Submitted:
10 October 2024
Posted:
11 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Socio-Demographic Data
2.2. Anthropometric Evaluation
2.3. Biochemical Evaluation
2.4. Dietary Evaluation
2.5. Microbiota Analysis
|
Control Group |
Nseqsa | Coverageb (%) | OTUc |
Chao1 (lcid-hcie) |
Inv-Simpson (lcid-hcie) |
Shannon (lcid-hcie) |
Pielou |
| CL-NW (n = 12) | |||||||
| M15 | 25575 | 99.75 | 278 | 412 (357-520) | 6 (5.94-6.12) | 2.81 (2.8-2.83) | 0.993 |
| M45 | 55419 | 99.82 | 437 | 594 (572-638) | 6.27 (6.21-6.34) | 4.75 (4.72-4.79) | 0.991 |
| M13 | 58542 | 99.86 | 612 | 857 (795-97) | 4.74 (4.70-4.78) | 3.45 (3.44-3.47) | 0.994 |
| M23 | 57984 | 99.86 | 1029 | 1297 (1245-1391) | 6.44 (6.41-6.47) | 4.44 (4.43-4.45) | 0.998 |
| M29 | 98637 | 99.86 | 906 | 1264 (1162-1374) | 5.29 (5.26-5.31) | 4.12 (4.11-4.13) | 0.997 |
| M32 | 56648 | 99.86 | 1125 | 1468 (1436-1666) | 4.39 (4.35-4.44) | 3.10 (3.09-3.11) | 0.998 |
| M33 | 45032 | 99.88 | 832 | 1160 (1059-1363) | 5.73 (5.71-5.75) | 4.60 (4.59-4.61) | 0.997 |
| M34 | 64036 | 99.83 | 1081 | 1396 (1344-1508) | 6.55 (6.5-6.6) | 4.99 (4.98-5) | 0.996 |
| M37 | 50042 | 99.86 | 458 | 597 (505-641) | 4.66 (4.62-4.71) | 2.46 (2.45-2.47) | 0.996 |
| M38 | 5021 | 99.66 | 1191 | 1490 (1488-1688) | 6.52 (6.38-6.67) | 4.86 (4.82-4.87) | 0.998 |
| M43 | 46370 | 99.87 | 883 | 1239 (1217-1288) | 6.03 (5.98-6.09) | 4.60 (4.58-4.61) | 0.997 |
| M46 | 39738 | 99.83 | 950 | 1290 (1188-1334) | 6.32 (6.23-6.40) | 4.55 (4.53-4.56) | 0.998 |
| CL-OW (n = 8) | |||||||
| M48 | 6333 | 99.82 | 1146 | 1503 (1471-1704) | 5.59 (5.51-5.68) | 4.75 (4.73-4.76) | 0.997 |
| M49 | 14430 | 99.83 | 998 | 1330 (1318-1434) | 5.41 (5.36-5.45) | 4.85 (4.84-4.88) | 0.994 |
| M6 | 8343 | 99.75 | 1037 | 1345 (1253-1410) | 5.71 (5.63-5.8) | 4.84 (4.82-4.86) | 0.997 |
| M16 | 59311 | 99.85 | 681 | 908 (849-1009) | 5.99 (5.95-6) | 4.9 (4.8-5) | 0.979 |
| M17 | 52056 | 99.84 | 1015 | 1332 (1267-1450) | 4.87 (4.82-4.92) | 3.43 (3.42-3.44) | 0.997 |
| M19 | 40529 | 99.81 | 861 | 1128 (1072-1229) | 6.14 (6-6.2) | 4.75 (4.74-4.76) | 0.999 |
| M25 | 64872 | 99.88 | 822 | 1091 (1080-1202) | 5.43 (5.41-5.45) | 4.59 (4.58-4.61) | 0.778 |
| M30 | 21765 | 99.74 | 625 | 884 (872-919) | 4.94 (4.86-5) | 2.14 (2.13-2.16) | 0.992 |
|
T2DM Group |
Nseqsa | Coverageb (%) | OTUc |
Chao1 (lcid-hcie) |
Inv-Simpson (lcid-hcie) |
Shannon (lcid-hcie) |
Pielou |
| T2DM-NW (n = 12) | |||||||
| DM5 | 44662 | 99.86 | 715 | 944 (897-1037) | 5.65 (5.6-5.7) | 4.3 (4.29-4.32) | 0.995 |
| DM38 | 28103 | 99.84 | 1130 | 1471 (1431-1559) | 5.62 (5.54-5.7) | 4 (4.05-4.08) | 0.995 |
| DM1 | 42462 | 99.81 | 703 | 890 (830-998) | 6.24 (6.17-6.32) | 3.47 (3.46-3.48) | 0.997 |
| DM2 | 63930 | 99.84 | 1047 | 1346 (1276-1454) | 5.98 (5.92-6) | 3.53 (3.52-3.54) | 0.997 |
| DM6 | 51875 | 99.86 | 879 | 1172 (1125-1257) | 3.77 (3.74-3.8) | 3 (3-3.03) | 0.995 |
| DM8 | 53503 | 99.85 | 790 | 1057 (993-1175) | 4.27 (4.22-4.31) | 3.89 (3.88-3.9) | 0.997 |
| DM9 | 63090 | 99.82 | 908 | 1168 (1090-1299) | 4.62 (4.56-4.68) | 4.53 (4.52-4.54) | 0.999 |
| DM17 | 37353 | 99.83 | 1187 | 1517 (1460-1629) | 6.53 (6.46-6.61) | 4.87 (4.86-4.88) | 0.998 |
| DM19 | 10255 | 99.74 | 738 | 1097 (1073-1161) | 4.72 (4.6-4.86) | 3.95 (3.92-3.97) | 0.994 |
| DM27 | 33597 | 99.79 | 680 | 943 (866-1093) | 7.1 (7-7.19) | 5 (5.01-5.03) | 0.998 |
| DM50 | 40129 | 99.82 | 931 | 1278 (1203-1425) | 6.95 (6.88-7) | 4 (4.06-4.08) | 0.998 |
| DM51 | 38123 | 99.81 | 799 | 1056 (981-1198) | 6.64 (6.59-6.7) | 4.48 (4.47-5) | 0.894 |
| T2DM-OW (n = 8) | |||||||
| DM16 | 46862 | 99.84 | 1009 | 1328 (1275-1425) | 5.37 (5.32-5.43) | 4.82 (4.81-4.83) | 0.999 |
| DM20 | 67321 | 99.86 | 799 | 1125 (1052-1253) | 4.87 (4.84-4.9) | 3.33 (3.32-3.34) | 0.996 |
| DM21 | 58523 | 99.85 | 1250 | 1609 (1531-1749) | 6.23 (6.18-6.28) | 5.23 (5.22-5.24) | 0.997 |
| DM22 | 58044 | 99.87 | 1011 | 1390 (1333-1495) | 4.85 (4.81-4.9) | 4 (4.06-4.08) | 0.997 |
| DM23 | 39135 | 99.84 | 981 | 1270 (1220-1368) | 5.57 (5.52-5.63) | 4.75 (4.74-4.77) | 0.996 |
| DM28 | 37674 | 99.85 | 922 | 1366 (1327-1443) | 4.75 (4.69-4.82) | 2.92 (2.9-2.93) | 0.996 |
| DM42 | 18130 | 99.74 | 933 | 1228 (1166-1367) | 5.14 (5-5.25) | 3.84 (3.82-3.86) | 0.994 |
| DM42 | 71836 | 99.86 | 646 | 879 (809-1000) | 5.96 (5.92-6) | 4 (4.04-4.06) | 0.998 |
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Study Design
4.3. Study Subjects
4.4. Anthropometric Evaluation
4.5. Determination of Biochemical Profile
4.6. Dietary Evaluation and Patient Categorization
4.7. Collection of Feces
4.8. Analysis of the Fecal Microbiota
4.10. Illumina Sequencing by Amplicons of the V4-V5 Region of the 16s rRNA Gene
4.11. Bioinformatic Analysis of the Sequences
4.12. Statistical analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- NORMA Oficial Mexicana 015-SSA2-2010, Para la prevención, tratamiento y control de la diabetes mellitus. Secretaria de Gobernación. Diario Oficial de la Federación. Diabetes Tipo 2. Available from: http://www.salud.gob.mx/unidades/cdi/nom/m015ssa24.html (Update of January 20, 2022).
- Zimmet, P.; Alberti, K.G.; Magliano, D.J.; Bennett, P.H. Diabetes mellitus statistics on prevalence and mortality: facts and fallacies. Nat. Rev. Endocrinol. 2016, 12, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Instituto Nacional de Estadística y Geografía (INEGI) (2015). Estadísticas de mortalidad 2015. Consulta interactiva de datos. Available from: https://www.inegi.org.mx/contenidos/programas/enasem/2018/doc/enasem_2018_presentacion.pdf. (Update of January 20, 2022).
- Zheng, Y.; Ley, S.H.; Hu, F.B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat. Rev. Endocrinol. 2018, 14, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef]
- Candela, M.; Biagi, E.; Soverini, M.; Consolandi, C.; Quercia, S.; Severgnini, M.; Peano, C.; Turroni, S.; Rampelli, S.; Pozzilli, P.; et al. Modulation of gut microbiota dysbioses in type 2 diabetic patients by macrobiotic Ma-Pi 2 diet. Br. J. Nutr. 2016, 116, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Wen, L.; Duffy, A. Factors Influencing the Gut Microbiota, Inflammation, and Type 2 Diabetes. J. Nutr. 2017, 147, 1468S–1475S. [Google Scholar] [CrossRef]
- Harsch, I.A.; Konturek, P.C. The Role of Gut Microbiota in Obesity and Type 2 and Type 1 Diabetes Mellitus: New Insights into “Old” Diseases. Med Sci. 2018, 6, 32. [Google Scholar] [CrossRef]
- Bridgewater, L.C.; Zhang, C.; Wu, Y.; Hu, W.; Zhang, Q.; Wang, J.; Li, S.; Zhao, L. Gender-based differences in host behavior and gut microbiota composition in response to high fat diet and stress in a mouse model. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Rinninella, E.; Cintoni, M.; Raoul, P.; Lopetuso, L.R.; Scaldaferri, F.; Pulcini, G.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. Food Components and Dietary Habits: Keys for a Healthy Gut Microbiota Composition. Nutrients 2019, 11, 2393. [Google Scholar] [CrossRef]
- Muralidharan, J.; Galiè, S.; Hernández-Alonso, P.; Bulló, M.; Salas-Salvadó, J. Plant-Based Fat, Dietary Patterns Rich in Vegetable Fat and Gut Microbiota Modulation. Front. Nutr. 2019, 6, 157. [Google Scholar] [CrossRef]
- US Department of Health and Human Services and US Department of Agriculture. 2015–2020 Dietary Guidelines for Americans. 8th ed. December 2015. Available online: http://health.gov/dietaryguidelines/2015/guidelines/ (accessed on 21 March 2019).
- Townsend GE 2nd, Han W ND 3rd, V Raghavan, NA Barry, AL Goodman, EA Groisman. Dietary sugar silences a colonization factor in a mammalian gut symbiont. Proc Natl Acad Sci 2019; 116 (1):233-238.
- Sonnenburg, E.D.; Smits, S.A.; Tikhonov, M.; Higginbottom, S.K.; Wingreen, N.S.; Sonnenburg, J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature 2016, 529, 212–215. [Google Scholar] [CrossRef] [PubMed]
- World Medical Association [WMA], Declaration of Helsinki Ethical Principles for Medical Research Involving Human Subjects. Clin Rev Ed. 2013;310(20):2191-2194. Available from: https://www.wma.net/wp-content/uploads/2016/11/DoH-Oct2013-JAMA.pdf (Update of , 2022). 20 January.
- AB Pérez-Lizaur, B Palacios-González, AL Castro-Becerra, I Flores-Galicia, Sistema Mexicano de Alimentos Equivalentes, 4th ed. México: Porrúa; 2014. P168.
- Chávez-Carbajal, A.; Pizano-Zárate, M.L.; Hernández-Quiroz, F.; Ortiz-Luna, G.F.; Morales-Hernández, R.M.; De Sales-Millán, A.; Hernández-Trejo, M.; García-Vite, A.; Beltrán-Lagunes, L.; Hoyo-Vadillo, C.; et al. Characterization of the Gut Microbiota of Individuals at Different T2D Stages Reveals a Complex Relationship with the Host. Microorganisms 2020, 8, 94. [Google Scholar] [CrossRef]
- Akkermans ADL, van Elsas JD, de Bruijn F. Manual molecular microbial ecology. 1rst ed. eBook: Springer, Kluwer Academic Publishers, Dordrecht; 1996. Chapter 1.4.4, Ramírez-Saad H, Akkermans WM, Akkermans ADL. DNA extraction from actinorhizal nodules; p1-11.
- Aguirre-Garrido, J.F.; Ramírez-Saad, H.C.; Toro, N.; Martínez-Abarca, F. Bacterial Diversity in the Soda Saline Crater Lake from Isabel Island, Mexico. Microb. Ecol. 2016, 71, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Marañón, M.; Miralles, I.; Aguirre-Garrido, J.F.; Anguita-Maeso, M.; Millán, V.; Ortega, R.; García-Salcedo, J.A.; Martínez-Abarca, F.; Soriano, M. Changes in the soil bacterial community along a pedogenic gradient. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.A. Sugar-Sweetened and Artificially-Sweetened Beverages in Relation to Obesity Risk. Adv. Nutr. Int. Rev. J. 2014, 5, 797–808. [Google Scholar] [CrossRef]
- American Diabetes Association (ADA) (2019) Making sense of food labels. Available from:https://www.diabetes.org/nutrition/understanding-food-labels/making-sense-of-food-labels (Update of January 20, 2022).
- World Health Organization (WHO). Sugar intake for adults and children. 2015 WHO reference number: WHO/NMH/NHD/15.2. Available from: https://www.who.int/publications/i/item/9789241549028 (update of January 20, 2022).
- Murugesan, S.; Ulloa-Martínez, M.; Martínez-Rojano, H.; Galván-Rodríguez, F.M.; Miranda-Brito, C.; Romano, M.C.; Piña-Escobedo, A.; Pizano-Zárate, M.L.; Hoyo-Vadillo, C.; García-Mena, J. Study of the diversity and short-chain fatty acids production by the bacterial community in overweight and obese Mexican children. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1337–1346. [Google Scholar] [CrossRef]
- Leite, A.Z.; Rodrigues, N.d.C.; Gonzaga, M.I.; Paiolo, J.C.C.; de Souza, C.A.; Stefanutto, N.A.V.; Omori, W.P.; Pinheiro, D.G.; Brisotti, J.L.; Junior, E.M.; et al. Detection of Increased Plasma Interleukin-6 Levels and Prevalence of Prevotella copri and Bacteroides vulgatus in the Feces of Type 2 Diabetes Patients. Front. Immunol. 2017, 8, 1107. [Google Scholar] [CrossRef]
- MetaHIT consortium; Forslund, K. ; Hildebrand, F.; Nielsen, T.; Falony, G.; Le Chatelier, E.; Sunagawa, S.; Prifti, E.; Vieira-Silva, S.; Gudmundsdottir, V.; et al. Erratum: Corrigendum: Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2017, 545, 116–116. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An Obesity-Associated Gut Microbiome with Increased Capacity for Energy Harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Ahmad, A.; Yang, W.; Chen, G.; Shafiq, M.; Javed, S.; Zaidi, S.S.A.; Shahid, R.; Liu, C.; Bokhari, H. Analysis of gut microbiota of obese individuals with type 2 diabetes and healthy individuals. PLOS ONE 2019, 14, e0226372. [Google Scholar] [CrossRef] [PubMed]
- Larsen, N.; Vogensen, F.K.; Van Den Berg, F.W.J.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut Microbiota in Human Adults with Type 2 Diabetes Differs from Non-Diabetic Adults. PLoS ONE 2019, 5, e9085. [Google Scholar] [CrossRef] [PubMed]
- Lippert, K.; Kedenko, L.; Antonielli, L.; Kedenko, I.; Gemeier, C.; Leitner, M.; Kautzky-Willer, A.; Paulweber, B.; Hackl, E. Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Benef. Microbes 2017, 8, 545–556. [Google Scholar] [CrossRef]
- Chávez-Carbajal, A.; Nirmalkar, K.; Pérez-Lizaur, A.; Hernández-Quiroz, F.; Ramírez-Del-Alto, S.; García-Mena, J.; Hernández-Guerrero, C. Gut Microbiota and Predicted Metabolic Pathways in a Sample of Mexican Women Affected by Obesity and Obesity Plus Metabolic Syndrome. Int. J. Mol. Sci. 2019, 20, 438. [Google Scholar] [CrossRef]
- Barczynska, R.; Kapusniak, J.; Litwin, M.; Slizewska, K.; Szalecki, M. Dextrins from Maize Starch as Substances Activating the Growth of Bacteroidetes and Actinobacteria Simultaneously Inhibiting the Growth of Firmicutes, Responsible for the Occurrence of Obesity. Plant Foods Hum. Nutr. 2016, 71, 190–196. [Google Scholar] [CrossRef]
- Jang, C.; Hui, S.; Lu, W.; Cowan, A.J.; Morscher, R.J.; Lee, G.; Liu, W.; Tesz, G.J.; Birnbaum, M.J.; Rabinowitz, J.D. The Small Intestine Converts Dietary Fructose into Glucose and Organic Acids. Cell Metab. 2018, 27, 351–361. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Hamaguchi, M.; Kaji, A.; Sakai, R.; Osaka, T.; Inoue, R.; Kashiwagi, S.; Mizushima, K.; Uchiyama, K.; Takagi, T.; et al. Intake of sucrose affects gut dysbiosis in patients with type 2 diabetes. J. Diabetes Investig. 2020, 11, 1623–1634. [Google Scholar] [CrossRef] [PubMed]
- Maya-Lucas, O.; Murugesan, S.; Nirmalkar, K.; Alcaraz, L.D.; Hoyo-Vadillo, C.; Pizano-Zárate, M.L.; García-Mena, J. The gut microbiome of Mexican children affected by obesity. Anaerobe 2019, 55, 11–23. [Google Scholar] [CrossRef]
- Dong, T.S.; Mayer, E.A.; Osadchiy, V.; Chang, C.; Katzka, W.; Lagishetty, V.; Gonzalez, K.; Kalani, A.; Stains, J.; Jacobs, J.P.; et al. A Distinct Brain-Gut-Microbiome Profile Exists for Females with Obesity and Food Addiction. Obesity 2020, 28, 1477–1486. [Google Scholar] [CrossRef] [PubMed]
- Chevrot R, Carlotti A, Sopena V, Marchand P, Rosenfeld E. Megamonas rupellensis sp. nov., an anaerobe isolated from the caecum of a duck. Int J Syst Evol Microbiol 2008; 58(Pt 12): 2921–2914.
- Sakon H, Nagai F, Morotomi M, Tanaka R Sutterella parvirubra sp. nov. and Megamonas funiformis sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 2008; 58(Pt4):970–975.
- Wu X, Mac Han L, Nawaz M, Gao F, Zhang X, et.al. Molecular characterization of the fecal microbiota in patients with type II diabetes. Curr Microbiol 2010; 61(1):69–78.
- Anhê FF, Jensen BAH, Varin TV, Servant F, Van Blerk S, Richard D, & Schertzer JD. Type 2 diabetes influences bacterial tissue compartmentalization in human obesity. Nat Metab 2020; 2(3):233-242.
- Larsen, J.M. The immune response toPrevotellabacteria in chronic inflammatory disease. Immunology 2017, 151, 363–374. [Google Scholar] [CrossRef]
- Leite, A.Z.; Rodrigues, N.d.C.; Gonzaga, M.I.; Paiolo, J.C.C.; de Souza, C.A.; Stefanutto, N.A.V.; Omori, W.P.; Pinheiro, D.G.; Brisotti, J.L.; Junior, E.M.; et al. Detection of Increased Plasma Interleukin-6 Levels and Prevalence of Prevotella copri and Bacteroides vulgatus in the Feces of Type 2 Diabetes Patients. Front. Immunol. 2017, 8, 1107. [Google Scholar] [CrossRef]
- Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, et.al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018; 555(7695):210-215.
- Leylabadlo, H.E.; Ghotaslou, R.; Feizabadi, M.M.; Farajnia, S.; Moaddab, S.Y.; Ganbarov, K.; Khodadadi, E.; Tanomand, A.; Sheykhsaran, E.; Yousefi, B.; et al. The critical role of Faecalibacterium prausnitzii in human health: An overview. Microb. Pathog. 2020, 149, 104344. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Halder, C.V.; de Sousa Faria, A.V.; Andrade, S.S. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 643–648. [Google Scholar] [CrossRef]
- Lopez-Siles, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 2017, 11, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Tapia, M.; Martínez-Medina, J.; Tovar, A.R.; Torres, N. Natural and Artificial Sweeteners and High Fat Diet Modify Differential Taste Receptors, Insulin, and TLR4-Mediated Inflammatory Pathways in Adipose Tissues of Rats. Nutrients 2019, 11, 880. [Google Scholar] [CrossRef] [PubMed]
- Könner, A.C.; Brüning, J.C. Toll-like receptors: linking inflammation to metabolism. Trends Endocrinol. Metab. 2011, 22, 16–23. [Google Scholar] [CrossRef]
- Kasprzak-Drozd, K.; Oniszczuk, T.; Stasiak, M.; Oniszczuk, A. Beneficial Effects of Phenolic Compounds on Gut Microbiota and Metabolic Syndrome. Int. J. Mol. Sci. 2021, 22, 3715. [Google Scholar] [CrossRef]
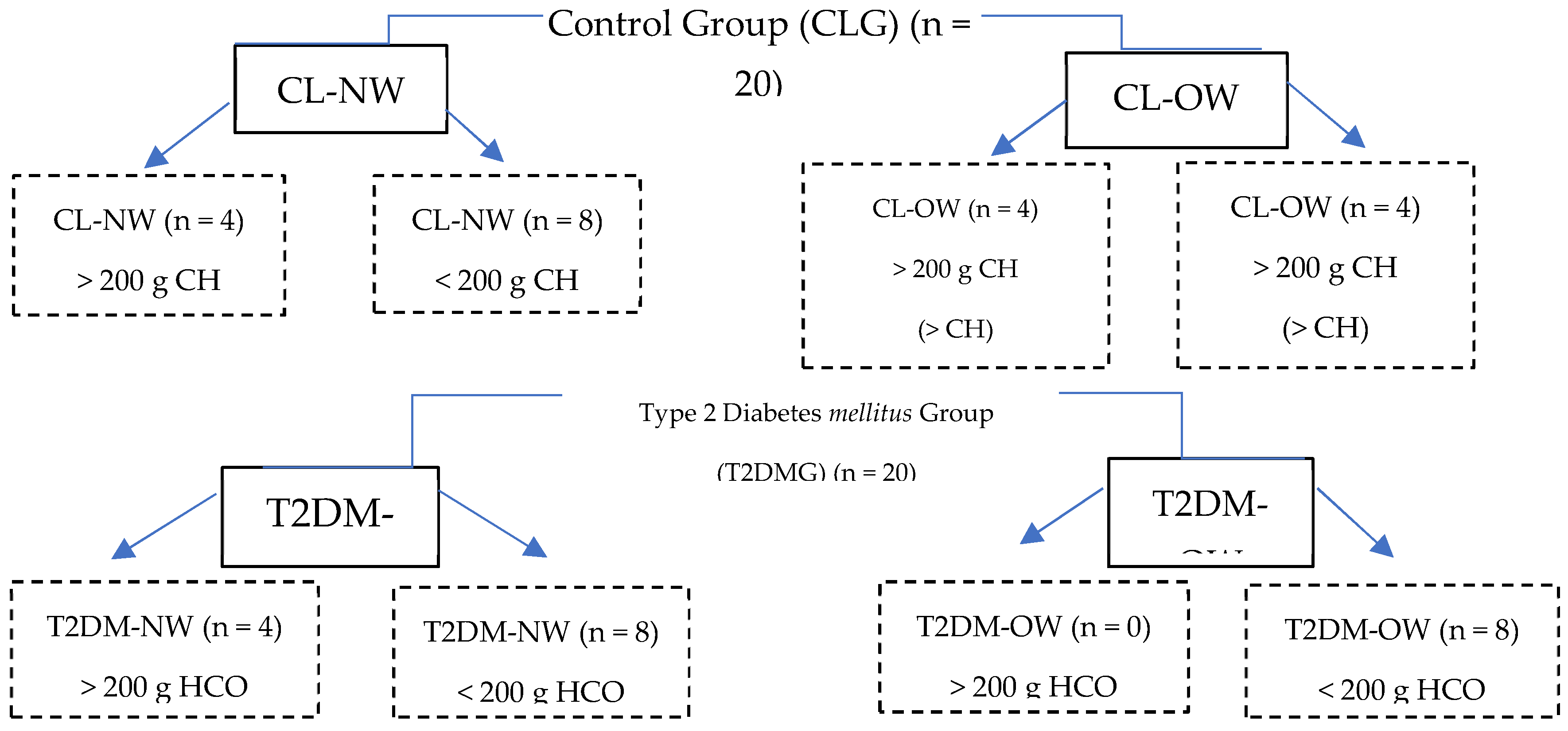
|
CL-NW n = 12 |
CL-OW n = 8 |
T2DM-NW n = 12 |
T2DM-OW n = 8 |
||
| Mean ±SD | Mean ±SD | Mean ±SD | Mean ±SD | P value | |
| Serum glycaemia | 81.1 ±1.7 | 83.3 ±5.9 | 150 ± 7.1** | 135 ±4.3** | 0.001* |
| Hb1Ac | 5.1 ±0.206 | 5.4 ±0.471 | 9.2 ±1.5** | 8.4 ±3.3** | 0.001* |
| TG | 128 ±3.6 | 155 ±8.3 | 208 ± 1.5 | 251 ±1.5 | 0.119 |
| CT | 175.2 ±2.3 | 174 ±2.4 | 195 ±6 | 192 ±3.9 | 0.519 |
|
CLNW n = 12 |
% |
CLOW n = 8 |
% |
T2DMNW n = 12 |
% |
T2DMOW n = 8 |
||
|
>200 g HCO |
Faecalibacterium Prevotella Roseburia Blautia Roseburia Bacteroides Less than 5% |
23.92 18.48 8.47 8.11 8.47 5.85 26.7 |
Faecalibacterium Bacteroides Prevotella Blautia Ruminococcus Parabacteroides Less than 5% |
23.3 21.23 15.23 11.09 7.5 4.83 16.82 |
Bacteroides Prevotella Faecalibacterium Roseburia Blautia Less than 5% |
29.75 14.19 10.13 8.47 6.82 30.64 |
No patient consumed more than 200 g of CHO per day, then we don’t have samples in this group. |
-- |
| n = 4 | n = 4 | n = 4 | n = 0 | -- | ||||
|
< 200 g HCO |
Faecalibacterium Bacteroides Prevotella Blautia Roseburia Streptococcus Less than 5% |
20.49 16.79 10.87 9.32 8.36 4.10 15.67 |
Faecalibacterium Roseburia Prevotella Megamones Bacteroides Blautia Less than 5% |
16.3 15.52 11.41 10.89 10.3 10.28 25.3 |
Faecalibacterium Prevotella Roseburia Bacteroides, Blautia Lachnospiracea_ incertae_sedis Less than 5% |
16.63 12.15 12.1 9.85 8.22 5.71 35.34 |
Prevotella Faecalobacterium Blautia Roseburia Less than 5% |
52.13 13.57 5.94 5.03 23.33 |
| n = 8 | n = 4 | n = 8 | n = 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





