Velez, F.J.; Temperton, N.J.; et al., Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest, 2010, 120, 1663–73.
- Wang, W., Sun, X.; Li, Y.; Su, J.; Ling, Z.; Zhang, T.; Wang, F.; Zhang, H.; Chen, H.; Ding, J.; et al., Human antibody 3E1 targets the HA stem region of H1N1 and H5N6 influenza A viruses. Nature Communications. 2016, 7, 13577.
- Wu, Y.; Cho, M.; Shore, D.; Song, M.; Choi, J.; Jiang, T.; Deng, Y.-Q.; Bourgeois, M.; Almli, L.; Yang, H.; et al. , A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus. Nature Communications, 2015, 6, 7708. [Google Scholar] [CrossRef]
- Joyce, M.G., Wheatley, A.K.; Thomas, P.V.; Chuang, G.-Y.; Soto, C.; Bailer, R.T.; Druz, A.; Georgiev, I.S.; Gillespie, R.A.; Kanekiyo, M.; et al., Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses. Cell. 2016, 166, 609–623.
- Beukenhorst, A.L., Frallicciardi, J.; Koch, C.M.; Klap, J.M.; Phillips, A.; Desai, M.M.; Wichapong, K.; Nicolaes, G.A.F.; Koudstaal, W.; Alter, G.; et al., Corrigendum: The influenza hemagglutinin stem antibody CR9114: Evidence for a narrow evolutionary path towards universal protection. Frontiers in Virology, 2023, 3.
- Beukenhorst, A.L.; Frallicciardi, J.; Rice, K.L.; Koldijk, M.H.; Moreira de Mello, J.C.; Klap, J.M.; Hadjichrysanthou, C.; Koch, C.M.; da Costa, K.A.S.; Temperton, N.; et al. , A pan-influenza monoclonal antibody neutralizes H5 strains and prophylactically protects through intranasal administration. Sci. Rep., 2024, 14, 3818. [Google Scholar] [CrossRef] [PubMed]
- Prachanronarong, K.L. , Canale, A.S.; Liu, P.; Somasundaran, M.; Hou, S.; Poh, Y.-P.; Han, T.; Zhu, Q.; Renzette, N.; Zeldovich, K.B.; et al., Mutations in Influenza A Virus Neuraminidase and Hemagglutinin Confer Resistance against a Broadly Neutralizing Hemagglutinin Stem Antibody. J Virol, 2019, 93(2).
- Ali, S.O.; Takas, T.; Nyborg, A.; Shoemaker, K.; Kallewaard, N.L.; Chiong, R.; Dubovsky, F.; Mallory, R.M. Evaluation of MEDI8852, an Anti-Influenza A Monoclonal Antibody, in Treating Acute Uncomplicated Influenza. Antimicrob Agents Chemother, 2018, 62(11).
- Paules, C.I., Lakdawala, S.; McAuliffe, J.M.; Paskel, M.; Vogel, L.; Kallewaard, N.L.; Zhu, Q.; Subbarao, K. The Hemagglutinin A Stem Antibody MEDI8852 Prevents and Controls Disease and Limits Transmission of Pandemic Influenza Viruses. J Infect Dis. 2017, 216, 356–365.
- Mark Throsby, R.H. , Edward Friesen, Theodorus Hendrikus, Jacobus Kwaks, Mandy Antonia, Catharina Jongeneelen, Human binding molecules capable of neutralizing influenza virus H3N2 and uses thereof. 2010: United States.
- Tan, G.S., Lee, P.S.; Hoffman, R.M.B.; Mazel-Sanchez, B.; Krammer, F.; Leon, P.E.; Ward, A.B.; Wilson, I.A.; Palese, Characterization of a broadly neutralizing monoclonal antibody that targets the fusion domain of group 2 influenza A virus hemagglutinin. J Virol. 2014, 88, 13580–92.
- Myers, M.L.; Gallagher, J.R.; Kim, A.J.; Payne, W.H.; Maldonado-Puga, S.; Assimakopoulos, H.; Bock, K.W.; Torian, U.; Moore, I.N.; Harris, A.K. Commercial influenza vaccines vary in HA-complex structure and in induction of cross-reactive HA antibodies. Nature Communications, 2023, 14, 1763. [Google Scholar] [CrossRef]
- Yu, X. , Tsibane, T. ; McGraw, P.A.; House, F.S.; Keefer, C.J.; Hicar, M.D.; Tumpey, T.M.; Pappas, C.; Perrone, L.A.; Martinez, O.; et al, Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature, 2008, 455, 532–6. [Google Scholar]
- Yoshida, R. , Igarashi, M. ; Ozaki, H.; Kishida, N.; Tomabechi, D.; Kida, H.; Ito, K.; Takada, A. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog, 2009, 5, e1000350. [Google Scholar]
- Ekiert, D.C.; Kashyap, A.K.; Steel, J.; Rubrum, A.; Bhabha, G.; Khayat, R.; Lee, J.H.; Dillon, M.A.; O'Neil, R.E.; Faynboym, A.M.; et al. Cross-neutralization of influenza A viruses mediated by a single antibody looNature, 2012, 489, 526-32.
- Dreyfus, C. , Laursen, N. S.; Kwaks, T.; Zuijdgeest, D.; Khayat, R.; Ekiert, D.C.; Lee, J.H.; Metlagel, Z.; Bujny, M.V.; Jongneelen, M.; et al. Highly conserved protective epitopes on influenza B viruses. Science, 2012, 337, 1343–8. [Google Scholar]
- Whittle, J.R.R. , Zhang, R. ; Khurana, S.; King, L.R.; Manischewitz, J.; Golding, H.; Dormitzer, P.R.; Haynes, B.F.; Walter, E.B.; Moody, M.A.; et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proceedings of the National Academy of Sciences, 2011, 108, 14216–14221. [Google Scholar]
- Lee, P.S. , Ohshima, N. ; Stanfield, R.L.; Yu, W.; Iba, Y.; Okuno, Y.; Kurosawa, Y.; Wilson, I.A. Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat. Commun. 2014, 5, 3614. [Google Scholar]
- Krause, J.C. , Tsibane, T. ; Tumpey, T.M.; Huffman, C.J.; Basler, C.F.; Crowe Jr, J.E. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol, 2011, 85, 10905–8. [Google Scholar]
- Li, T. , Chen, J.; Zheng, Q.; Xue, W.; Zhang, L.; Rong, R.; Zhang, S.; Wang, Q.; Hong, M.; Zhang, Y.; et al. Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses. Nature Commun, 2022, 13, 5182. [Google Scholar] [CrossRef] [PubMed]
- Guthmiller, J.J. , Han, J.; Li, L.; Freyn, A.W.; Liu, S.T.H.; Stovicek, O.; Stamper, C.T.; Dugan, H.L.; Tepora, M.E.; Utset, H.A.; et al. First exposure to the pandemic H1N1 virus induced broadly neutralizing antibodies targeting hemagglutinin head epitopes. Sci Transl Med, 2021, 13(596).
- Raymond, D.D. , Bajic, G. ; Ferdman, J.; Suphaphiphat, P.; Settembre, E.C.; Moody, M.A.; Schmidt, A.G.; Harrison, S.C. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc Natl Acad Sci U S A, 2018, 115, 168–173. [Google Scholar] [PubMed]
- Jia, M.; Zhao, H.; Morano, N.C.; Lu, H.; Lui, Y.-M.; Du, H.; Becker, J.E.; Yuen, K.-Y.; Ho, D.D; Kwong, P.D.; et al. Human neutralizing antibodies target a conserved lateral patch on H7N9 hemagglutinin head. Nature Communications, 2024, 15, 4505. [Google Scholar] [CrossRef]
- Zheng, Z., Paul, S.S.; Mo, X.; Yuan, Y.-R.; Tan, Y.J. The Vestigial Esterase Domain of Haemagglutinin of H5N1 Avian Influenza A Virus: Antigenicity and Contribution to Viral Pathogenesis. Vaccines (Basel), 2018, 6(3).
- Bangaru, S. , Zhang, H.; Gilchuk, I.M.; Voss, T.G.; Irving, R.P.; Gilchuk, P.; Matta, P.; Zhu, X.; Lang, S.; Nieusma, T.; et al. A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA. Nature Communications, 2018, 9, 2669. [Google Scholar] [CrossRef] [PubMed]
- Crowe, J.E. , Antibody Determinants of Influenza Immunity. J Infect Dis, 2019, 219(Suppl_1): S21-s29.
- Gao, R.; Sheng, Z.; Sreenivasan, C.C.; Wang, D.; Li, F. Influenza A Virus Antibodies with Antibody-Dependent Cellular Cytotoxicity Function. Viruses, 2020, 12, 276. [Google Scholar] [CrossRef]
- Chai, N., Swem, L.R.; Park, S.; Nakamura, G.; Chiang, N.; Estevez, A.; Fong, R.; Kamen, L.; Kho, E.; Reichelt, M.; et al. A broadly protective therapeutic antibody against influenza B virus with two mechanisms of action. Nat Commun, 2017, 8: 14234.
- Yan, L. , Sun, L. ; Guo, C.; Li, L.; Sun, J.; Huang, X.; Zhao, P.; Xie, X.; Hu, J. Neutralizing antibody PR8-23 targets the footprint of the sialoglycan receptor binding site of H1N1 hemagglutinin. J Med Virol, 2021, 93, 3508–3515. [Google Scholar]
- Li, J. , Wang, Y. ; Liang, Y.; Ni, B.; Wan, Y.; Liao, Z.; Chan, K.-h.; Yuen, K.-y.; Fu, X.; Shang, X.; et al. Fine antigenic variation within H5N1 influenza virus hemagglutinin's antigenic sites defined by yeast cell surface display. Eur J Immunol, 2009, 39, 3498–510. [Google Scholar]
- Yewdell, J.W. , Taylor A. ; Yellen, A.; Caton, A.; Gerhard, W.; Bächi, T. Mutations in or near the fusion peptide of the influenza virus hemagglutinin affect an antigenic site in the globular region. J Virol, 1993, 67, 933–42. [Google Scholar]
- Bangaru, S. , Lang, S. ; Schotsaert, M.; Vanderven, H.A.; Zhu, X.; Kose, N.; Bombardi, R.; Finn, J.A.; Kent, S.J.; Gilchuk, P.; et al. A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface. Cell, 2019, 177, 1136–1152. [Google Scholar]
- Watanabe, A. , McCarthy, K. R.; Kuraoka, M.; Schmidt, A.G.; Adachi, Y.; Onodera, T.; Tonouchi, K.; Caradonna, T.M.; Bajic, G.; Song, S.; et al. Antibodies to a Conserved Influenza Head Interface Epitope Protect by an IgG Subtype-Dependent Mechanism. Cell, 2019, 177, 1124–1135. [Google Scholar]
- Bajic, G. , Maron, M. J.; Adachi, Y.; Onodera, T.; McCarthy, K.R.; McGee, C.E.; Sempowski, G.D.; Takahasi, Y.; Kelsoe, G.; Kuraoka, M.; et al. Influenza Antigen Engineering Focuses Immune Responses to a Subdominant but Broadly Protective Viral Epitope. Cell Host Microbe, 2019, 25, 827–835. [Google Scholar]
- Iba, Y. , Fujii, Y. ; Ohshima, N.; Sumida, T.; Kubota-Koketsu, R.; Ikeda, M.; Wakiyama, M.; Shirouzu, M.; Okada, J.; Okuno, Y.; et al. Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses. J Virol, 2014, 88, 7130–44. [Google Scholar]
- Lee, P.S.; Yoshida, R.; Ekiert, D.C.; Sakai, N.; Suzuki, Y.; Takada, A.; Wilson, I.A. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci U S A, 2012, 109, 17040–5. [Google Scholar] [CrossRef]
- He, W.; Tan, G.S.; Mullarkey, C.E.; Lee, A.J.; Lam, M.M.W.; Krammer, F.; Henry, C.; Wilson, P.C.; Ashkar, A.A.; Palese, et al. Epitope specificity plays a critical role in regulating antibody-dependent cell-mediated cytotoxicity against influenza A virus. Proc Natl Acad Sci U S A. 2016, 113, 11931–11936.
- McCarthy, K.R.; Watanabe, A.; Kuraoka, M.; Do, K.T.; McGee, C.E.; Sempowski, G.D.; Kepler, T.B.; Schmidt, A.G.; Kelsoe, G.; Harrison, S.C. Memory B Cells that Cross-React with Group 1 and Group 2 Influenza A Viruses Are Abundant in Adult Human Repertoires. Immunity, 2018, 48, 174–184. [Google Scholar] [CrossRef]
- Krause, J.C.; Tsibane, T.; Tumpey, T.M.; Huffman, C.J.; Albrecht, R.; Blum, D.L. ; Ramos, I:; Fernandez-Sesma, A. ; Edwards, K.M.; García-Sastre, A.; et al. Human monoclonal antibodies to pandemic 1957 H2N2 and pandemic 1968 H3N2 influenza viruses. J Virol, 2012, 86, 6334–40. [Google Scholar]
- Tsibane, T.; Ekiert, D.C.; Krause, J.C.; Martinez, O.; Crowe Jr., J. E. ; Wilson, I.A.; Basler, C.F. Influenza human monoclonal antibody 1F1 interacts with three major antigenic sites and residues mediating human receptor specificity in H1N1 viruses. PLoS Pathog, 2012, 8, e1003067. [Google Scholar]
- Schmidt, A.G.; Therkelsen, M.D.; Stewart, S.; Kepler, T.B.; Liao, H.-X.; Moody, M.A.; Haynes, B.F.; Harrison, S.C. Viral receptor-binding site antibodies with diverse germline origins. Cell, 2015, 161, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Yan, S.; Zhu, L.; Wang, F.X.C.; Liu, F.; Cheng, L.; Yao, H.; Wu, N.; Lu, R.; Wu, H. Evaluation of panel of neutralising murine monoclonal antibodies and a humanised bispecific antibody against influenza A(H1N1)pdm09 virus infection in a mouse model. Antiviral Res, 2022, 208: 105462.
- Portnoff, A.D.; Patel, N.; Massare, M.J.; Zhou, H.; Tian, J.-H.; Zhou, B.; Shinde, V.; Glenn, G.M.; Smith, G. Influenza Hemagglutinin Nanoparticle Vaccine Elicits Broadly Neutralizing Antibodies against Structurally Distinct Domains of H3N2 HA. Vaccines (Basel), 2020, 8(1).
- Lin, Q.; Li, T.; Chen, Y.; Lau, S.-Y.; Wei, M.; Zhang, Y.; Zhang, Z.; Yao, Q.; Li, J.; Li, Z.; et al. Structural Basis for the Broad, Antibody-Mediated Neutralization of H5N1 Influenza Virus. J Virol, 2018, 92(17).
- Zuo, Y. , Wang, P.; Sun, J.; Guo, S.; Wang, G.; Zuo, T.; Fan, S.; Zhou, P.; Liang, M.; Shi, X.; et al. Complementary recognition of the receptor-binding site of highly pathogenic H5N1 influenza viruses by two human neutralizing antibodies. J Biol Chem, 2018, 293, 16503–16517. [Google Scholar] [CrossRef]
- Wu, R.; Li, X.; Leung, H.-C.; Cao, Z.; Qiu, Z.; Zhou, Y.; Zheng, B.-J.; He, Y A novel neutralizing antibody against diverse clades of H5N1 influenza virus and its mutants capable of airborne transmission. Antiviral Res 2014, 106: 13-23.
- Winarski, K.L.; Thornburg, N.J.; Yu, Y.; Sapparapu, G.; Crowe Jr., J. E. ; Spiller, B.W. Vaccine-elicited antibody that neutralizes H5N1 influenza and variants binds the receptor site and polymorphic sites. Proceedings of the National Academy of Sciences, 2015, 112, 9346–9351. [Google Scholar]
- Zhu, X.; Guo, Y.-H.; Jiang, T.; Wang, Y.-D.; Chan, K.-H.; Li, X.-F. , Yu, W. ; McBride, R.; Paulson, J.C.; Yuen, K.-Y., et al. A unique and conserved neutralization epitope in H5N1 influenza viruses identified by an antibody against the A/Goose/Guangdong/1/96 hemagglutinin. J Virol, 2013, 87, 12619–35. [Google Scholar]
- Paul, S.S.; Mok, C.-K.; Mak, T.-M.; Ng, O.-W.; .Aboagye, J.O.; Wohlbold, T.J.; Krammer, F.; Tan, Y.-J. A cross-clade H5N1 influenza A virus neutralizing monoclonal antibody binds to a novel epitope within the vestigial esterase domain of hemagglutinin. Antiviral Res, 2017, 144: 299-310.
- Zheng, Z.; Teo, S.H.C.; Arularasu, S.C.; Liu, Z.; Mohd-Ismail, N.K.; Mok, C.K.; Ong, C.B.; Chu, J.J.-h.; Tan, Y.-J. Contribution of Fc-dependent cell-mediated activity of a vestigial esterase-targeting antibody against H5N6 virus infection. Emerg Microbes Infect, 2020, 9: 95-110.
- Qian, M.; Hu, H.; Zuo, T.; Wang, G.; Zhang, L. ; Zhou, Unraveling of a neutralization mechanism by two human antibodies against conserved epitopes in the globular head of H5 hemagglutinin. J Virol, 2013, 87, 3571–7. [Google Scholar] [CrossRef]
- Wang, S.; Ren, H.; Jiang, W.; Chen, H.; Hu, H.; Chen, Z. ; Zhou, Divergent Requirement of Fc-Fcγ Receptor Interactions for In Vivo Protection against Influenza Viruses by Two Pan-H5 Hemagglutinin Antibodies. J Virol, 2017, 91(11).
- Zhang, X.; Qi, X.; Zhang, Q.; Zeng, X.; Shi, Z. ; Jin, Q:; Zhan, F. ; Xu, Y.; Liu, Z.; Feng, Z.; Jiao, Y. Human 4F5 single-chain Fv antibody recognizing a conserved HA1 epitope has broad neutralizing potency against H5N1 influenza A viruses of different clades. Antiviral Res 2013, 99, 91–99. [Google Scholar]
- Jin, Q.; Yao, Z.; Liu, F.; Di, Y.; Gao, J.; Zhang, X. The protective effect of a combination of human intracellular and extracellular antibodies against the highly pathogenic avian influenza H5N1 virus. Hum Vaccin Immunother, 2022, 18, 2035118. [Google Scholar] [CrossRef]
- Tan, G.S. , et al. , Broadly-Reactive Neutralizing and Non-neutralizing Antibodies Directed against the H7 Influenza Virus Hemagglutinin Reveal Divergent Mechanisms of Protection. PLOS Pathogens, 2016, 12, e1005578. [Google Scholar]
- Chai, N.; Swem, L.R.; Park, S.; Nakamura, G.; Chiang, N.; Estevez, A.; Fong, R.; Kamen, L.; Kho, E.; Reichelt, M.; et al. A broadly protective therapeutic antibody against influenza B virus with two mechanisms of action. Nat Commun, 2017, 8: 14234.
- Raymond, D.D. , Bajic, G. ; Ferdman, J.; Suphaphiphat, P.; Settembre, E.C.; Moody, M.A.; Schmidt, A.G.; Harrison, S.C. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc Natl Acad Sci U S A, 2018, 115, 168–173. [Google Scholar]
- Henry Dunand, C.J.; Leon, P.E.; Huang, M.; Choi, A.; Chromikova, V.; Ho, I.Y.; Tan, G.S.; Cruz, J.; Hirsh, A.; Zheng, N.-Y.; et al. Both Neutralizing and Non-Neutralizing Human H7N9 Influenza Vaccine-Induced Monoclonal Antibodies Confer Protection. Cell Host Microbe, 2016, 19, 800–13. [Google Scholar] [CrossRef]
- Jia, M.; Zhao, H.; Morano, N.C.; Lu, H.; Lui, Y.-M.; Du, H.; Becker, J.E.; Yuen, K.-Y.; Ho, D.D; Kwong, P.D.; et al. Human neutralizing antibodies target a conserved lateral patch on H7N9 hemagglutinin head. Nature Communications, 2024, 15, 4505. [Google Scholar] [CrossRef]
- Dong, J.; Gilchuk, I.; Li, S.; Irving, R.; Goff, M.T.; Turner, H.L.; Ward, A.B.; Carnahan, R.H. ; Anti-influenza H7 human antibody targets antigenic site in hemagglutinin head domain interface. J Clin Invest, 2020, 130, 4734–4739. [Google Scholar] [CrossRef]
- Turner, H.L.; Pallesen, J.; Lang, S.; Bangaru, S.; Urata, S. ; Li; S. ; Cottrell, C.A.; Bowman, C.A.; Crowe, Jr., J.E.; Wilson, I.A.; et al. Potent anti-influenza H7 human monoclonal antibody induces separation of hemagglutinin receptor-binding head domains. PLoS Biol, 2019, 17, e3000139. [Google Scholar]
- Kohler, G. ; C. Milstein. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature, 1975, 256, 495–7. [Google Scholar]
- Vidarsson, G.; G. Dekkers, T.; Rispens. IgG Subclasses and Allotypes: From Structure to Effector Functions. Frontiers in Immunology, 2014, 5.
- Brezski, R.J. ; G. Georgiou. Immunoglobulin isotype knowledge and application to Fc engineering. Curr Opin Immunol, 2016, 40: 62-9.
- Tharakaraman, K.; Subramanian, V.; Cain, D.; Sasisekharan, V.; Sasisekharan, R. Broadly neutralizing influenza hemagglutinin stem-specific antibody CR8020 targets residues that are prone to escape due to host selection pressure. Cell Host Microbe, 2014, 15, 644–51. [Google Scholar] [CrossRef]
- Yi, K.S.; Choi, J.-A.; Kim, P.; Ryu, D.-K.; Yang, E.; Son, D.; Shin, J.; Park, H.; Lee, S.; Lee, H.; et al. Broader neutralization of CT-P27 against influenza A subtypes by combining two human monoclonal antibodies. PLOS ONE, 2020, 15, e0236172. [Google Scholar] [CrossRef]
- Kallewaard, N.L.; Corti, D.; Collins, P.J.; Neu, U.; McAuliffe, J.M.; Benjamin, E.; Wachter-Rosati, L.; Palmer-Hill, F.J.; Yuan, A.Q.; Walker, P.A.; et al. Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes. Cell, 2016, 166, 596–608. [Google Scholar] [CrossRef]
- Tharakaraman, K.; Subramanian, V.; Viswanathan, K.; Sloan, S.; Yen, H.-L.; Barnard, D.L.; Leung, Y.H.C.; Szretter, K.J.; Koch, T.J.; Delaney, J.C.; et al. A broadly neutralizing human monoclonal antibody is effective against H7N9. Proceedings of the National Academy of Sciences, 2015, 112, 10890–10895. [Google Scholar] [CrossRef]
- Izadi, A.; Hailu, A.; Godzwon, M.; Wrighton, S.; Olofsson, B.; Schmidt, T.; Söderlund-Strand, A.; Elder, E.; Appelberg, S.; Valsjö, M.; et al. Subclass-switched anti-spike IgG3 oligoclonal cocktails strongly enhance Fc-mediated opsonization. Proceedings of the National Academy of Sciences, 2023, 120(15).
- Bolton, M.J.; Arevalo, C.P.; Griesman, T. ;Li; S. H.; Bates, P.; Wilson, P.C.; Hensley, S.E. IgG3 subclass antibodies recognize antigenically drifted influenza viruses and SARS-CoV-2 variants through efficient bivalent binding. Proc Natl Acad Sci U S A, 2023, 120, e2216521120. [Google Scholar]
- Bowles, J.A.; Wang, S.-Y.; Link, B.K; Allan, B.; Beuerlein, G.; Campbell, M.-A.; Marquis, D.; Ondek, B.; Wooldridge, J.E.; Smith, B.J.; et al. Anti-CD20 monoclonal antibody with enhanced affinity for CD16 activates NK cells at lower concentrations and more effectively than rituximab. Blood, 2006, 108, 2648–2654. [Google Scholar] [CrossRef]
- Forero-Torres, A.; de Vos, S.; Pohlman, B.L.; Pashkevich, M.; Cronier, D.M.; Dang, N.H.; Carpenter, S.P.; Allan, B.W.; Nelson, J.G.; Slapak, C.A.; et al. Results of a Phase 1 Study of AME-133v (LY2469298), an Fc-Engineered Humanized Monoclonal Anti-CD20 Antibody, in FcγRIIIa-Genotyped Patients with Previously Treated Follicular Lymphoma. Clinical Cancer Res, 2012, 18, 1395–1403. [Google Scholar] [CrossRef]
- Van Der Horst, H.J.; Nijhof, I.S.; Mutis, T.; Chamuleau, M.E.D. Fc-Engineered Antibodies with Enhanced Fc-Effector Function for the Treatment of B-Cell Malignancies. Cancers, 2020, 12, 3041. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.; Park, S.; Sohn, M.H.; Jo, M.; Ko, B.J.; Na, J.-H.; Yoo, H.; Jeong, A.L.; Ha, K.; Woo, J.R.; et al. An Fc variant with two mutations confers prolonged serum half-life and enhanced effector functions on IgG antibodies. ExMol. Med., 2022, 54, 1850–1861. [Google Scholar] [CrossRef] [PubMed]
- Natsume, A.; In, M.; Takamura, H.; Nakagawa, T.; Shimizu, Y.; Kitajima, K.; Wakitani, M.; Ohta, S.; Satoh, M.; Shitara, K.; et al. Engineered antibodies of IgG1/IgG3 mixed isotype with enhanced cytotoxic activities. Cancer Res, 2008, 68, 3863–72. [Google Scholar] [CrossRef]
- Chu, T.H. ; E. F. Patz; M.E. Ackerman. Coming together at the hinges: Therapeutic prospects of IgG3. mAbs, 2021, 13, 1882028. [Google Scholar]
- Ali, S.O. , et al., Takas, T.; Nyborg, A.; Shoemaker, K.; Kallewaard, N.L.; Chiong, R.; Dubovsky, F.; Mallory, R.M. Evaluation of MEDI8852, an Anti-Influenza A Monoclonal Antibody, in Treating Acute Uncomplicated Influenza. Antimicrob Agents Chemother, 2018, 62(11).
- Sloan, S.E.; Szretter, K.J.; Sundaresh, B.; Narayan, K.M.; Smith, P.F.; Skurnik, D.; Bedard, S.; Trevejo, J.M.; Oldach, D.; Shriver, Z. Clinical and virological responses to a broad-spectrum human monoclonal antibody in an influenza virus challenge study. Antiviral Res, 2020, 184: 104763.
- Hershberger, E.; Sloan, S.; Narayan, K.; Hay, C.A.; Smith, P.; Engler, F.; Jeeninga, R.; Smits, S.; Trevejo, J.; Shriver, Z.; et al. Safety and efficacy of monoclonal antibody VIS410 in adults with uncomplicated influenza A infection: Results from a randomized, double-blind, phase-2, placebo-controlled study. EBioMedicine, 2019, 40: 574-582.
- Lim, J.J.; Nilsson, A.C.; Silverman, M.; Assy, N.; Kulkarni, P.; McBride, J.M.; Deng, R.; Li, C.; Yang, X.; Nguyen, A.; et al. A Phase 2 Randomized, Double-Blind, Placebo-Controlled Trial of MHAA4549A, a Monoclonal Antibody, plus Oseltamivir in Patients Hospitalized with Severe Influenza A Virus Infection. Antimicrob Agents Chemother, 2020, 64(7).
- Han, A.; Czajkowski, L; Rosas, L.A.; Cervantes-Medina, A.; Xiao, Y.; Gouzoulis, M.; Lumbard, K.; Hunsberger, S.; Reed, S.; Athota, R.; et al. Safety and Efficacy of CR6261 in an Influenza A H1N1 Healthy Human Challenge Model. Clinical Infectious Diseases. 2021, 73, e4260–e4268.
- Impagliazzo, A.; Milder, F.; Kuipers, H.; Wagner, M.V.; Zhu, X.; Hoffman, R.M.B.; van Meersbergen, R.; Huizingh, J.; Wanningen, P.; Verspuij, J.; et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science, 2015, 349, 1301–1306. [Google Scholar] [CrossRef]
- Van Der Lubbe, J.E.M.; Huizingh, J.; Verspuij, J.W.A.; Tettero, L.; Schmit-Tillemans, S.P.R.; Mooij, P.; Mortier, D.; Koopman, G.; Bogers, W.M.J.M.; et al. , Mini-hemagglutinin vaccination induces cross-reactive antibodies in pre-exposed NHP that protect mice against lethal influenza challenge. NPJ Vaccines, 2018, 3(1).
- Folschweiller, N.; Abeele, C.V.; Chu, L.; Van Damme, P.; García-Sastre, A.; Krammer, F.; Nachbagauer, R.; Palese, P; Solórzano, A.; Bi, D.; et al. Reactogenicity, safety, and immunogenicity of chimeric haemagglutinin influenza split-virion vaccines, adjuvanted with AS01 or AS03 or non-adjuvanted: a phase 1-2 randomised controlled trial. Lancet Infect Dis. 2022, 22, 1062–1075.
- Nishiyama, A.; Adachi, Y.; Tonouchi, K.; Moriyama, S.; Sun, L.; Aoki, M.; Asanuma, H.; Shirakura, M.; Fukushima, A.; Yamamoto, T.; et al. Post-fusion influenza vaccine adjuvanted with SA-2 confers heterologous protection via Th1-polarized, non-neutralizing antibody responses. Vaccine, 2023, 41, 4525–4533. [Google Scholar] [CrossRef]
- Atmar, R.L.; Bernstein, D.I.; Winokur, P.; Frey, S.E.; Angelo, L.S.; Bryant, C. ; Ben-Yedida-T. ; Roberts, P.C.; El Sahly, H.M.; Keitel, W.A. Safety and immunogenicity of Multimeric-001 (M-001) followed by seasonal quadrivalent inactivated influenza vaccine in young adults - A randomized clinical trial. Vaccine, 2023, 41, 2716–2722. [Google Scholar]
- Corbett, K.S.; Moin, S.M.; Yassine, H.M.; Cagigi, A.; Kanekiyo, M.; Boyoglu-Barnum, S.; Myers, S.I.; Tsybovsky, Y.; Wheatley, A.K.; Schramm, C.A.; et al. Design of Nanoparticulate Group 2 Influenza Virus Hemagglutinin Stem Antigens That Activate Unmutated Ancestor B Cell Receptors of Broadly Neutralizing Antibody Lineages. mBio, 2019, 10(1).
- Lee, P.S.; Yoshida, R.; Ekiert, D.C.; Sakai, N.; Suzuki, Y.; Takada, A.; Wilson, I.A. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci U S A, 2012, 109, 17040–5. [Google Scholar] [CrossRef]
- Ekiert, D.C.; Kashyap, A.K.; Steel, J.; Rubrum, A.; Bhabha, G.; Khayat, R.; Lee, J.H.; Dillon, M.A.; O'Neil, R.E.; Faynboym, A.M.; et al. Cross-neutralization of influenza A viruses mediated by a single antibody looNature, 2012, 489, 526-32.
- Sarah, F. Andrews, Y. H., Kaval Kaur, Lyubov I. Popova, Irvin Y. Ho, Noel T. Pauli, Carole J. Henry Dunand, William M Taylor, Samuel Lim, Min Huang, Xinyan Qu, Jane-Hwei Lee, Marlene Salgado-Ferrer, Florian Krammer, Peter Palese, Jens Wrammert, Rafi Ahmed, and Patrick C. Wilson. Immune history profoundly affects broadly protective B cell responses to influenza. Science Translational Medicine, 2015, 7, 316ra192. [Google Scholar]
- Bajic, G.; van der Poel, C.E.; Kuraoka, M.; Schmidt, A.G.; Carroll, M.C.; Kelsoe, G.; Harrison, S.C. Autoreactivity profiles of influenza hemagglutinin broadly neutralizing antibodies. Scientific Reports, 2019, 9(1).
- Willey, S.; Aasa-Chapman, M.M.I.; O'Farrell, S.; Pellegrino, P.; Williams, I.; Weiss, R.A.; Neil, S.J.D. Extensive complement-dependent enhancement of HIV-1 by autologous non-neutralising antibodies at early stages of infection. Retrovirology, 2011, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Monsalvo, A.C.; Batalle, J.P.; Lopez, M.F.; Krause, J.C.; Klemenc, J.; Hernandez, J.Z.; Maskin, B.; Bugna, J.; Rubinstein, C.; Aguilar, L.; et al. Severe pandemic 2009 H1N1 influenza disease due to pathogenic immune complexes. Nature Medicine, 2011, 17, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Wheatley, A.K.; Kent, S.J.; DeKosky, B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol., 2020, 5, 1185–1191. [Google Scholar] [CrossRef] [PubMed]
- Rao, G.K.; Prell, R.A.; Laing, S.T.; Burleson, S.C.M.; Nguyen, A.; McBride, J.M.; Zhang, C.; Sheinson, D.; Halpern, W.G. In Vivo Assessment of Antibody-Dependent Enhancement of Influenza B Infection. Toxicol Sci, 2019, 169, 409–421. [Google Scholar] [CrossRef]
- Winarski, K.L.; Tang, J.; Klenow, L.; Lee, J.; Coyle, E.M.; Manischewitz, J.; Turner, H.L.; Takeda, K.; Ward, A.B.; Golding, H.; et al. Antibody-dependent enhancement of influenza disease promoted by increase in hemagglutinin stem flexibility and virus fusion kinetics. Proc Natl Acad Sci U S A, 2019, 116, 15194–15199. [Google Scholar] [CrossRef]
| Name | In vitro Binding | In vitro Neutralisation | In vivo Protection | Germline Encoded IGHV | CDR Recognition Mode | ADCC activity | Source | Escape Mutants | IgG-type in Studies | Ref | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Central Stem | C179 | H1, H2, H5, H6, H9 | H1, H2, H5, H6, H9 | H1, H5 | - | - | Yes | Mouse | T332K, V395E * |
IgG2a | [9,27,28] | ||
| 27F3 | H1, H2, H5, H6, H9, H11, H12, H13, H16, H3, H7, H10, FluB | H1, H5, H6, H3, H7, H10 | - | IGHV1–69 | CDRH2 | - | Humans | - | IgG1 | [29,30] | |||
| FI6 | H1-H16 | H1, H5, H3, H7 | H1, H5, H3 | IGHV3–30 | CDRH3 CDRL1 |
Yes | Humans | R62K, D239G, R240Q T333K, A388T ° | - | [21,22,23,31,32] | |||
| CR6261 | H1, H2, H5, H6, H8, H9 | H1, H2, H5, H6, H8, H9 | H1, H5 | IGHV1–69 | CDRH2 | Weak | Humans | A388V | IgG1 | [18,30,33,34,35] | |||
| CR6323 | H1, H2, H5, H6, H8, H9 | H1, H2, H5, H6, H8, H9 | - | IGHV1–69 | HCDR2 | - | Humans | H357L/T* | IgG1 | [34] | |||
| 09-2A06 | H1 | H1 | - | IGHV1–69 | - | - | Humans | - | - | [36] | |||
| 09-3A01 | H1 | H1 | - | IGHV4–39 | - | - | Humans | - | - | ||||
| 05-2G02 | H1, H3, H5 | H1, H3, H5 | - | IGHV1–18 | - | - | Humans | - | - | ||||
| A06 | H1, H5 | H1, H5 | H1 | IGHV1–69 | - | - | Humans | - | IgG1 | [37] | |||
| 39.18 | H1, H2 | H1, H2 | - | IGHV1–69 | - | - | Humans | - | - | [38,39] | |||
| 39.29 | H1, H2, H3 | H1, H2, H3 | H1, H3 | IGHV3-30 | CDRH3 | - | Humans | G387K, D391Y/G | - | ||||
| 81.39 | H1, H2, H3 | H1, H2, H3 | - | IGHV3-15 | - | - | Humans | - | - | ||||
| 36.89 | H3 | H3 | - | IGHV1–18 | - | - | Humans | - | - | ||||
| FE43 | H1, H5, H6, H9 | H1, H5, H6, H9 | H1, H5, H6 | IGHV1–69 | - | - | Humans | None found | IgG1 | [40] | |||
| FB110 | H1, H2, H5 | H1, H2, H5 | - | IGHV3-23 | - | - | Humans | None found | IgG3 | ||||
| 3Е1 | H1, H5, H9, H3, H7 | H1, H5, H9, H3, H7 | H1, H5 | IGHV4-4 | Mostly Heavy Chain | - | Humans | - | IgG1 | [41] | |||
| CT149 | H1, H5, H9, H3, H7 | H5, H9, H3, H7 | H1, H5, H3, H7 | IGHV1–18 | CDRH3CDRH2 | Yes | Humans | - | IgG1 | [42] | |||
| 31.a.83 | H1, H2, H5, H9, H3, H7 | H1, H2, H5, H9, H3, H7 | - | IGHV3–23 | Mostly CDRH3 CDRH2 |
- | Humans | - | - | [43] | |||
| 56.a.09 | H1, H5, H3, H7 | H1, H5, H3, H7 | - | IGHV6–1 | Mostly CDRH3 CDRH2 |
- | Humans | - | - | ||||
| CR9114 | H1, H2, H5, H6, H8, H9, H12, H13, H16, H3, H4, H7, H10, H15, FluB | H1, H2, H5, H6, H8, H9, H12, H3, H4, H7, H10 | H1, H2, H3, H5, H9, FluB | IGHV1–69 | CDRH2 | Weak | Humans | R62K, D239G, R240Q, L335V, D363G, A388T ° | IgG1 | [30,31,33,44,45] | |||
| F10 | H1, H2, H5, H6, H8, H9, H11, H13, H16 | H1, H2, H5, H6, H8, H9, H11 | H1, H5 | IGHV1–69 | CDRH2 | Yes | Humans | N460, S123, E190D+G225D, N203VHA + E329KNA* | IgG1 | [19,30,32,46] | |||
| MEDI8852 | H1-H18 | H1, H2, H5, H6, H9, H3, H7 | H1, H5, H3 | IGHV6-1 | CDRH2 CDRH3 CDRL1 |
Yes | Humans | - | IgG1 | [47,48] | |||
| CR9117 | Mouse homologue of CR9114, presumed to have similar neutralization capacity | - | Yes | Mouse | - | IgG2a | [33] | ||||||
| Anchor Domain | Polyclonal response (FISW84 / 222-1C06 were named) | H1, H2, H5 | H1, H2, H5 | H1 | IGHV3-23 IGHV3-30 IGHV3-30-3 IGHV3-48 |
CDRk3 CDRH2 CDRH3 |
No | Humans | - | IgG1 | [26] | ||
| Fusion Peptide | CR8020 | H3, H4, H7, H10, H14, H15 | H3, H7, H10 | H3, H7 | IGHV1–18 | CDRH1 CDRH3 |
Weak | Humans | D372N, G376E * | IgG1 | [20,25,49,50] | ||
| CR8043 | H3, H4, H7, H10, H14, H15 | H3, H7, H10 | H3, H7 | IGHV1–3 | CDRH1 CDRH3 |
- | Humans | R378M, Q380R/T * | IgG1 | [25,50] | |||
| 9H10 | H3, H9 | H3, H10 | H3 | - | - | - | Mice | R378M T385R Q387R/T G386E * |
- | [50] | |||
| Name | Type and Target | Dosage/ Infection Model | Result | Trial Registry ID/ Reference |
|---|---|---|---|---|
| CT-P27 | CT-120 & CT-149 mAb’s targeting the stem region of group 1 and group 2 influenza hemagglutinin | 10 mg/kg CT-P27, 20 mg/kg CT-P27, or placebo in an influenza challenge model | Reduction of AUC of Viral Load, as measured by Quantitative PCR of Nasopharyngeal Swab for patients who received CT-P27 | NCT02071914, [105] |
| 90 mg/kg CT-P27, 45 mg/kg CT-P27, or placebo | NCT03511066 was terminated due to CT-P27 inactivation | NCT03511066. | ||
| MEDI8852 | Human IgG1 kappa monoclonal antibody (MAb) targeting H1N1 and H3N2 viruses, as well as subtypes such as H2, H5, H6, H7, and H9 via the stem region | 750 mg or 3000mg MEDI8852 given with oseltamivir or 3000 mg MEDI8852 on its own to patients with acute, uncomplicated influenza caused by Type A strains. | MEDI8852 provided no statistically significant improvement over oseltamivir alone, potentially worsened disease in combination compared to oseltamivir alone | NCT02603952,[116] |
| Low dose and high dose of MEDI8852 and oseltamivir in comparison to oseltamivir and placebo | Withdrawn due to company decision | NCT03028909 | ||
| VIS410 | Human immunoglobulin IgG1 monoclonal antibody engineered to bind to the stem region of group 1 and 2 influenza A hemagglutinins | Influenza challenge with H1N1 followed by a single administration of VIS410 or placebo | No results posted | NCT02468115, [117] |
| 2000mg or 4000mg VIS410 was given to patients with uncomplicated influenza A infection and compared to a placebo | Statistically significant improvement in signs and symptoms of influenza infection on day 3 and 4 with VIS410 compared to placebo. Statistically significant reduction in time to resolution of peak viral load when patients were given VIS410. | NCT02989194, [118] | ||
| 3600 mg or 8400 mg VIS410 combined with oral oseltamivir or placebo with oseltamivir in patients hospitalised with influenza A infection | No statistically significant reduction in time to cessation of oxygen, or reduction of viral load in nasopharyngeal samples | NCT03040141 | ||
| MHAA4549A | Human monoclonal antibody, IgG1, targeting the influenza A virus hemagglutinin stem across multiple subtypes | Influenza challenge with H3N2 influenza virus followed by a dose of 400 mg, 1200 mg or 3600 mg | Statistically significant reduction in AUC of virus in nasopharyngeal samples was seen at 3600mg compared to placebo. Influenza symptom scores, mucus weight, and inflammatory biomarkers were also reduced. | NCT01980966 |
| 3600mg or 8400mg given either on its own or with oseltamivir to patients hospitalised with severe influenza infection | MHAA4549A did not improve clinical outcomes over OTV alone. MHAA4549A+OTV did not further reduce viral load versus placebo+OTV.MHAA4549A did not alleviate symptoms quicker than a placebo. | NCT02293863, [119] | ||
| 3600mg or 8400mg given to patients with uncomplicated seasonal influenza A infection | 3600mg dose was able to statistically reduce the number of days to alleviate symptoms compared to the control | NCT02623322 | ||
| CR8020 | A mAb targeting the stem region of group 2 influenza A hemagglutinin | 15 mg/kg CR8020 given before challenge with a H3N2 influenza virus. | No results | NCT01938352 |
| CR6261 | mAb that targets the stem region of group 1 and group 2 influenza hemagglutinin | 50 mg/kg administered one day after challenge with H1N1 | Statistically reduced percentage of participants who experienced influenza symptoms. No statistically significant reduction in AUC or viral shedding. | NCT02371668, [120] |
| CR8020/ CR6261 | Withdrawn due to preliminary efficacy results from an influenza challenge trial | NCT01992276 |
| Phase | Name of vaccine | Target/ Type of vaccine | Dosage/ Infection model | Results | Trial Registry ID/ Reference |
|---|---|---|---|---|---|
| Recruiting | fH1/DSP-0546LP | Post-fusion hemagglutinin antigen | Combination of 2 dose levels of fH1 (2 and 8 μg), 3 dose levels of DSP-0546LP (2.5, 5, and 10 μg), and placebo. Each dose level of fH1 will be combined with the low, medium, and high dose level of DSP-0546LP to assess safety, tolerability, and immunogenicity | Active | NCT06460064, [124] |
| Phase 1 | EBS-UFV-001 | Induction of antibodies against conserved stem antigens across group 1 and 2 via a hemagglutinin stabilized stem nanoparticle vaccine | Testing the safety, tolerability and immunogenicity of 20 µg or 60 µg of UFluA as single dose or as two dose | No results posted | NCT05155319, [126] |
| H1ssF | HA stem domain from Influenza A/New Caledonia/20/1999 (H1N1) genetically fused to the ferritin protein from H. pylori. | 20 mcg was given to group 1, group 2 received 60 mcg on a prime boost schedule. | All regimes generated an increased IC80 concentration when tested in a pseudoviral neutralization assay against the homologous H1N1 A/New Caledonia/20/99 virus | NCT03814720 | |
| GSK3816302A | Chimeric vaccines of D-SUIV cH8/1 N1, D-SUIV cH5/1 N1, and D-SUIV cH11/1 N1 to induce cross reactive stem targeting antibodies against H1 stem | Chimeric H5, H8 and H11 with and without adjuvants AS03 or AS01 were tested for their reactogenicity, safety and immunogenicity. H8 and H5 were given with a placebo second dose, or all three were given. | An increase in anti H1 stem antibodies, as measured by ELISA and MN assay, was seen across all dose schedules with adjuvant AS03 providing a statistically significant increase in humoral immune response for anti-H1 stem antibody by ELISA at Day 29 and Day 85. Increases in antibody titres against H2 and H18 were also identified. | NCT03275389, [123] | |
| Phase 1/2 | G1 mHA | Mini-hemagglutinin stem-derived protein vaccine antigen | Single dose of influenza G1 mHA with or without Al(OH)3 adjuvant at two dose levels to evaluate safety, reactogenicity and immunogenicity | Active | NCT05901636, [121,122] |
| Phase 3 | (M-001) | A recombinant 45 kDa protein produced in Escherichia coli. consisting of three repetitions of nine linear, conserved influenza A and B epitopes to form a single recombinant protein. Epitopes were derived from: M1 matrix protein, NP and HA | Vaccination with 1mg dose of M-001 twice: Once at Day 0, and once at Day 21 then followed for 2 years | No statistical difference in prevention of influenza infection. Did not statistically reduce the number of patients with influenza like symptoms, or a reduction of severity of either qRT-PCR or culture-confirmed influenza illness | NCT03450915, [125] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




