1. Introduction
Polycystic ovary syndrome (PCOS) is the most common endocrinopathy in women of childbearing age. The prevalence of PCOS varies according to region, race/ethnicity, and criteria used for diagnosis [
1]. Worldwide, it has been reported to affect between 5% to 15% of women [
2]. Diagnostic criteria for PCOS include irregular menstrual cycles, hyperandrogenism, and polycystic ovarian morphology [
3]. Given the complexity of this disorder, women frequently present associated clinical features such as endometrial cancer, reproductive abnormalities, metabolic and mental disorders, and an increased risk of developing cardiovascular disease [
4].
The etiology of PCOS has attempted to be explained by environmental and genetic factors. Being multifactorial, this disorder has been reported to be polygenic, with individual genes, gene-gene interactions, gene-environment interactions, and epigenetic events influencing the risk for PCOS development [
5]. PolyCystic Ovary Syndrome KnowledgeBase (PCOSKBR2), a manually curated database, consolidates information from 533 genes, 34 miRNAs, 145 single nucleotide polymorphisms (SNPs), 1150 pathways, 3874 ontologies, and 1230 diseases associated with PCOS [
6].
Genome-wide association studies (GWAS) contribute significantly to the knowledge of the genetic factor of this disorder by identifying SNPs in candidate genes that increase risk [
7]. However, the susceptibility of polymorphisms varies according to the population studied. Therefore, it is necessary to evaluate the SNPs identified in GWAS in different populations [
8]. The second GWAS performed in the Chinese population identified eight new risk loci including SNPs in
INSR (insulin receptor),
TOX3 (high mobility group box family member 3),
YAP1 (Yes1 associated transcriptional regulator),
HMGA2 (high mobility group AT-hook 2) and
ERBB3 (erb-b2 receptor tyrosine kinase 3) genes. These genes are involved in metabolic changes underlying the syndrome and in the development of associated comorbidities [
9].
The
INSR gene is located on chromosome 19p13.2 and consists of 22 exons, 21 introns, and 1382 amino acids [
10]. In PCOS women, it has been associated with dysfunctions associated with the insulin signaling pathway [
11]. In turn, the
TOX3 gene is located on chromosomal region 16q12.1 and consists of 10 exons and 576 amino acids [
12]. SNPs in this gene have been associated with insulin resistance [
13], hyperandrogenemia, oligomenorrhea, and ovarian morphology [
14]. The
YAP1 gene is located on chromosome 11q22.1, consists of 11 exons and 504 amino acids [
12]. By participating in the Hippo signaling pathway that contributes to follicle growth, it has been associated with infertility, a prevalent feature in women with PCOS [
15].
According to the UniProt database, the
HMGA2 gene is cytogenetically mapped at 12q14.3 and is composed of eight exons and 109 amino acids. In women with PCOS, it has been associated with type II diabetes mellitus, vascular tumors including angiomyxomas and pulmonary hamartomas [
9], oligomenorrhea, and hyperandrogenism [
14]. Finally, the
ERBB3 gene is located at position 12q13.2 and is composed of 30 exons and 1342 amino acids [
16]. Polymorphisms in
ERBB3 have been associated with insulin and glucose resistance [
17], and type 1 diabetes mellitus due to its immune regulation and beta cell apoptosis [
18].
For Colombia, as far as we know, there is no published research studying the relationship between these genes and PCOS. Therefore, the aim of this study is to evaluate the genetic contribution of five SNPs (rs2059807 in INSR, rs4784165 in TOX3, rs1894116 in YAP1, rs2272046 in HMGA2, and rs2292239 in ERBB3) to PCOS in a sample of Colombian women.
2. Results
2.1. Characteristics of Study Subjects
The clinical characteristics, hormonal, and biochemical features of PCOS patients and control group were previously described [
19]. Among the statistically significant differences between groups, it stands out that women with PCOS had higher weight, longer menstrual cycles, higher levels of anti-mullerian hormone (AMH), luteinizing hormone (LH), LH/ follicle-stimulating hormone (FSH) ratio, Estradiol (E
2), greater ovarian volume, and number of antral follicles. In addition, a greater number of women with the syndrome reported a family history of polycystic ovaries and endometriosis. In relation to reproductive characteristics, women with PCOS reported fewer pregnancies and a greater number of early pregnancy losses.
More than 50% of women with PCOS presented acne, hair loss, facial and abdominal hair, fatty discharge from the scalp and facial, menstrual bleeding stopped for more than three months, and dysmenorrhea.
2.2. Allelic and Genotypic Frequencies
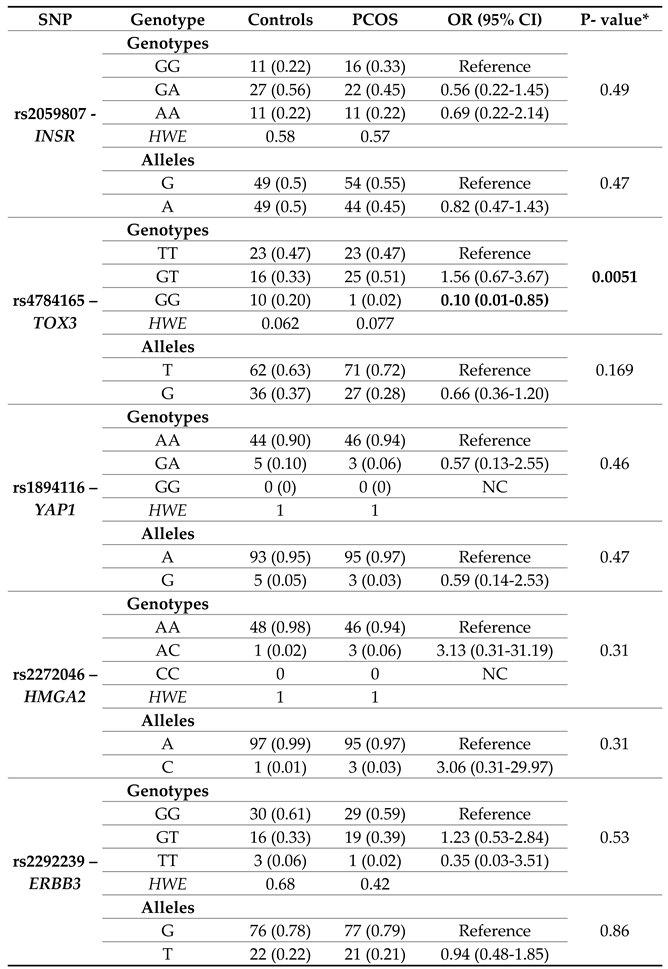
According to the National Center for Biotechnology Information (NCBI), for Latin American individuals with mostly European and Native American Ancestry, the nucleotide change for rs2059807-INSR is A>G,T, for rs4784165-TOX3 is T>A,G, for rs1894116-YAP1 is A>G, for rs2272046-HMGA2 is A>C, and for rs2292239-ERBB3 is T>C,G (
Supplementary Table S1). Our results were coincident for the polymorphisms in the TOX3, YAP1, and HMGA2 genes. The reference allele for the polymorphisms in INSR and ERRB3 was G.
Table 1 shows the distribution of genotypic and allelic frequencies in both groups. The genotypes distribution complied with HWE. No statistically significant differences were observed between groups for the rs2059807, rs1894116, rs2272046, and rs2292239 polymorphisms. Statistically significant differences were evident in the homozygous genotype for the lowest frequency allele GG of rs4784165–TOX3 (OR=0.10; CI=0.01-0.85; p=0.0051).
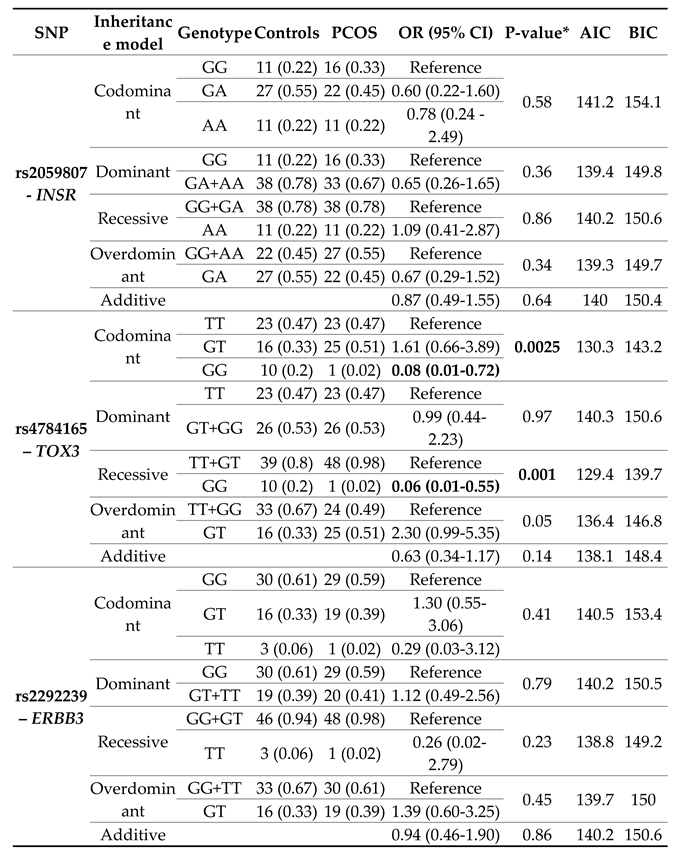
Table 2 details the risk to PCOS of the polymorphisms in each gene under the codominant, dominant, recessive, overdominant, and additive inheritance models. It should be noted that this analysis was not performed for the rs1894116 polymorphisms of YAP1 and rs2272046 of HMGA2, because no woman presented the homozygous genotype for the lowest frequency allele (GG and CC, respectively).
No associations were observed between PCOS and the genotypes of rs2059807-INSR, and rs2292239-ERBB3 polymorphisms under any inheritance model. However, with the logistic regression adjustment for age and BMI, statistically significant differences were evident between groups for the GG genotype of rs4784165 of the TOX3 gene, under the codominant (OR=0.008; CI=0.01-0.72; p=0.0025), and recessive models (OR=0.06; CI=0.01-0.55; p=0.001).
According to the Akaike’s information criterion (AIC) and Bayesian information criterion (BIC) values, the best inheritance model for rs2059807-INSR was overdominant (AIC:139.3; BIC:149.7), and recessive for rs4784165-TOX3 (AIC:129.4; BIC:139.7), and rs2292239-ERBB3 (AIC:138.8; BIC:149.2). For the polymorphisms rs1894116-YAP1 and rs2272046-HMGA2, the codominant model was assumed.
3.3. Genotype/Phenotype Association Analyses
Supplementary Tables S2–S6 detail the results obtained from the genotype/phenotype analysis for each SNP in PCOS women. The p-value for rs4784165-TOX3 and rs2292239-ERBBB3 was not calculated since there were genotypes with a single patient.
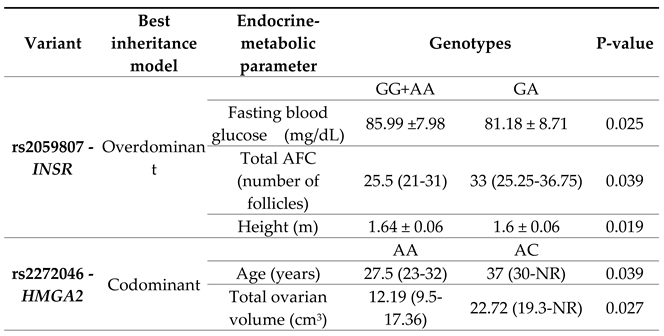
Table 3 shows a summary of the statistically significant associations.
Differences in rs2059807-INSR were identified using the overdominant model (GG+AA vs GA). Women with GG+AA genotypes had greater height (p=0.019) and higher fasting blood glucose levels (p=0.025), while women with GA genotype had a greater number of antral follicles (p=0.039).
In rs2272046–HMGA2, according to the codominant model (AA vs AC vs CC), differences in age and total ovarian volume were observed. PCOS women with the AC genotype presented greater ovarian volume compared to women with the homozygous genotype for the most frequent allele AA (p=0.027).
3. Discussion
In this study, we evaluated the individual effects of five polymorphisms in the INSR, TOX3, YAP1, HMGA2, and ERBB3 genes associated with PCOS. In our results, the lowest frequency allele for SNPs rs4784165-TOX3, rs1894116-YAP1, and rs2272046-HMGA2, matched that reported by NCBI for the Latino population. However, the lowest frequency allele for SNPs rs2059807-INSR and rs2292239-ERBB3 were not matched with the database. This may be due to the small sample size of our study.
The analysis of allelic, genotypic frequencies, OR, and CI (95%) showed a significant negative association between the GG genotype of the SNP rs4784165–
TOX3 and the risk of PCOS (OR=0.10; CI=0.01-0.85; p=0.0051), but at the genotype not at the allele level. This association was confirmed in the logistic regression adjustment under the co-dominant (OR=0.008; CI=0.01-0.72; p=0.0025), and recessive models (OR=0.06; CI=0.01-0.55; p=0.001).
TOX3 was first identified as a candidate gene for PCOS in a GWAS in a Chinese population, where the minor allele G of rs4784165 was positively associated with the syndrome [
9]. A Cross-Ethnic Meta-Analysis in women from China, United States, and The Netherlands reported an association between the SNP and PCOS (OR=1.18; p=8.1x10-3) [
20]. Furthermore, this polymorphism has been linked with clinical characteristics of the syndrome such as homeostasis model assessment of insulin resistance (HOMA-IR) [
13], hyperandrogenism [
21], and is involved in inflammation processes and its consequences [
22].
In a previous SNP interaction study, we identified that rs4784165–
TOX3 interacts with rs11692782–
FSHR and rs2268361–
FSHR, conferring a considerable increase in PCOS risk (OR=11.29; p<0.0001). This highlights the contrasting results between analyzing individual genetic effects versus gene-gene interactions [
23].
No statistically significant differences were observed between groups for the rs2059807-
INSR, rs1894116-
YAP1, rs2272046-
HMGA2, and rs2292239-
ERBB3 polymorphisms. Similar results, indicating no associations for rs2059807-
INSR, have been reported in studies involving women from Korea [
24], Iran [
25], and The Netherlands [
20]. Few studies have evaluated the genetic contribution of the SNPs rs1894116-
YAP1, rs2272046-
HMGA2, and rs2292239-
ERBB3 and PCOS.
The genotype–phenotype analysis demonstrated a significant association between rs2059807-
INSR, and rs2272046-
HMGA2 and PCOS features. Using the overdominant model (GG+AA vs GA) in
INSR, it was observed that women with the GG+AA genotypes presented higher levels of fasting blood glucose compared to women with the heterozygous GA genotype (85.99 vs 81.18 mg/dL; p=0.025). It should be noted that these data are within the normal reference values for this variable. The
INSR gene plays a fundamental role in the insulin signaling pathway, therefore, SNPs such as rs2059807 may influence metabolic disorders associated with the syndrome, since increased glucose levels may indicate an inefficient response to insulin [
26]. These results may help prevent future complications in women at higher risk. In turn, the heterozygous GA genotype was associated with a higher number of antral follicles compared to the GG+AA genotypes (33 vs 25.5; p=0.039). A similar result was reported by Cui et al., who identified an association between rs2059807 and women with anovulation, a diagnostic criterion for PCOS characterized by an increased number of follicles in the ovaries due to failure to release an egg for fertilization [
27].
In rs2272046-
HMGA2, associations with age and total ovarian volume (OV) were observed. Women with the heterozygous AC genotype compared to women with the homozygous genotype for the most frequent allele AA, presented higher age (27.5 vs. 37 years; p=0.039) and total OV (12.19 vs. 22.72 cm3; p=0.027). Although Alsamarai et al., concluded that OV decreases with age in women with PCOS [
28], other authors have shown that there is no relationship between OV and age, and, therefore, there are no significant changes in OV during the reproductive ages up to perimenopause [
29]. Meanwhile, overexpression of
HMGA2 in granulosa cells has been identified to contribute to increased cell proliferation, and thus to the polycystic ovarian phenotype [
30]. The above may be related to increased OV in these women.
The main limitation of the research is associated with the small sample size, which restricts adequate statistical power. However, considering the relationship between the population studied and the risk of PCOS, our results correspond to the first approximation of the study of these genes in women with PCOS in the country and represent a baseline for future population studies.
4. Materials and Methods
4.1. Subjects
This pilot study included 49 control women and 49 women with a confirmed diagnosis of PCOS. The inclusion and exclusion criteria were previously detailed [
19]. The diagnosis was made according to the Rotterdam criteria [
31]. All women signed the informed consent. The study was conducted in compliance with The Code of Ethics of the World Medical Association, and the study protocol was approved by the research and ethics committees of the Universidad Pedagógica y Tecnológica de Colombia (UPTC) (Reference number: SGI code 2386-VIE 05 of 2018; SGI code 2677-VIE 06 of 2019) and Universidad de Boyacá (Reference number: 011–2019CB, 29/03/2019).
4.2. Clinical Measurements and Biochemical Analyses
Data collected included age, height, weight, body mass index (BMI), menstrual features, family background, reproductive features, and presence of signs associated with PCOS.
Between 07:00 and 09:00 a.m. following an overnight fast, venous blood samples were taken. Blood biochemical analyses included measuring AMH, and androstenedione using ELISA immunoassay (MyBiosource, San Diego, USA, MBS2023458, and DiaMetra, Italy, DKO008, respectively). Levels of FSH, dehydroepiandrosterone sulfate (DHEAS), and LH were determined using chemiluminescence techniques. Plasma insulin, E2, and thyroid-stimulating hormone (TSH) were analyzed using the amplified enzyme chemiluminescence technique (SIEMENS IMMULITE, Germany). Free testosterone was measured using the radioimmunoassay (RIA) technique. Plasma glucose levels were determined using the hexokinase method (GLUC3 GLUCOSE HK GEN.3, 04404483190, Roche Diagnostics), and glycosylated hemoglobin (HbA1C) was measured using the HbA1C monoclonal antibody technique (MyBiosource, San Diego, USA, MBS2031845), following the manufacturer’s instructions.
Calculation of the HOMA-IR was: fasting insulin (mIU/L)*fasting glucose (mmol/L)/22.5, and for homeostatic model assessment for insulin sensitivity (HOMA-IS) was: 1/fasting insulin (mIU/L)*fasting glucose (mmol/L).
4.3. Polymorphisms Genotyping
The Invisorb R Spin Universal Kit (Stratec Molecular) was used for DNA extraction from the blood. The DNA concentration was read by EPOCHTM2 Microplate Spectrophotometer (Biotek). The DNA was stored at -20 °C until use. Using Assay Design Suite (ADS) software, specific primers were designed for each gene (
INSR, TOX3, YAP1, HMGA2, and
ERBB3). Design data is presented in
Supplementary Table S7. SNPs typing was performed with the iPLEX Assay and the MassARRAY system (Agena Bioscience) which employs matrix-assisted laser desorption/ionization mass spectrometry (MALDI-TOF MS). We followed the previously established protocol [
32].
4.4. Statistical Analyses
Analyses were performed using the IBM-SPSS statistical package (version 25.0). Categorical variables are described as number of cases and absolute frequencies. Continuous variables with a normal distribution are expressed as mean ± standard deviation (mean± SD). Continuous variables that did not follow a normal distribution are expressed as median (interquartile range). Differences in categorical variables between groups were estimated using a chi-square test. The Student’s T-test was used to compare quantitative normal variables between groups, and the Mann–Whitney U-test to compare non-normal variables.
Genotype and allelic frequencies of target SNPs were calculated in both groups. All SNPs were tested for Hardy– Weinberg equilibrium (HWE) with the chi-square test. The odds ratios (ORs) between groups and 95% confidence intervals (95% CI) were evaluated. A logistic regression analysis for age and BMI was designed to evaluate the risk of PCOS under the codominant, dominant, recessive, overdominant, and additive inheritance models. AIC and BIC were used to determine the best genetic model for each SNP [
33]. The genotype/phenotype association analyses were performed with the best inheritance model. Significant statistical differences were assumed in all cases showing p<0.05.
5. Conclusions
The present pilot study has shown that the SNP rs4784165-TOX3 gene is potentially negatively associated with PCOS in a sample of Colombian women. In addition, we identified some SNP genotypes associated with PCOS features. In the rs2059807-INSR, women with GG+AA genotypes had higher levels of fasting blood glucose, and women with GA genotypes had a higher number of total antral follicles. Women with AC genotype at rs2272046-HMGA2 presented higher age and total ovarian volume. Considering the SNP-population-PCOS relationship, this research represents the first generation of knowledge on this topic for the country. However, a population study is suggested to confirm these findings.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org.
Table S1: SNPs genotyped in the
INSR, TOX3, YAP1, HMGA1, and
ERBB3 genes; Table S2: Clinical and endocrine-metabolic parameters of women with PCOS according to the overdominant model of rs2059807 in
INSR gene; Table S3: Clinical and endocrine-metabolic parameters of women with PCOS according to the recessive model of rs4784165 in
TOX3 gene; Table S4: Clinical and endocrine-metabolic parameters of women with PCOS according to the genotypes of rs1894116 in
YAP1 gene; Table S5: Clinical and endocrine-metabolic parameters of women with PCOS according to the genotypes of rs2272046 in
HMGA2 gene; Table S6: Clinical and endocrine-metabolic parameters of women with PCOS according to the recessive model of rs2292239 in
ERBBB3 gene; Table S7: MassArray design details for SNPs (rs2059807 in
INSR, rs4784165 in
TOX3, rs1894116 in
YAP1, rs2272046 in
HMGA2, and rs2292239 in
ERBB3).
Author Contributions
Conceptualization, all authors; methodology, all authors; validation, all authors; formal analysis, MCAG and MFC; investigation, all authors; resources, all authors; data curation, MCAG, HMO, MFC; writing—original draft preparation, MCAG; writing—review and editing, MFC; visualization, all authors; supervision, MFC and GECV; project administration, MCAG, AFC, GECV and MFC; funding acquisition, MFC and GECV. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded through an inter-institutional cooperation between the UPTC and Universidad de Boyacá (SGI code 2386-VIE 05 of 2018; SGI code 2677-VIE 06 of 2019).
Institutional Review Board Statement
This study was approved by the Ethics Committee of the Universidad Pedagógica y Tecnológica de Colombia (reference number: SGI code 2386 - VIE 05 of 2018; SGI code 2677-VIE 06 of 2019) and Universidad de Boyacá (reference number: 011–2019 CB, 29/03/2019) and followed the guidelines of the Declaration of Helsinki. Additionally, each participant signed an informed consent form.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Acknowledgments
This research was supported by projects SGI 2386 and SGI 2677, established through a collaboration between the Research Group in Biomedical Sciences at Universidad Pedagógica y Tecnológica de Colombia (GICBUPTC) and the Public Health Research Group (HYGEA) at Universidad de Boyacá. Genotyping services were provided by CEGEN-PRB3-ISCIII, with funding from grant PT17/0019, part of the PE I+D+i 2013-2016 initiative, financed by ISCIII and the European Regional Development Fund (ERDF)
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wolf, W.M.; Wattick, R.A.; Kinkade, O.N.; Olfert, M.D. Geographical Prevalence of Polycystic Ovary Syndrome as Determined by Region and Race/Ethnicity. International Journal of Environmental Research and Public Health 2018, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ajmal, N.; Khan, S.Z.; Shaikh, R. Polycystic Ovary Syndrome (PCOS) and Genetic Predisposition: A Review Article. European Journal of Obstetrics & Gynecology and Reproductive Biology X 2019, 3, 100060. [Google Scholar] [CrossRef]
- Abruzzese, G.A.; Velazquez, M.E.; Cerrone, G.E.; Motta, A.B. Polycystic Ovary Syndrome in Latin American Populations: What Is Known and What Remains Unresolved. The Journal of Steroid Biochemistry and Molecular Biology 2023, 225, 106195. [Google Scholar] [CrossRef] [PubMed]
- Dapas, M.; Dunaif, A. Deconstructing a Syndrome: Genomic Insights Into PCOS Causal Mechanisms and Classification. Endocr Rev 2022, 43, 927–965. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.J.; Ullah, A.; Basit, S. Genetic Basis of Polycystic Ovary Syndrome (PCOS): Current Perspectives. Application of Clinical Genetics 2019, 12, 249–260. [Google Scholar] [CrossRef]
- Sharma, M.; Barai, R.S.; Kundu, I.; Bhaye, S.; Pokar, K.; Idicula-Thomas, S. PCOSKBR2: A Database of Genes, Diseases, Pathways, and Networks Associated with Polycystic Ovary Syndrome. Sci Rep 2020, 10, 14738. [Google Scholar] [CrossRef]
- Nautiyal, H.; Imam, S.S.; Alshehri, S.; Ghoneim, M.M.; Afzal, M.; Alzarea, S.I.; Güven, E.; Al-Abbasi, F.A.; Kazmi, I. Polycystic Ovarian Syndrome: A Complex Disease with a Genetics Approach. Biomedicines 2022, 10, 540. [Google Scholar] [CrossRef]
- Sendur, S.N.; Yildiz, B.O. Influence of Ethnicity on Different Aspects of Polycystic Ovary Syndrome: A Systematic Review. Reproductive BioMedicine Online 2021, 42, 799–818. [Google Scholar] [CrossRef]
- Shi, Y.; Zhao, H.; Shi, Y.; Cao, Y.; Yang, D.; Li, Z.; Zhang, B.; Liang, X.; Li, T.; Chen, J.; et al. Genome-Wide Association Study Identifies Eight New Risk Loci for Polycystic Ovary Syndrome. Nature Genetics 2012, 44, 1020–1025. [Google Scholar] [CrossRef]
- Daghestani, M.H. Rs1799817 in INSR Associates with Susceptibility to Polycystic Ovary Syndrome. Journal of Medical Biochemistry 2019. [Google Scholar] [CrossRef]
- Rasool, S.U.A.; Ashraf, S.; Nabi, M.; Masoodi, S.R.; Fazili, K.M.; Amin, S. Clinical Manifestations of Hyperandrogenism and Ovulatory Dysfunction Are Not Associated with His1058 C/T SNP (Rs1799817) Polymorphism of Insulin Receptor Gene Tyrosine Kinase Domain in Kashmiri Women with PCOS. Int J Endocrinol 2021, 2021, 7522487. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.-Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving Coverage, Classification and Access to Protein Sequence Annotations. Nucleic Acids Res 2019, 47, D351–D360. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Li, J.; Su, S.; Cao, Y.; Wang, Z.; Zhao, S.; Zhao, H. PCOS-GWAS Susceptibility Variants in THADA, INSR, TOX3, and DENND1A Are Associated With Metabolic Syndrome or Insulin Resistance in Women With PCOS. Frontiers in Endocrinology 2020, 11, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bakhashab, S.; Ahmed, N. Genotype Based Risk Predictors for Polycystic Ovary Syndrome. Bioinformation 2019, 15, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, K.; Cheng, Y.; Suzuki, N.; Deguchi, M.; Sato, Y.; Takae, S.; Ho, C.; Kawamura, N.; Tamura, M.; Hashimoto, S.; et al. Hippo Signaling Disruption and Akt Stimulation of Ovarian Follicles for Infertility Treatment. Proc Natl Acad Sci U S A 2013, 110, 17474–17479. [Google Scholar] [CrossRef]
- Sithanandam, G.; Anderson, L.M. The ERBB3 Receptor in Cancer and Cancer Gene Therapy. Cancer Gene Ther 2008, 15, 413–448. [Google Scholar] [CrossRef]
- Day, F.R.; Hinds, D.A.; Tung, J.Y.; Stolk, L.; Styrkarsdottir, U.; Saxena, R.; Bjonnes, A.; Broer, L.; Dunger, D.B.; Halldorsson, B.V.; et al. Causal Mechanisms and Balancing Selection Inferred from Genetic Associations with Polycystic Ovary Syndrome. Nature Communications 2015. [Google Scholar] [CrossRef]
- Wang, D.; Pan, G. The Association between Rs2292239 Polymorphism in ERBB3 Gene and Type 1 Diabetes: A Meta-Analysis. Biomed Res Int 2019, 2019, 7689642. [Google Scholar] [CrossRef]
- Alarcón-Granados, M.C.; Moreno-Ortíz, H.; Esteban-Pérez, C.I.; Ferrebuz-Cardozo, A.; Camargo-Villalba, G.E.; Forero-Castro, M. Assessment of THADA Gene Polymorphisms in a Sample of Colombian Women with Polycystic Ovary Syndrome: A Pilot Study. Heliyon 2022, 8, e09673. [Google Scholar] [CrossRef]
- Louwers, Y.V.; Stolk, L.; Uitterlinden, A.G.; Laven, J.S.E. Cross-Ethnic Meta-Analysis of Genetic Variants for Polycystic Ovary Syndrome. Journal of Clinical Endocrinology and Metabolism 2013, 98, 2006–2012. [Google Scholar] [CrossRef]
- Cui, Y.; Zhao, S.; Zhao, H.; Lv, Y.; Yu, M.; Wang, Y.; Chen, Z.-J. Mutational Analysis of TOX3 in Chinese Han Women with Polycystic Ovary Syndrome. Reprod Biomed Online 2014, 29, 752–755. [Google Scholar] [CrossRef] [PubMed]
- Pau, C.T.; Mosbruger, T.; Saxena, R.; Welt, C.K. Phenotype and Tissue Expression as a Function of Genetic Risk in Polycystic Ovary Syndrome. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Alarcón-Granados, M.C.; Camargo-Villalba, G.E.; Forero-Castro, M. Exploring Genetic Interactions in Colombian Women with Polycystic Ovarian Syndrome: A Study on SNP-SNP Associations. International Journal of Molecular Sciences 2024, 25, 9212. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.-J.; Oh, B.; Lee, J.-Y.; Kimm, K.; Lee, S.-H.; Baek, K.-H. A Novel Single Nucleotide Polymorphism of INSR Gene for Polycystic Ovary Syndrome. Fertility and Sterility 2008, 89, 1213–1220. [Google Scholar] [CrossRef]
- Ranjzad, F.; Mahmoudi, T.; Shemirani, A.I.; Mahban, A.; Nikzamir, A.; Vahedi, M.; Ashrafi, M.; Gourabi, H. A Common Variant in the Adiponectin Gene and Polycystic Ovary Syndrome Risk. Molecular Biology Reports 2012, 39, 2313–2319. [Google Scholar] [CrossRef]
- Shaaban, Z.; Khoradmehr, A.; Amiri-Yekta, A.; Nowzari, F.; Jafarzadeh Shirazi, M.R.; Tamadon, A. Pathophysiologic Mechanisms of Insulin Secretion and Signaling-Related Genes in Etiology of Polycystic Ovary Syndrome. Genet Res (Camb) 2021, 2021, 7781823. [Google Scholar] [CrossRef]
- Cui, L.; Li, G.; Zhong, W.; Bian, Y.; Su, S.; Sheng, Y.; Shi, Y.; Wei, D.; Zhang, W.; Zhao, H.; et al. Polycystic Ovary Syndrome Susceptibility Single Nucleotide Polymorphisms in Women with a Single PCOS Clinical Feature. Human Reproduction 2015, 30, 732–736. [Google Scholar] [CrossRef]
- Alsamarai, S.; Adams, J.M.; Murphy, M.K.; Post, M.D.; Hayden, D.L.; Hall, J.E.; Welt, C.K. Criteria for Polycystic Ovarian Morphology in Polycystic Ovary Syndrome as a Function of Age. J Clin Endocrinol Metab 2009, 94, 4961–4970. [Google Scholar] [CrossRef]
- Han, Y.S.; Lee, A.R.; Song, H.K.; Choi, J.I.; Kim, J.H.; Kim, M.R.; Kim, M.J. Ovarian Volume in Korean Women with Polycystic Ovary Syndrome and Its Related Factors. J Menopausal Med 2017, 23, 25–31. [Google Scholar] [CrossRef]
- Li, M.; Zhao, H.; Zhao, S.-G.; Wei, D.-M.; Zhao, Y.-R.; Huang, T.; Muhammad, T.; Yan, L.; Gao, F.; Li, L.; et al. The HMGA2-IMP2 Pathway Promotes Granulosa Cell Proliferation in Polycystic Ovary Syndrome. J Clin Endocrinol Metab 2019, 104, 1049–1059. [Google Scholar] [CrossRef]
- Christ, J.P.; Cedars, M.I. Current Guidelines for Diagnosing PCOS. Diagnostics 2023, 13, 1113. [Google Scholar] [CrossRef]
- Gabriel, S.; Ziaugra, L.; Tabbaa, D. SNP Genotyping Using the Sequenom massARRAY iPLEX Platform. Current Protocols in Human Genetics 2009. [Google Scholar] [CrossRef]
- Li, W.; Nyholt, D.R. Marker Selection by Akaike Information Criterion and Bayesian Information Criterion. Genet Epidemiol 2001, 21 Suppl 1, S272–S277. [Google Scholar] [CrossRef]
Table 1.
Genotypic and allelic frequencies of polymorphisms in the INSR, TOX3, YAP1, HMGA1, and ERBB3 genes.
Table 1.
Genotypic and allelic frequencies of polymorphisms in the INSR, TOX3, YAP1, HMGA1, and ERBB3 genes.
Table 2.
Variant risk analysis according to inheritance models adjusted for age and body mass index.
Table 2.
Variant risk analysis according to inheritance models adjusted for age and body mass index.
Table 3.
Associations identified in the genotype/phenotype analyses.
Table 3.
Associations identified in the genotype/phenotype analyses.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).