Introduction
Enamel formation is an intricate process that proceeds in three major consecutive developmental stages: presecretory, secretory and maturation. Ameloblasts are highly specialized cells that dictate the progression of enamel development by altering their function and shape. In the presecretory stage, preameloblasts differentiate and extend cellular projections through the disintegrating basement membrane and into the underlying predentin [
1]. Once the dentin has formed near the ameloblasts, the secretory stage begins. Ameloblasts elongate, move away from the dentin as they secrete proteins and lengthen enamel ribbons that become crystallites. Each ameloblast secretes approximately 10,000 crystallites that will merge into a single enamel rod [
2]. The mineralized enamel rod embodies the migratory path of the moving ameloblasts, resulting in a decussating rod pattern in rodent incisors. Once the enamel layer has reached full thickness, mass secretion of the enamel proteins and movement of ameloblasts ceases as secretory ameloblasts transition into the maturation stage where, enamel matrix proteins are removed and enamel hardens into its final form [
3].
Matrix metalloproteinases are zinc-dependent endopeptidases that cleave extracellular matrix molecules. Matrix metalloproteinase 20 (MMP20; enamelysin) is a tooth specific proteinase essential for healthy enamel development as mutations in
MMP20 are known to cause enamel malformation [
4]. Due to its activity on cell-cell junction proteins, MMP20 was suggested to facilitate ameloblast cell migration [
5]. However, because of the complexity of ameloblast cell movement, other proteinases are likely to be involved.
A Disintegrin
And
Metalloproteinases (ADAMs) are a family of membrane-bound zinc-dependent proteinases that are classified as sheddases because they cleave/shed extracellular domains of membrane bound proteins [
6]. Previously, we reported a positive expression screen of six proteolytically active ADAM family members in the developing mouse enamel organ. Among the six ADAMs, expression of
Adam10 was observed in the apical loop containing the stem cells that become pre-ameloblasts and then ameloblasts as they migrate with the continuously erupting incisor. Adam10 expression persists throughout the secretory stage in rodent incisors. Strikingly, expression of
Adam10 predominantly ceases at the end of secretory stage when ameloblasts movement also ends [
7]. Notably ADAM10 sheds about a 100 substrates [
8], including cell-cell adhesion proteins [
9,
10] and ADAM10 can facilitate cell migration/invasion [
6], which makes it a candidate for facilitating ameloblast migration. Additionally, ADAM10 sheds cell signaling proteins from cell surfaces. Notch is the best characterized of these factors. However, Notch-1, -2 and -3 proteins are not expressed on pre-ameloblasts or ameloblasts [
11]. But, among the 100 substrates ADAM10 sheds [
8] and that pre-ameloblasts and ameloblasts express, are the hyaluronic acid receptor CD44 [
12], EGFR [
13], ERBB2 [
14], RELT [
15], and COL17A1 [
16,
17]. Therefore, ADAM10 is also a candidate for regulating cell signaling during enamel development.
A previous study [
18] used Keratin14
Cre/+;
Adam10fl/fl/ to demonstrate
Notch 1 downregulation in dental epithelium with subsequent loss of epithelial cell boundaries. However, over 95% of the Keratin14
Cre/+;
Adam10fl/fl/ mice die within the first 24 hours of life, which is likely due to perturbed skin barrier function causing transdermal water loss [
19] (pg 479). Since developing enamel defects result from as little stress as occurs with a fever [
20,
21], we generated an
Adam10 conditional knockout mouse using an amelogenin promoter-driven
Cre recombinase (
Amelx-i
Cre; Adam10fl/fl) that we demonstrate is only expressed in ameloblasts and therefore avoids the stress of lethal transdermal water loss associated with the Keratin14
Cre/+;
Adam10fl/fl/ mice. Provided here are detailed analyses of enamel formation when
Adam10 is ablated specifically from ameloblasts.
2. Results
2.1. Amelx-iCre Mice Express Cre Recombinase Only in Ameloblasts
Previously, five different
Amelx promoter driven Cre mouse lines were developed that expressed improved
Cre (i
Cre) recombinase within a large (250 kb) bacterial artificial chromosome DNA vector [
22]. After preliminary testing, we chose the Tg(
Amelx-i
Cre)872pap mice for further analyses by assessing the tissue specific expression of this i
Cre construct. We bred the
Amelx-i
Cre mice with ROSA
mTmG mice to generate
Amelx-i
Cre; mTmG mice. Without
Cre recombination, these mice express tdTomato (mT) that generates red fluorescence in tissues. When the
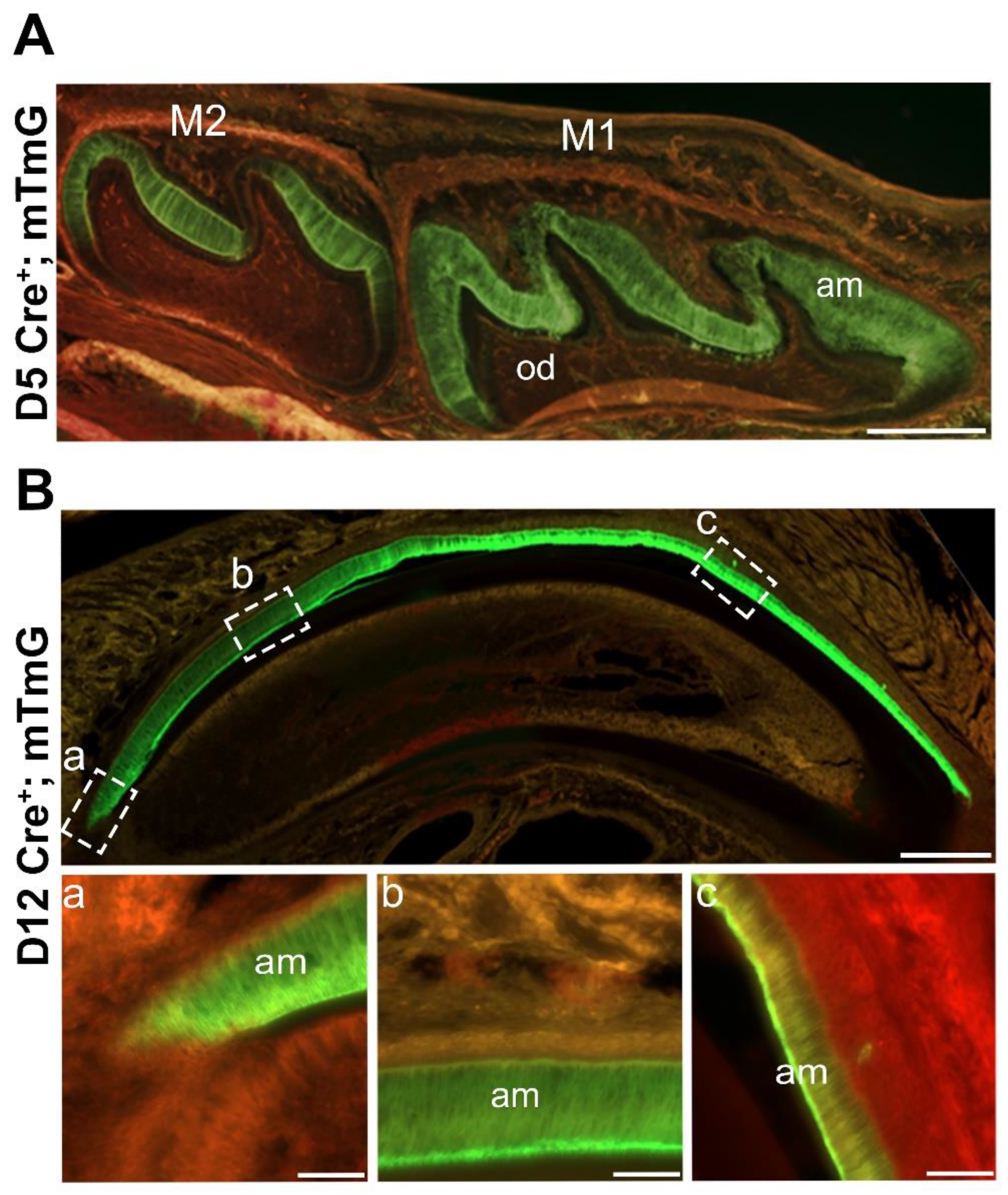
Cre recombinase is expressed, mT is spliced out and tissues stain with green fluorescent protein (mG). Fluorescent imaging of post-natal day 5 molars revealed strong mG staining in ameloblasts from first and second mandibular molars (
Figure 1A). In contrast, expression of mT was detected in odontoblasts, alveolar bone and remaining layers (stratum intermedium, stellate reticulum, outer epithelium) of the enamel organ indicating that
Cre-mediated recombination did not occur in these tissues. Murine incisors at post-natal day 12 yielded similar results. Positive mG expression was observed in pre-ameloblasts, which persisted in secretory and maturation-stage ameloblasts (
Figure 1B).
2.2. Assessment of Amelx-iCre Expression in Multiple Tissues and Quantification of Adam10 Expression in the Amelx-iCre; Adam10fl/fl (Adam10 cKo) Mouse Incisors
To confirm ameloblast-specific i
Cre expression, we performed genomic PCR analyses in multiple tissues using primers that can amplify a 376 base pair mG product only if i
Cre recombination has occurred [
23]. A total of 20 tissues were tested (alveolar bone, aorta, brain, muscle, colon, eye, heart, kidney, knee, liver, lung, ovary, rib, intestine, skull, spleen, thymus, tongue, trachea, and whole incisor).
Cre activity was detected only in the incisor (
Figure 2A) demonstrating that
Amelx-i
Cre activation is tooth-specific. To further confirm tooth-specific i
Cre expression, we performed genomic PCR analyses in the same tissues using primers that can amplify a 200 base pair mT product only if i
Cre-recombination has not occurred [
23]. The presence of the 200 base pair mT product revealed that in the tissues examined, including whole incisors containing bone dentin and enamel, i
Cre recombination did not occur (
Figure 2B). Taken together with the results of
Figure 1, whole incisor ameloblasts displayed
Cre recombination, but the associated incisor tissues (bone and dentin) did not. These data confirm that i
Cre recombination occurs only in the ameloblasts of the mouse enamel organ and not in the other 19 tissues examined.
The
Amelx-i
Cre; mTmG mice confirmed that ameloblasts express the
Amelx-i
Cre transgene. We therefore crossed the
Amelx-i
Cre mice with
Adam10fl/fl mice to generate
Amelx-i
Cre;
Adam10fl/fl mice (
Adam10 cKO). qPCR analyses of 5-day old first molars that were predominantly in the secretory stage of enamel development, revealed that
Adam10 expression was significantly downregulated in the
Adam10 cKO enamel organs indicating that the preponderance of the
Adam10 genes in the ameloblast layer were successfully excised (
Figure 2C). Note that the pulp organ was not completely removed from the day five enamel organs, which means the pulp contributed to the
Adam10 expression observed in the
Adam10 cKO mice. To assess whether the
Amelx-i
Cre construct affected expression of the endogenous amelogenin gene in
Adam10 cKO mice, we performed qPCR of amelogenin gene expression in the cKO mice and controls. No significant difference was observed in endogenous amelogenin gene expression between the two genotypes assessed (
Figure 2C). Therefore, any observed phenotype discovered in the cKO mice would be attributed to the loss of
Adam10 expression by ameloblasts.
2.3. Phenotypic Assessment of Teeth by Genotype
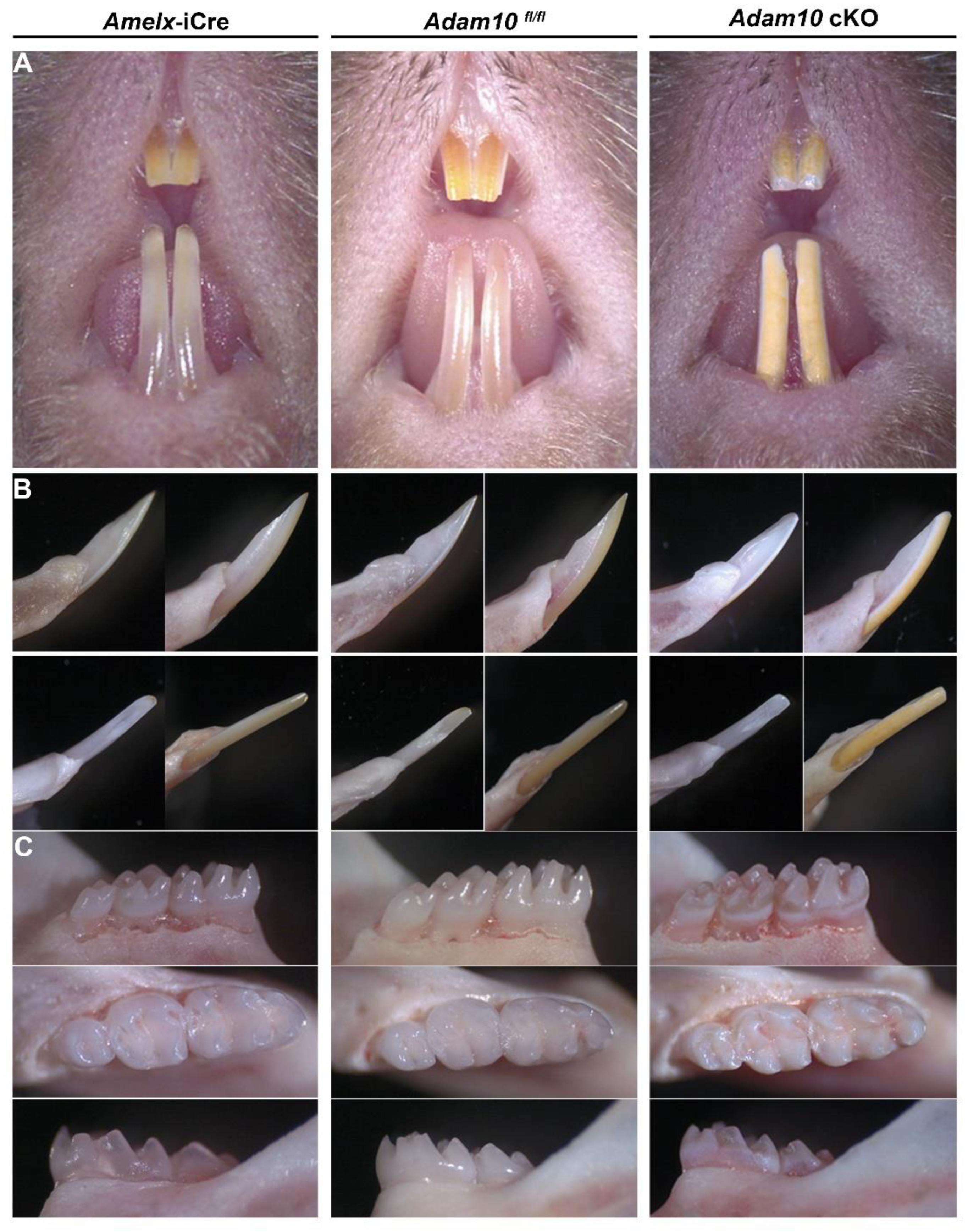
Adam10 cKO incisors at 7 weeks show abrasion on the labial surface of the maxillary incisors (
Figure 3A). The mandibular incisors were blunted, display a yellow discoloration, and had a rough sandpaper-like surface when compared to the smooth translucent enamel observed in both
Amelx-i
Cre and
Adam10fl/fl mice (
Figure 3B)
. Adam10 cKO molars also presented with a rough enamel texture with signs of occlusal wear on the cusps. A distinct chalky white band was present near the cervical margin of the cKO molars. In contrast, the
Amelx-i
Cre and
Adam10fl/fl molars appeared normal (
Figure 3C).
2.4. Quantification of Incisor Enamel Hardness
Nanoindentation was performed on mandibular hemi-mandibles sectioned at the buccal alveolar crest (
Figure 4A). These analyses showed that, compared to
Amelx-i
Cre and
Adam10fl/fl incisors,
Adam10 cKO incisors were significantly softer (**
P < 0.01) than normal (
Figure 4B). The average hardness value of the cKO incisors was 2.7 GPa, which reflects a 40% decrease compared to hardness values of
Amelx-i
Cre and
Adam10fl/fl incisors (4.2 and 4.6 GPa respectively). Among the three genotypes of mandibular incisors, no statistically significant nanohardness differences were observed in dentin or alveolar bone (
Figure 4B) demonstrating that only the enamel was affected in the
Adam10 cKO mice.
2.5. µ. CT Analyses
Next, we performed µCT to determine if volumetric and mineralization defects were present in the cKO incisor enamel. High-resolution sagittal images (
Figure 4C-E) of mandibular incisors revealed a small decrease in incisor enamel volume with a 10% decrease in mineral density plus a 10% reduced enamel thickness in cKO incisors at the buccal alveolar crest compared to
Amelx-i
Cre and
Adam10fl/fl controls (
Figure 4F-H).
We also measured enamel density and volume in
Adam10 cKO mandibular first molars compared to molars from the control
Amelx-i
Cre and
Adam10fl/fl mice. At 7-weeks of age, signs of occlusal wear were observed on cusps tips from cKO first molars, indicating loss of enamel through abrasion. The enamel layer was hypomineralized and could not be distinguished from dentin by µCT settings at the 1,600 mg HA/cm
3 threshold (
Figure 5A). Using a combination of semi-automatic and manual corrections, the
Adam10 cKO enamel layer was traced so that we could identify a 27% reduction in enamel volume and a 14% decrease in mineral density (
Figure 5B).
Molar crowns were then subdivided into cervical and occlusal sections to quantify enamel volume and density at these locations. Occlusal enamel volume was significant reduced by 23% in the cKO molars compared to controls and the cervical enamel volume in cKO molars had a greater reduction of 32% relative to controls. Assessment of segmented mineral density revealed that the cKO mice had an 8% reduction in occlusal mineral density and that the cKO cervical enamel had a 21% reduced mineral density compared to the corresponding densities of control
Amelx-i
Cre and
Adam10fl/fl molars (
Figure 5B). This demonstrates that the chalky white banding pattern observed in the cervical region of cKO molars (
Figure 3C) has both a decreased volume and density relative to the occlusal region of the same cKO molars.
Next, we quantified volume and density in mandibular first molars at post-natal day 15 (P15). At P15, enamel formation on the first molars is virtually complete but the molars have not fully erupted into the oral cavity. Unlike erupted molars, no signs of attrition were observed in the unerupted P15 molars. Like erupted molars, unerupted cKO molars exhibited a significant reduction of total enamel volume (16%) and density (15%) versus controls (
Figure 5C, D). Also, as with the erupted molars, the differences versus controls were reduced more in the cervical area of the unerupted molars (volume 17% and density 17%) than the occlusal regions (volume 14% and density 13%) compared to the corresponding control molar locations (
Figure 5D).
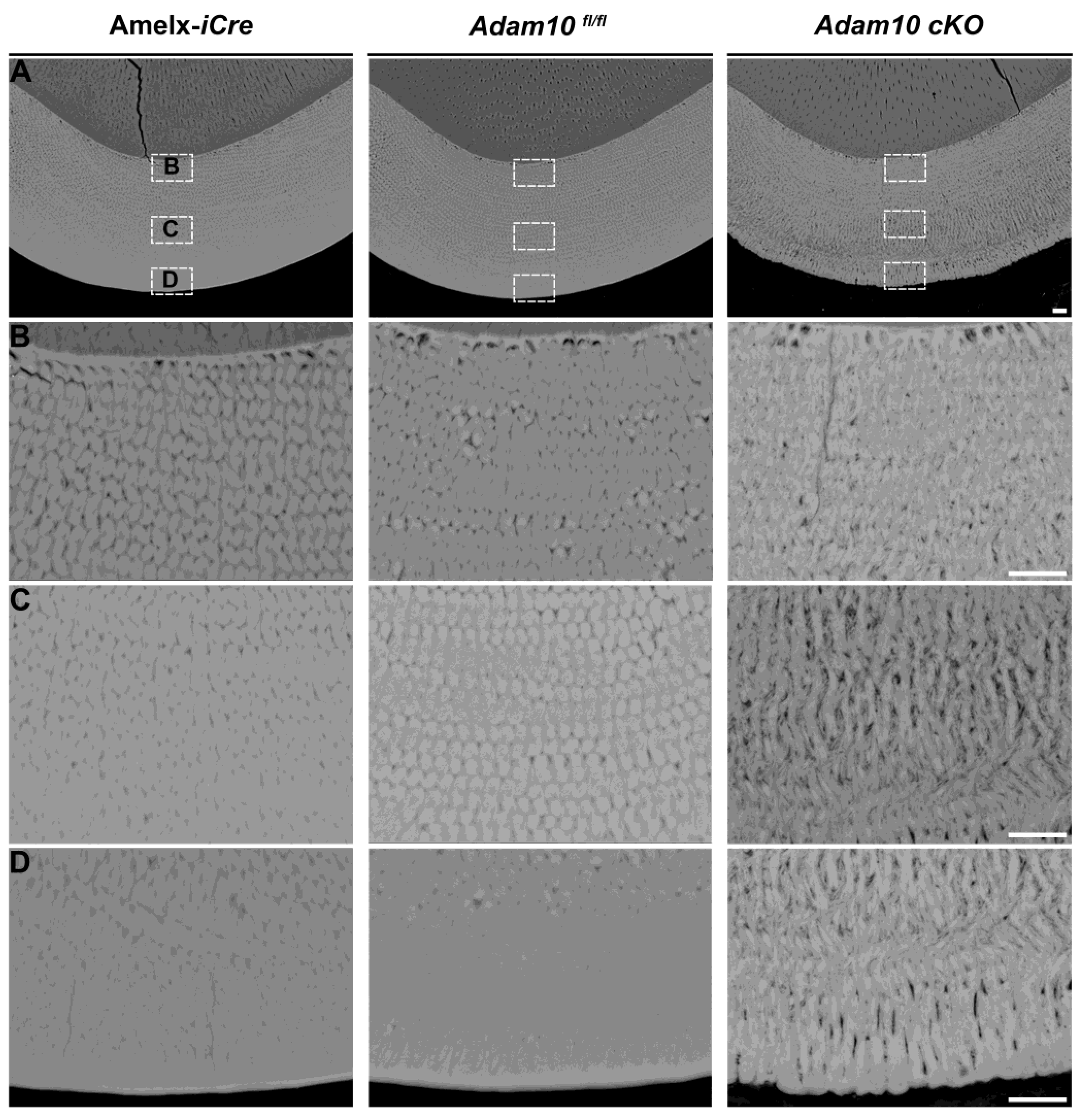
2.6. Backscatter Scanning Electron Microscopy (bSEM) Assessment of Ultrastructural Changes in Adam10 cKO Incisors
Incisor enamel at the level of buccal alveolar crest was examined by bSEM. The enamel layers from
Amelx-i
Cre and
Adam10fl/fl control incisors were highly mineralized and of normal thickness. In contrast, the enamel from
Adam10 cKO incisors was less thick and appeared porous (
Figure 6A). Distinctively organized and decussating enamel rods were observed in
Amelx-i
Cre and
Adam10fl/fl incisors, whereas the rod-interrod pattern in cKO incisors was loosely arranged and disorganized with apparent large spaces between adjacent rods. This rod pattern disorganization became progressively more severe in cKO incisors from the dentin-enamel junction (DEJ) to the incisal edge (
Figure 6B - D).
2.7. Quantification of Col17a1, Relt, Ambn Enam and Mmp20 Gene Expression Levels by Genotype
To determine if ADAM10-mediated cell signaling effects the expression of genes that when mutated can cause enamel defects, we performed a preliminary study by use of qPCR to quantify the expression levels of secretory stage enamel matrix genes (
Ambn, Enam,
Mmp20) and quantify expression levels of genes encoding cell surface proteins (
Col17a1,
Relt) that ADAM10 is known to cleave. Day-5 first molar mRNA from Adam10
fl/fl controls was extracted to compare gene expression levels with mRNA from
Adam10 cKO mouse first molars. Compared to the
Adam10fl/fl control, no significant difference in expression was observed among the genes encoding enamel matrix or cell surface proteins when compared to the expression levels observed in the
Adam10 cKO mice (
Figure S3). Further investigations are necessary to determine if ADAM10-mediated cell signaling plays a role in the enamel phenotype observed in
ADAM10 cKO mice.
3. Discussion
ADAM10 is a multifunctional protease that attaches to the cell surface via a transmembrane domain. Its catalytic domain is positioned extracellularly so that it can shed proteins from the cell surface [
24]. RELT is a receptor of the tumor necrosis factor super family that when mutated can cause AI [
15]. Previously we demonstrated
in vitro that ADAM10 cleaves the RELT extracellular domain and we suggested that absence of ADAM10 may cause RELT-mediated enamel malformation [
7]. However, an engineered RELT mutation in mice caused a more mild enamel phenotype than we observed here in
Adam10 cKO mice [
15]. ADAM10 also cleaves COL17A1, which likewise is a transmembrane protein that when mutated causes enamel defects in human teeth [
17]. Like
Adam10 cKO mice,
Col17a1-/- mice have an irregular enamel rod pattern and abraded teeth indicating defective mineralization [
16]. However, it is difficult to make conclusions about a protease’s contribution to enamel formation by examining the functional loss of a potential substrate. If ADAM10 cleaves the substrate, ADAM10 absence allows the substrate to remain on the cell surface and this phenotype may be different from what occurs when the substrate is rendered nonfunctional.
ADAM10 is transported to the cell surface by a class of tetraspanins (Tspans) termed TspanC8s because they have 8 cysteines in their extracellular domain. Six different TspanC8s are capable of transporting ADAM10 to the cell surface. Once there, the TspanC8 remains complexed with ADAM10 at the cell surface. So, each TspanC8 selects the ADAM10 location and thereby modulates ADAM10 substrate selectivity [
25,
26]. Additionally, each TspanC8 may influence ADAM10 substrate specificity, perhaps through conformational interactions, which likely contributes to the ability of ADAM10 to cleave approximately 100 substrates [
8]. Previously, we screened mouse enamel organs for the presence of ADAM proteinases.
Adam10 was the only ADAM expressed during the secretory stage when ameloblasts move relative to each other, but not during the maturation stage when ameloblasts stop moving. Enamel rods are the mineralized path of ameloblast cell movement and since ameloblasts lacking ADAM10 have malformed rod patterns, this suggests that ADAM10 plays a role in ameloblast movement during the secretory stage of enamel development. Therefore, it is possible that a TspanC8 transports ADAM10 to the ends of each ameloblast cohort to detachment them from adjacent cohorts. This may facilitate groups of ameloblasts to slide by one another, which is necessary to form the decussating enamel rod pattern characteristic of rodent incisor enamel [
27,
28]. We did examine ameloblast incisor and molar histology sections and found that the cKO ameloblasts were apparently normally aligned (
Figure S4). However, at least some level of ameloblast alignment would likely be necessary to generate a rod pattern even if that pattern was disorganized compared the wild-type rod pattern.
Since ADAM10 plays a significant role in regulating cell signaling [
29,
30,
31], another possibility is that lack of ADAM10 resulted in disrupted cell signaling within the enamel organ, which caused the dysplastic phenotype observed in the cKO mice. The finding that the cervical region of the cKO molars had less volume and reduced density compared to the occlusal region was unexpected. However, since enamel development begins at the cusp tip and progresses to the cervical regions, this is consistent with latest formed enamel becoming more adversely affected by a lack of ADAM10 than the earliest formed enamel. Note that the latest formed outer layers of incisor enamel were more disrupted than the inner layers (
Figure 6). This suggests that ADAM10 function becomes increasingly more important as enamel development progresses and lends support to the possibility that cell signaling is disrupted in the enamel organ. This is because the loss of appropriate cell signaling could become cumulative as enamel development progresses resulting in a more dysplastic phenotype near the end of enamel development as compared to the beginning. However, our preliminary investigations into gene regulation of enamel matrix proteins and ADAM10 cell surface substrates (
Figure S3) revealed no significant differences in their transcript levels between control and cKO mouse enamel organs. Further studies are necessary to identify the mechanistic function of ADAM10 during enamel development.
In addition to perinatal lethality [
19], another difference between this and the previous
Adam10 conditional knockout study [
18] is the timing of when
Adam10 was knocked out. Keratin14 is expressed in the first branchial arch of the oral and tooth ectoderm [
32]. Therefore, Keratin14
Cre/+; Adam10fl/fl mice delete
Adam10 at the earliest stages of tooth development in all epithelial tissues. However, our study showed that when
Adam10 is deleted specifically from pre-ameloblast and ameloblasts, the resulting enamel is malformed. Notably, in contrast to our findings, the Keratin14
Cre/+; Adam10fl/fl study [
18] also examined
AmelxCre/+;
Adam10fl/fl mice and described the enamel as forming normally. However, the authors did not identify which of 5 available
AmelxCre/+ constructs they used, and a figure in their supplementary data showing
AmelxCre/+; R26
mTmG results, indicated that very few ameloblasts would have deleted
Adam10 from their nuclei because only a few were colored green due to excision of the TdTomato region. Regardless, this did not significantly affect their Notch study results as both their and our results examined different developmental time points and epithelial tissues.
4. Conclusion
The Tg(Amelx-iCre)872pap construct was expressed only in pre-ameloblasts and ameloblasts demonstrating its suitability to eliminate Adam10 expression in these cells. We demonstrated that Adam10 cKO mice have teeth that are discolored, abraded and their enamel is softer than normal. Notably, the cKO enamel density and enamel volume were significantly reduced in both incisors and molars. Moreover, the enamel rod pattern became progressively more disorganized moving from the DEJ to the outer enamel surface and this disorganized rod structure created large gaps between and among the rods, suggesting that the ameloblasts do not signal or perhaps move properly to form a normal rod pattern. Future studies are necessary to interrogate the mechanisms of how ADAM10 functions to facilitate healthy dental enamel formation. Perhaps by examining ADAM10 shedding of critical receptors such as RELT or cell-cell contact proteins such as COL17A1 and E-, N-cadherins, additional mechanistic clues will be discovered.
5. Materials and Methods
5.1. Mice
All animals used in this study were housed in Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) accredited facilities and were treated humanely based on protocols approved by the Ohio State University Institutional Animal Care and Use Committee. The
Amelx-iCre mice were generated as described previously [
22]. These mice were backcrossed into C57BL/6 (wild-type, WT) mice for at least eight generations to obtain a homogeneous genetic background. To confirm
Cre recombinase activity,
Amelx-i
Cre mice were crossed with B6.129(Cg)-
Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (ROSA
mT/mG) mice obtained from Jackson Laboratory (JAX:007676) [
33]. To study the effects of
Adam10 deletion in ameloblasts,
Amelx-iCre mice were mated with B6;129S6-
Adam10tm1Zhu/J (
Adamfl/fl ) obtained from Jackson Laboratory (JAX:009357) [
34] to generate
Amelx-iCre; Adam10fl/fl (
Adam10 cKO) mice.
5.2. Detection of Cre-Recombination by genomic PCR and Assessment of Gene Expression Levels by qPCR
Total DNA was extracted from
Amelx-i
Cre; mTmG mouse tissues using DNeasy Blood & Tissue Kit (Qiagen). PCR primer pairs (mT-F: GCAACGTGCTGGTTATTGTG ; mT-R: TGATGACCTCCTCTCCCTTG) and (mG-F: GTTCGGCTTCTGGCGTGT; mG-R: TGCTCACGGATCCTACCTTC) amplified non-recombined and
Cre-recombined alleles generating 200bp and 376bp DNA products, respectively [
23].
For quantitative polymerase chain reaction (qPCR), molar enamel organs were extracted from 5-day Adam10
fl/fl and Adam10 cKO mice. Total RNA from enamel organs was isolated using the miRNeasy Micro Kit (Qiagen) and cDNA was synthesized by High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR amplification was performed using the TaqMan Fast Advanced Master Mix (Applied Biosystems) with specific TaqMan assays (
Table S1). Relative expression levels of assessed genes were calculated by 2−ΔΔCt method using
Gapdh as the housekeeping gene control. Five samples per group were analyzed in triplicate.
5.3. Tissue Preparation and Fluorescence Microscopy
Mouse incisors were harvested from post-natal day 12 mice and first molars were harvested from post-natal day 5 Amelx-iCre; mTmG pups. All other tissues were collected from 7-week-old Amelx-iCre; mTmG mice. Tissues were fixed in 4% paraformaldehyde at room temperature overnight. Following fixation, maxillae and mandibles were decalcified in EDTA for 5-12 days, embedded in optimal cutting temperature, snap frozen and sectioned at 10 µm with a cryostat. Soft tissue samples were embedded and sectioned directly after fixation. Sections were rinsed in 1x PBS and stained with DAPI (Invitrogen, D1306, 1:5000). Sections were imaged with Lionheart LX microscope and Zeiss AXIO Imager.Z2.
5.4. Phenotypic Assessment of Incisors and Molars
Mandibles from 7-week-old mice were stripped of soft tissues and subsequently immersed in 10% protease solution (Sigma) diluted in 1x phosphate-buffered saline (PBS) at 56ºC for 10 minutes. Teeth were rinsed with 1x PBS, air dried and photographed using a Leica S9i stereomicroscope.
5.5. Nanohardness Testing
Hemi-mandibles from Amelx-iCre, Adam10fl/fl and Amelx-iCre; Adam10fl/fl mice were collected at 7 weeks. After removing soft tissues, the right-side mandibles were fixed in 4% PFA overnight and dehydrated with acetone gradient (30, 50, 70, 80, 90 and 100%) for 30 minutes each and embedded in resin (Embed 812). The samples were cured at 60ºC for 48 hours. Sample discs were cut transversely at the level of labial crest of the alveolar bone. Samples were re-embedded in Castolite AC in 25-mm SeriForm molds (Struers). Incisor cross sections were polished with waterproof carbide papers and 1-micron diamond paste. Samples were then nanoindented using a Hysitron 950 Triboindenter with a nanoDMA transducer and Berkovich probe. Indentations were analyzed using Triboscan 9 software.
5.6. Micro-Computed Tomography (µCT)
Left mandibles from adult 7-week-old and post-natal day 15 mice were fixed in 10% neutral-buffered formalin and scanned in a μCT 50 (Scanco) at 70 kVp, 85 μA, 0.5 mm Al filter, 900 ms integration time, and 6 μm voxel size. Following reconstruction, DICOM files were calibrated to 5 known densities of hydroxyapatite (mg HA/cm
3) and analyzed with AnalyzeDirect 14 software.
Incisor enamel was segmented at >1,600 mg HA/cm3 for all groups. Molar enamel was manually traced for Amelx-i
Cre; Adam10fl/fl mice [
35]
.
5.7. Backscattered Scanning Electron Microscopy (bSEM)
Sample preparation for bSEM was identical to hardness testing. Incisor cross sections were sputter coated with carbon and imaged using JEOL-JSM-7800FLV field-emission scanning electron microscope at a voltage of 20 kV and a working distance of 10 mm.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Figure S1: Assessment of mouse organs for Cre expression; Figure S2: Assessment of mouse tissue sections for Cre expression; Figure S3: Quantification of Ambn, Enam Col17a1, Mmp20 and Relt gene expression levels in Adam10fl/fl (control) and Adam10 cKO (experimental) mice; Figure S4: Histological comparison of incisor ameloblasts from Adam10fl/fl (control) and Adam10 cKO (experimental) mice; Table S1: TaqMan probes for qPCR analyses of gene expression.
Author Contributions
Conceptualization, S.S, B.L.F., J.P.S. and J.D.B; methodology, S.S., Y.H., F.M., L.R. M.C.L. and D.T.F.; software, S.S., Y.H. and F.M.; validation, S.S., Y.H., F.M., J.P.S. B.L.F. and J.D.B.; formal analysis, S.S., Y.H., F. M., and J.D.B.; investigation, S.S., Y.H., F.M. L.R. M.C.L. and D.T.F.; resources, P.P., B.L.F. J.P.S. and J.D.B.; data curation, S.S., B.L.F., J.P.S. and J.D.B.; writing original draft preparation, S.S. and J.D.B.; writing—review and editing, S.S., Y.H., F.M., L.R. M.C.L., D.T.F., P.P., B.L.F., J.P.S. and J.D.B.; visualization, S.S. and JDB; Supervision, B.L.F., J.P.S. and J.D.B.; Project Administration J.D.B.; funding acquisition, J.P.S., B.L.F. and J.D.B. All authors have read and agreed to the published version of the manuscript. .
Funding
This research was funded by the National Institute of Dental and Craniofacial Research of the National Institutes of Health under award numbers R01DE028297 (JDB), R01DE015846 (JPS) and R01DE027639; R01DE032334 (BLF). The authors declare no conflicts of interest.
Data Availability Statement
Amelx-iCre; Adam10fl/fl frozen mouse sperm is awaiting approval for distribution at Mutant Mouse Resource & Research Centers (MMRRC). Additional data used to support the findings of this study are available from the corresponding author upon request.
Acknowledgments
We thank Ai (Andy) Ni for advising us about biostatistical analyses. Backscattered scanning electron microscopy was performed at the University of Michigan Robert B. Mitchell Electron Microbeam Analysis Lab and nanohardness testing was performed at the University of Michigan Center for Materials Characterization (Ann Arbor, MI, USA).
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Bartlett, J.D.; Smith, C.E.; Hu, Y.; Ikeda, A.; Strauss, M.; Liang, T.; Hsu, Y.H.; Trout, A.H.; McComb, D.W.; Freeman, R.C.; et al. MMP20-generated amelogenin cleavage products prevent formation of fan-shaped enamel malformations. Sci. Rep. 2021, 11, 10570. [Google Scholar] [CrossRef] [PubMed]
- Simmer, J.P.; Richardson, A.S.; Hu, Y.Y.; Smith, C.E.; Ching-Chun Hu, J. A post-classical theory of enamel biomineralization... and why we need one. Int J Oral Sci 2012, 4, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, J.D. Dental enamel development: proteinases and their enamel matrix substrates. ISRN dentistry 2013, 2013, 684607. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Simmer, J.P.; Hart, T.C.; Hart, P.S.; Ramaswami, M.D.; Bartlett, J.D.; Hu, J.C. MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J. Med. Genet. 2005, 42, 271–275. [Google Scholar] [CrossRef]
- Guan, X.; Bartlett, J.D. MMP20 modulates cadherin expression in ameloblasts as enamel develops. J. Dent. Res. 2013, 92, 1123–1128. [Google Scholar] [CrossRef]
- Reiss, K.; Saftig, P. The "a disintegrin and metalloprotease" (ADAM) family of sheddases: physiological and cellular functions. Semin. Cell Dev. Biol. 2009, 20, 126–137. [Google Scholar] [CrossRef]
- Ikeda, A.; Shahid, S.; Blumberg, B.R.; Suzuki, M.; Bartlett, J.D. ADAM10 is Expressed by Ameloblasts, Cleaves the RELT TNF Receptor Extracellular Domain and Facilitates Enamel Development. Sci. Rep. 2019, 9, 14086. [Google Scholar] [CrossRef]
- Kuhn, P.H.; Wang, H.; Dislich, B.; Colombo, A.; Zeitschel, U.; Ellwart, J.W.; Kremmer, E.; Rossner, S.; Lichtenthaler, S.F. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 2010, 29, 3020–3032. [Google Scholar] [CrossRef]
- Maretzky, T.; Reiss, K.; Ludwig, A.; Buchholz, J.; Scholz, F.; Proksch, E.; de Strooper, B.; Hartmann, D.; Saftig, P. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 9182–9187. [Google Scholar] [CrossRef]
- Reiss, K.; Maretzky, T.; Ludwig, A.; Tousseyn, T.; de Strooper, B.; Hartmann, D.; Saftig, P. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. EMBO J. 2005, 24, 742–752. [Google Scholar] [CrossRef]
- Mitsiadis, T.A.; Lardelli, M.; Lendahl, U.; Thesleff, I. Expression of Notch 1, 2 and 3 is regulated by epithelial-mesenchymal interactions and retinoic acid in the developing mouse tooth and associated with determination of ameloblast cell fate. J. Cell Biol. 1995, 130, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Felszeghy, S.; Modis, L.; Tammi, M.; Tammi, R. The distribution pattern of the hyaluronan receptor CD44 during human tooth development. Arch. Oral Biol. 2001, 46, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Davideau, J.L.; Sahlberg, C.; Blin, C.; Papagerakis, P.; Thesleff, I.; Berdal, A. Differential expression of the full-length and secreted truncated forms of EGF receptor during formation of dental tissues. Int. J. Dev. Biol. 1995, 39, 605–615. [Google Scholar]
- Casasco, A.; Casasco, M.; Corbett, I.P.; Calligaro, A. Expression of c-erbB-2 proto-oncogene-encoded protein (p185erbB-2) in functional rat ameloblasts. Arch. Oral Biol. 1994, 39, 917–919. [Google Scholar] [CrossRef]
- Kim, J.W.; Zhang, H.; Seymen, F.; Koruyucu, M.; Hu, Y.; Kang, J.; Kim, Y.J.; Ikeda, A.; Kasimoglu, Y.; Bayram, M.; et al. Mutations in RELT cause autosomal recessive amelogenesis imperfecta. Clin. Genet. 2019, 95, 375–383. [Google Scholar] [CrossRef]
- Asaka, T.; Akiyama, M.; Domon, T.; Nishie, W.; Natsuga, K.; Fujita, Y.; Abe, R.; Kitagawa, Y.; Shimizu, H. Type XVII collagen is a key player in tooth enamel formation. Am. J. Pathol. 2009, 174, 91–100. [Google Scholar] [CrossRef]
- Hany, U.; Watson, C.M.; Liu, L.; Smith, C.E.L.; Harfoush, A.; Poulter, J.A.; Nikolopoulos, G.; Balmer, R.; Brown, C.J.; Patel, A.; et al. Heterozygous COL17A1 variants are a frequent cause of amelogenesis imperfecta. J. Med. Genet. 2024, 61, 347–355. [Google Scholar] [CrossRef]
- Mitsiadis, T.A.; Jimenez-Rojo, L.; Balic, A.; Weber, S.; Saftig, P.; Pagella, P. Adam10-dependent Notch signaling establishes dental epithelial cell boundaries required for enamel formation. iScience 2022, 25, 105154. [Google Scholar] [CrossRef]
- Weber, S.; Niessen, M.T.; Prox, J.; Lullmann-Rauch, R.; Schmitz, A.; Schwanbeck, R.; Blobel, C.P.; Jorissen, E.; de Strooper, B.; Niessen, C.M.; et al. The disintegrin/metalloproteinase Adam10 is essential for epidermal integrity and Notch-mediated signaling. Development 2011, 138, 495–505. [Google Scholar] [CrossRef]
- Alvarado-Gaytan, J.; Saavedra-Marban, G.; Velayos-Galan, L.; Gallardo-Lopez, N.E.; de Nova-Garcia, M.J.; Caleya, A.M. Dental Developmental Defects: A Pilot Study to Examine the Prevalence and Etiology in a Population of Children between 2 and 15 Years of Age. Dent J (Basel) 2024, 12. [Google Scholar] [CrossRef]
- Tung, K.; Fujita, H.; Yamashita, Y.; Takagi, Y. Effect of turpentine-induced fever during the enamel formation of rat incisor. Arch. Oral Biol. 2006, 51, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Said, R.; Zheng, L.; Saunders, T.; Zeidler, M.; Papagerakis, S.; Papagerakis, P. Generation of Amelx-iCre Mice Supports Ameloblast-Specific Role for Stim1. J. Dent. Res. 2019, 98, 1002–1010. [Google Scholar] [CrossRef] [PubMed]
- Hann, S.; Kvenvold, L.; Newby, B.N.; Hong, M.; Warman, M.L. A Wisp3 Cre-knockin allele produces efficient recombination in spermatocytes during early prophase of meiosis I. PloS one 2013, 8, e75116. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, S.; Seipold, L.; Saftig, P. The metalloproteinase ADAM10: A useful therapeutic target? Biochimica et biophysica acta. Molecular cell research 2017, 1864, 2071–2081. [Google Scholar] [CrossRef]
- Jouannet, S.; Saint-Pol, J.; Fernandez, L.; Nguyen, V.; Charrin, S.; Boucheix, C.; Brou, C.; Milhiet, P.E.; Rubinstein, E. TspanC8 tetraspanins differentially regulate the cleavage of ADAM10 substrates, Notch activation and ADAM10 membrane compartmentalization. Cell. Mol. Life Sci. 2016, 73, 1895–1915. [Google Scholar] [CrossRef]
- Harrison, N.; Koo, C.Z.; Tomlinson, M.G. Regulation of ADAM10 by the TspanC8 Family of Tetraspanins and Their Therapeutic Potential. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef]
- Reith, E.J.; Ross, M.H. Morphological evidence for the presence of contractile elements in secretory ameloblasts of the rat. Arch. Oral Biol. 1973, 18, 445–448. [Google Scholar] [CrossRef]
- Smith, C.E.; Hu, Y.; Hu, J.C.; Simmer, J.P. Quantitative analysis of the core 2D arrangement and distribution of enamel rods in cross-sections of mandibular mouse incisors. J. Anat. 2019, 234, 274–290. [Google Scholar] [CrossRef]
- Dempsey, P.J. Role of ADAM10 in intestinal crypt homeostasis and tumorigenesis. Biochimica et biophysica acta. Molecular cell research 2017, 1864, 2228–2239. [Google Scholar] [CrossRef]
- Rosenbaum, D.; Saftig, P. New insights into the function and pathophysiology of the ectodomain sheddase A Disintegrin And Metalloproteinase 10 (ADAM10). FEBS J 2024, 291, 2733–2766. [Google Scholar] [CrossRef]
- Weber, S.; Saftig, P. Ectodomain shedding and ADAMs in development. Development 2012, 139, 3693–3709. [Google Scholar] [CrossRef] [PubMed]
- Dassule, H.R.; Lewis, P.; Bei, M.; Maas, R.; McMahon, A.P. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 2000, 127, 4775–4785. [Google Scholar] [CrossRef]
- Muzumdar, M.D.; Tasic, B.; Miyamichi, K.; Li, L.; Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 2007, 45, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Wu, X.; Chi, C.; Han, M.; Xu, T.; Zhuang, Y. ADAM10 is essential for proteolytic activation of Notch during thymocyte development. Int. Immunol. 2008, 20, 1181–1187. [Google Scholar] [CrossRef]
- Chavez, M.B.; Chu, E.Y.; Kram, V.; de Castro, L.F.; Somerman, M.J.; Foster, B.L. Guidelines for Micro-Computed Tomography Analysis of Rodent Dentoalveolar Tissues. JBMR Plus 2021, 5, e10474. [Google Scholar] [CrossRef]
Figure 1.
Amelx-iCre mice express Cre recombinase only in ameloblasts. Amelx-iCre mice were crossed with the mouse mTmG reporter strain so that Cre expression would cause tissues to stain with green fluorescent protein (GFP). (A) GFP staining was restricted to the developing ameloblasts in molars at postnatal day 5 whereas areas of red florescence indicate no Cre recombination. Scale bar 200 µm. (B) In postnatal day 12 maxillary incisor sections, amelogenin promoter driven iCre expression starts early at the apical loop (a) and persists throughout the length of the incisor where ameloblasts are present (b, c). Scale bar 1000 µm. M1, first molar; M2, second molar; am, ameloblasts.
Figure 1.
Amelx-iCre mice express Cre recombinase only in ameloblasts. Amelx-iCre mice were crossed with the mouse mTmG reporter strain so that Cre expression would cause tissues to stain with green fluorescent protein (GFP). (A) GFP staining was restricted to the developing ameloblasts in molars at postnatal day 5 whereas areas of red florescence indicate no Cre recombination. Scale bar 200 µm. (B) In postnatal day 12 maxillary incisor sections, amelogenin promoter driven iCre expression starts early at the apical loop (a) and persists throughout the length of the incisor where ameloblasts are present (b, c). Scale bar 1000 µm. M1, first molar; M2, second molar; am, ameloblasts.
Figure 2.
Genomic PCR analysis to detect the presence or absence of Amelx-iCre mediated recombination in multiple mouse tissues and quantification of Adam10 and amelogenin expression. (A) A 376 base pair mG product results from the Cre-recombined construct, while a (B) 200 base pair product results from the mT region that was not recombined by Amelx-iCre. Of the 20 tissues assess, only the whole incisor demonstrated Amelx-iCre mediated recombination. (C) Quantification of Adam10 and amelogenin (AmelX) expression by qPCR analyses of 5-day old first molar enamel organs. The Adam10 cKO was expressed at significantly lower levels than was the Adam10fl/fl control, demonstrating that the preponderance of the Adam10 genes in the ameloblast layer were successfully excised. In contrast, no significant difference was observed in native amelogenin gene expression between the Adam10 cKO and Adam10fl/fl control mice, indicating that the Amelx-iCre construct did not affect native ameloblast expression.
Figure 2.
Genomic PCR analysis to detect the presence or absence of Amelx-iCre mediated recombination in multiple mouse tissues and quantification of Adam10 and amelogenin expression. (A) A 376 base pair mG product results from the Cre-recombined construct, while a (B) 200 base pair product results from the mT region that was not recombined by Amelx-iCre. Of the 20 tissues assess, only the whole incisor demonstrated Amelx-iCre mediated recombination. (C) Quantification of Adam10 and amelogenin (AmelX) expression by qPCR analyses of 5-day old first molar enamel organs. The Adam10 cKO was expressed at significantly lower levels than was the Adam10fl/fl control, demonstrating that the preponderance of the Adam10 genes in the ameloblast layer were successfully excised. In contrast, no significant difference was observed in native amelogenin gene expression between the Adam10 cKO and Adam10fl/fl control mice, indicating that the Amelx-iCre construct did not affect native ameloblast expression.
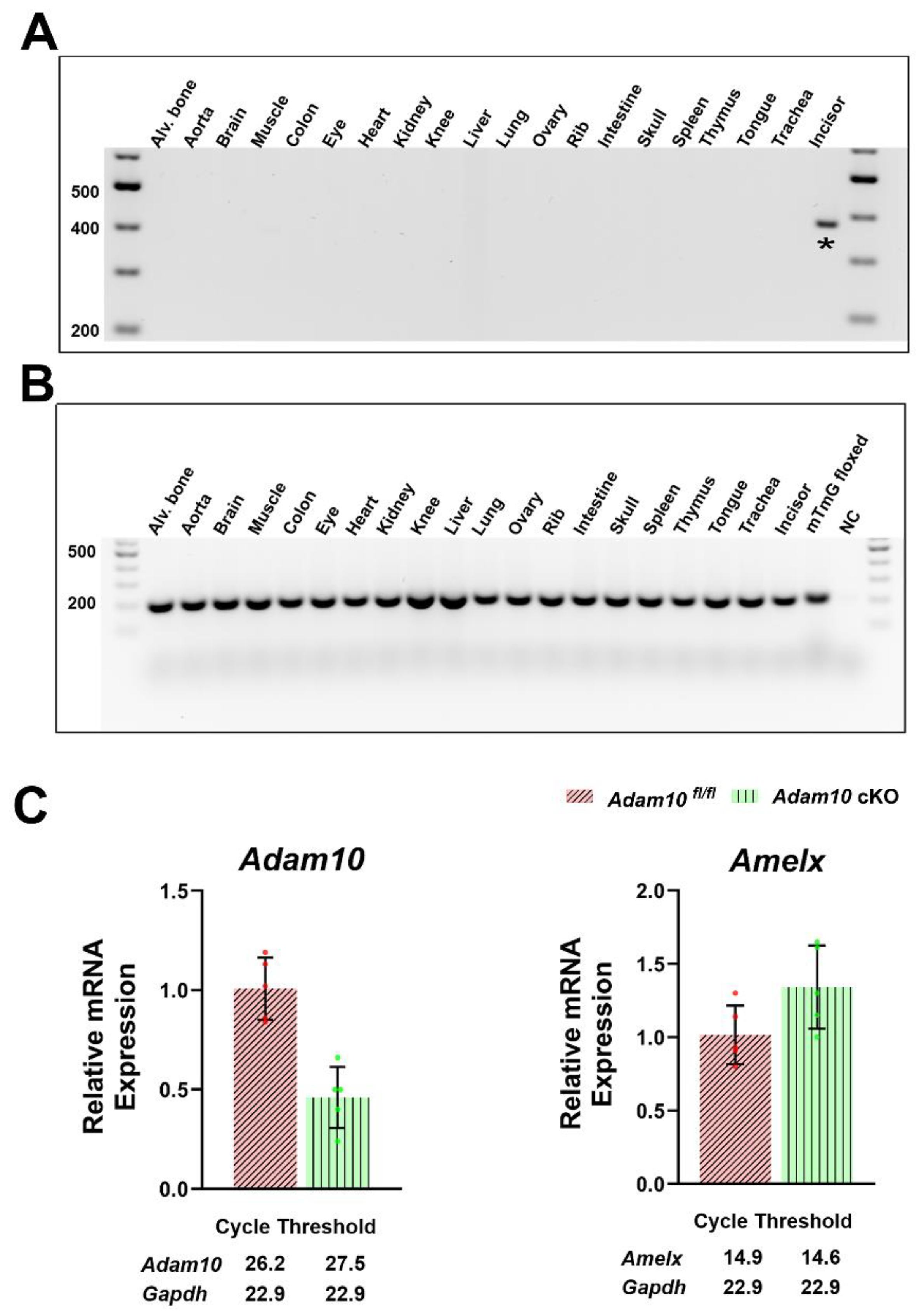
Figure 3.
Phenotypic assessment of Amelx-iCre, Adam10fl/fl, and Adam10 cKO incisors plus molars from mice at 7 weeks of age. (A) Frontal view of maxillary and mandibular incisors. The Adam10 cKO incisor enamel appears to be dull, rough, pigmented and abraded. (B) Mesial (upper left), lateral (upper right), lingual (lower right), and labial (lower left) views of mandibular incisors. The incisal edge of Adam10 cKO incisors were blunted due to attrition. (C) Buccal, occlusal, and lingual views of mandibular molars. Adam10 cKO molars appear rough, discolored and show distinct chalky banding near the cervical edge. These molar cusps are rounded and show signs of attrition. Teeth from Amelx-iCre and from Adam10fl/fl mice appear normal in shape and color.
Figure 3.
Phenotypic assessment of Amelx-iCre, Adam10fl/fl, and Adam10 cKO incisors plus molars from mice at 7 weeks of age. (A) Frontal view of maxillary and mandibular incisors. The Adam10 cKO incisor enamel appears to be dull, rough, pigmented and abraded. (B) Mesial (upper left), lateral (upper right), lingual (lower right), and labial (lower left) views of mandibular incisors. The incisal edge of Adam10 cKO incisors were blunted due to attrition. (C) Buccal, occlusal, and lingual views of mandibular molars. Adam10 cKO molars appear rough, discolored and show distinct chalky banding near the cervical edge. These molar cusps are rounded and show signs of attrition. Teeth from Amelx-iCre and from Adam10fl/fl mice appear normal in shape and color.
Figure 4.
Quantification of incisor enamel hardness, volume, density and thickness. (A) Shown are nanohardness indents of enamel, dentin and alveolar bone that were performed on Amelx-iCre, Adam10fl/fl, and Adam10 cKO mice. Each sample is indented at 3 positions in bone (positions 0-2), 12 positions in dentin (3-14) and 6 positions in enamel (15-20). (B) Bar graph of the mean nanohardness (GPa) values show the Adam10 cKO mice had significantly reduced enamel hardness compared to controls. Asterisks indicate a statistical difference of **P < 0.01. In contrast, dentin and bone nanohardness levels were not significantly different among the genotypes tested. (C-E) 2D renderings from µCT analyses of adult hemi-mandibles in sagittal view. The mineralized enamel appeared less dense in Adam10 cKO incisors compared to controls. (F-H) Compared to Amelx-iCre and Adam10fl/fl controls, Adam10 cKO mouse incisors exhibit 10% reduced enamel mineral density (***P = 0.001, **P < 0.01), and 10% reduced enamel thickness (***P< 0.001, **P < 0.01).
Figure 4.
Quantification of incisor enamel hardness, volume, density and thickness. (A) Shown are nanohardness indents of enamel, dentin and alveolar bone that were performed on Amelx-iCre, Adam10fl/fl, and Adam10 cKO mice. Each sample is indented at 3 positions in bone (positions 0-2), 12 positions in dentin (3-14) and 6 positions in enamel (15-20). (B) Bar graph of the mean nanohardness (GPa) values show the Adam10 cKO mice had significantly reduced enamel hardness compared to controls. Asterisks indicate a statistical difference of **P < 0.01. In contrast, dentin and bone nanohardness levels were not significantly different among the genotypes tested. (C-E) 2D renderings from µCT analyses of adult hemi-mandibles in sagittal view. The mineralized enamel appeared less dense in Adam10 cKO incisors compared to controls. (F-H) Compared to Amelx-iCre and Adam10fl/fl controls, Adam10 cKO mouse incisors exhibit 10% reduced enamel mineral density (***P = 0.001, **P < 0.01), and 10% reduced enamel thickness (***P< 0.001, **P < 0.01).
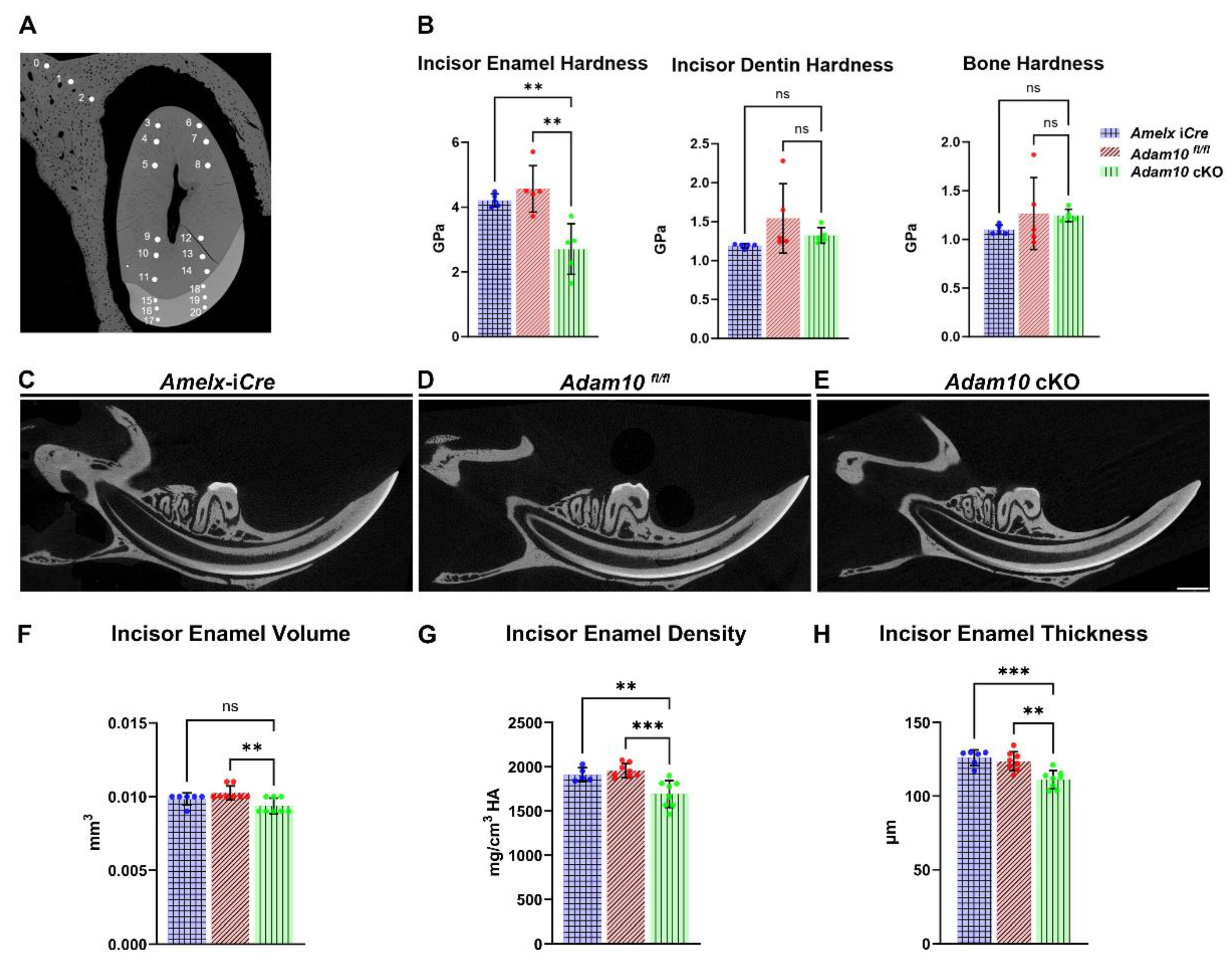
Figure 5.
2D and 3D renderings from µCT analyses of erupted and unerupted mandibular first molars in Adam10 cKO mice and controls. (A) Compared to controls, deletion of Adam10 leads to a poorly defined and under mineralized enamel layer. Sagittal and coronal views reveal flattening of Adam10 cKO cusps suggesting that these molars have undergone attrition. Occlusal views of Adam10 cKO molars show a severely hypomineralized enamel layer with reduced density as observed by the worn (3D) and darker (2D) enamel. (B) Quantitative analyses of total molar enamel volume and density show approximately a 27% decrease in volume and a 14% reduction in mineral density compared to controls (**P < 0.01). Segmentation of molars into occlusal and cervical regions reveal a greater reduction of volume and density at the cervical region of Adam10 cKO molars (**P < 0.01) when compared to the Amelx-iCre, and Adam10fl/fl molars. (C) 2D and 3D renderings from micro-CT analyses of unerupted mandibular molars at postnatal day 15 (P15). Like erupted molars, Adam10 cKO unerupted molars exhibit significantly reduced enamel volume and density. Enamel in the cervical region is present but is severely under mineralized. (D) For the unerupted molars, quantitative measurements confirmed an overall 16% and 15% reduction of total enamel volume and density respectively (**P < 0.01). A greater decrease in cervical enamel volume versus occlusal volume (17% versus 14%) and cervical density versus occlusal density (17% versus 13%) was also confirmed (**P < 0.01) in Adam10 cKO unerupted molars. Note that the total decrease in enamel volume for the Adam10 cKO erupted molars was 27% while that for the unerupted molars was 16%. This difference may represent the level of attrition in the 7-week-old erupted molars.
Figure 5.
2D and 3D renderings from µCT analyses of erupted and unerupted mandibular first molars in Adam10 cKO mice and controls. (A) Compared to controls, deletion of Adam10 leads to a poorly defined and under mineralized enamel layer. Sagittal and coronal views reveal flattening of Adam10 cKO cusps suggesting that these molars have undergone attrition. Occlusal views of Adam10 cKO molars show a severely hypomineralized enamel layer with reduced density as observed by the worn (3D) and darker (2D) enamel. (B) Quantitative analyses of total molar enamel volume and density show approximately a 27% decrease in volume and a 14% reduction in mineral density compared to controls (**P < 0.01). Segmentation of molars into occlusal and cervical regions reveal a greater reduction of volume and density at the cervical region of Adam10 cKO molars (**P < 0.01) when compared to the Amelx-iCre, and Adam10fl/fl molars. (C) 2D and 3D renderings from micro-CT analyses of unerupted mandibular molars at postnatal day 15 (P15). Like erupted molars, Adam10 cKO unerupted molars exhibit significantly reduced enamel volume and density. Enamel in the cervical region is present but is severely under mineralized. (D) For the unerupted molars, quantitative measurements confirmed an overall 16% and 15% reduction of total enamel volume and density respectively (**P < 0.01). A greater decrease in cervical enamel volume versus occlusal volume (17% versus 14%) and cervical density versus occlusal density (17% versus 13%) was also confirmed (**P < 0.01) in Adam10 cKO unerupted molars. Note that the total decrease in enamel volume for the Adam10 cKO erupted molars was 27% while that for the unerupted molars was 16%. This difference may represent the level of attrition in the 7-week-old erupted molars.
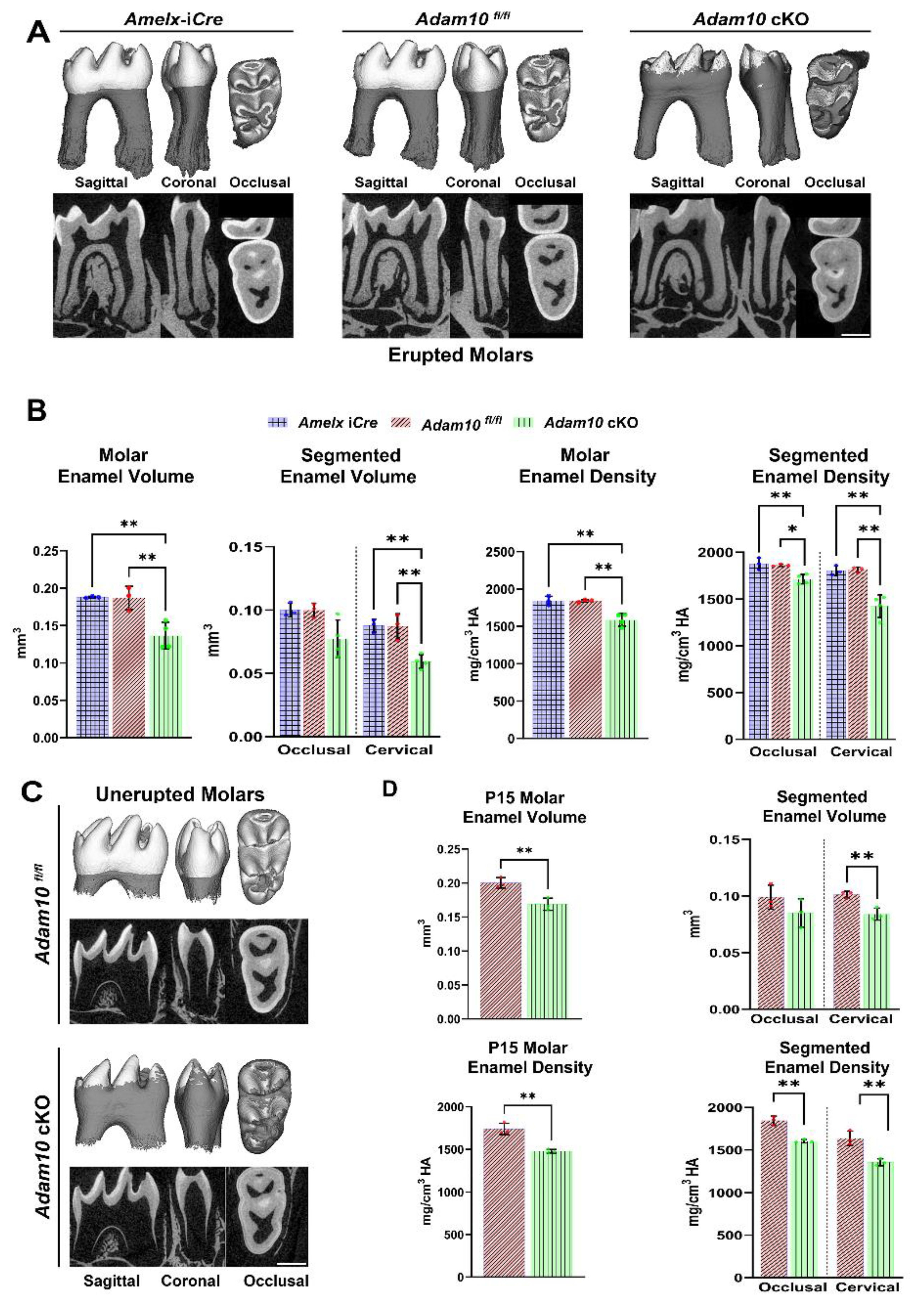
Figure 6.
Backscatter SEM of 7-week mandibular incisor cross sections from Amelx-iCre, Adam10fl/fl, and Adam10 cKO mice. (A) Panels show cross-sections of incisor enamel at the buccal alveolar crest where the enamel layer is near to erupting into the oral cavity. Normal enamel thickness is present in Amelx-iCre, and Adam10fl/fl incisors, but the Adam10 cKO enamel is darker indicative of increased protein content and appears thinner than normal. Scale bar 10 µm. (B-D) Higher magnification (2000x) reveals distinct enamel rod decussation patterns. Compared to the Amelx-iCre and Adam10fl/fl enamel, the rod/interrod pattern of Adam10 cKO enamel is somewhat disorganized near the dentin-enamel junction, but the enamel rods become progressively more loosely packed (porous) and disorganized towards the outer enamel edge. Scale bar 10 µm.
Figure 6.
Backscatter SEM of 7-week mandibular incisor cross sections from Amelx-iCre, Adam10fl/fl, and Adam10 cKO mice. (A) Panels show cross-sections of incisor enamel at the buccal alveolar crest where the enamel layer is near to erupting into the oral cavity. Normal enamel thickness is present in Amelx-iCre, and Adam10fl/fl incisors, but the Adam10 cKO enamel is darker indicative of increased protein content and appears thinner than normal. Scale bar 10 µm. (B-D) Higher magnification (2000x) reveals distinct enamel rod decussation patterns. Compared to the Amelx-iCre and Adam10fl/fl enamel, the rod/interrod pattern of Adam10 cKO enamel is somewhat disorganized near the dentin-enamel junction, but the enamel rods become progressively more loosely packed (porous) and disorganized towards the outer enamel edge. Scale bar 10 µm.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).