Submitted:
15 October 2024
Posted:
16 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
- Disconnected patient records negatively impact patients, clinicians, and population health planners. These gaps lead to suboptimal patient care, increased workloads for clinicians, and hinder the effectiveness of health planners and policymakers.
- Inflexible systems struggle to identify and adapt to changing healthcare needs and priorities.
- Manual data processing is not only resource-intensive but also susceptible to errors, further straining an already overwhelmed system.
- Structural Integration: A federated database design that preserves the autonomy of existing systems while facilitating varying levels of privacy and access.
- Semantic Integration: A record linkage module that complies with data governance policies, allowing for integration even without a universal identifier. This approach can be applied to systems such as HIPE, CDM, PCRS, and RetinaScreen.
- Adoption of Standards: A new framework based on the Fast Healthcare Interoperability Resources (HL7-FHIR) model [4], ensuring high levels of interoperability within integrated FHIR data, regardless of the participating healthcare systems.
2. Methodology
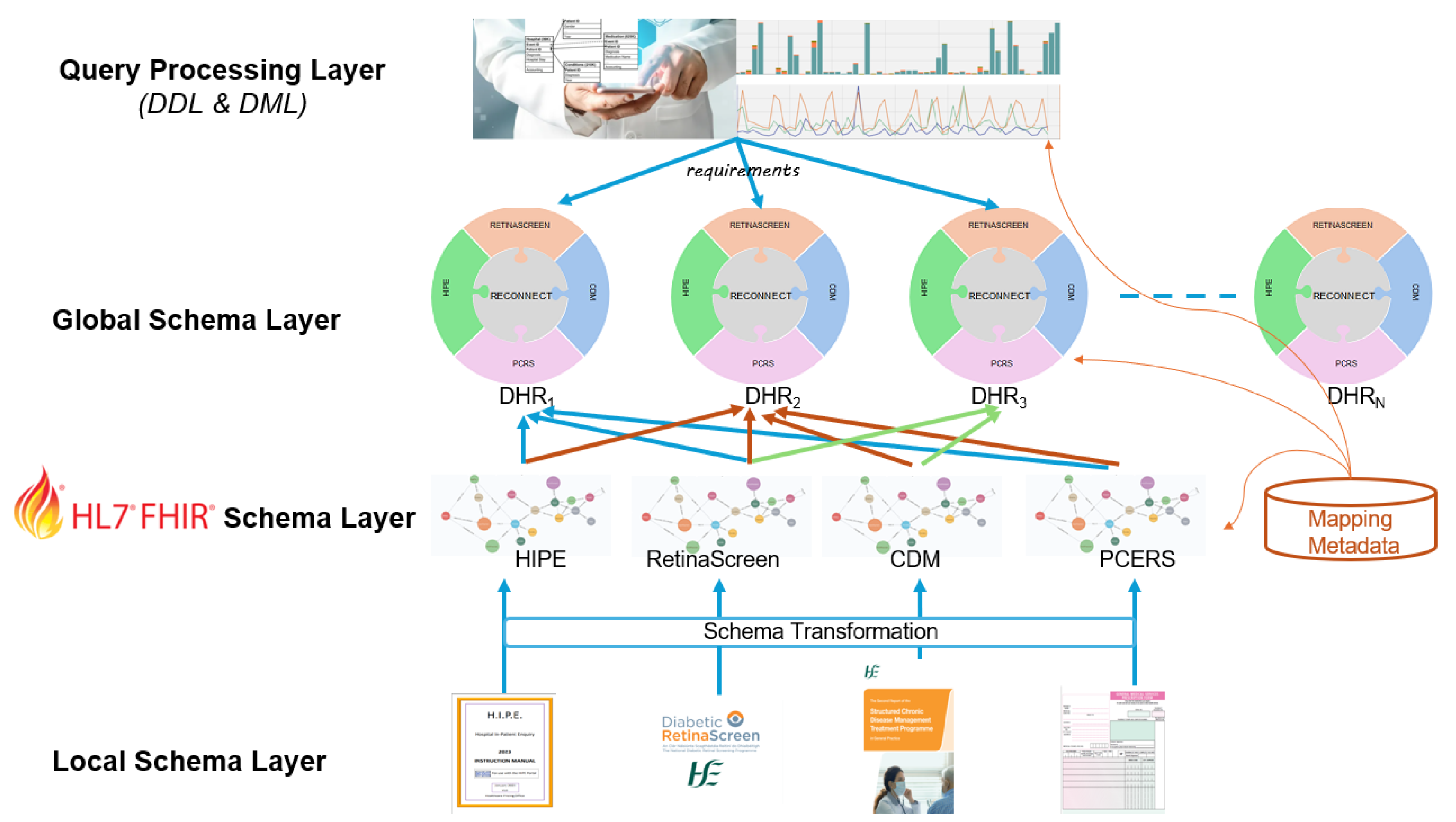
2.1. Local Schema Layer
- HIPE: Hospital In-Patient Enquiry
- CDM: Chronic Disease Management
- PCRS: Primary Care Reimbursement Service
- Retina Screen
2.2. FHIR Schema Layer
- ONE_TO_ONE mappings: Applied when there is a direct correspondence between source and FHIR properties, allowing the attribute value from the data source to be directly imported.
- MANY_TO_ONE mappings: Used when multiple source attributes are needed to populate a single FHIR property.
- INDIRECT mappings: Utilized to provide default values for FHIR resources that are absent in the source data.
- LOOKUP mappings: Indicate attributes that require record linkage to populate the corresponding FHIR property.
2.3. Global Schema Layer
2.4. Query Processing Layer
-
Uptake of Retina Screen among People with Diabetes. This case study refers to the percentage of people not participating in the prevention services led by the government but ending up in the hospital. Datasets used:
- (a)
- Retinascreen
- (b)
- CDM
- (c)
- HIPE
Diabetic retinopathy (DR) is the leading cause of preventable blindness. The independent risk factors for DR included diabetes duration, haemoglobin A1c, serum glucose, systolic blood pressure, and duration of diabetes. After 5 years, approximately 25% of type 1 diabetes patients will have retinopathy. After 10 years, almost 60% will have retinopathy, and after 15 years, 80% will have retinopathy. International guidelines for diabetic retinopathy (DR) screening, released by the International Council of Ophthalmology (ICO), specify that adequate DR screening should encompass a visual acuity test and a retinal examination.This case study aims to pull data from Retinascreen, CDM and HIPE databases. The data governance layer specifies the level of detail accessible by the system operator. -
Blood Pressure Control among People with DiabetesDatasets:
- (a)
- PCRS
- (b)
- CDM
- (c)
- HIPE
Randomized controlled trials have shown that lowering systolic blood pressure (SBP) to less than 140 mmHg and diastolic blood pressure (DBP) to less than 90 mmHg benefits people with diabetes. If SBP is 140 mmHg or more and/or DBP is 90 mmHg or more, drug therapy is necessary, preferably starting with a combination therapy. The use of renin-angiotensin system (RAS) inhibitors is strongly supported, especially in patients with evidence of end-organ damage. Controlling blood pressure often requires multiple drug therapies, and a combination of two or more drugs at fixed doses in a single pill should be considered to improve adherence and achieve earlier control of blood pressure. -
Amputations among People with DiabetesDatasets:
- (a)
- CDM
- (b)
- HIPE
Diabetes can lead to foot or leg amputation, with a limb amputated every 20 seconds globally due to diabetes. 85% of these amputations are preceded by a foot ulcer. The HSE introduced the National Diabetes Footcare program in 2010, recommending annual foot screenings for people with diabetes to assess their risk of lower extremity amputation. Those at risk should be referred to foot protection services in the community or hospital setting.
3. Generating Healthcare Assets
- Integrating HIPE, CDM, PCRS, and Retinascreen systems for an Individual: This asset involves the comprehensive integration of health data from HIPE, CDM, PCRS, and RetinaScreen for individual patients. The goal is to provide a holistic view of a patient’s health status and enhance care coordination, as shown in Figure 4.
- Uptake of Retina Screen among People with Diabetes: Since retinopathy affects individuals with both Type 1 and Type 2 diabetes, this dataset tracks the number of people who have undergone RetinaScreen based on their type of diabetes. Understanding these numbers can help identify gaps in care and improve screening practices.
- Uptake of Retina Screen among People on Type of Diabetes: Since retinopathy affects individuals with both Type 1 and Type 2 diabetes, this dataset tracks the number of people who have undergone RetinaScreen, categorized by their type of diabetes. Analyzing these numbers can help identify gaps in care and improve screening practices.
- Multimorbidity (Prevalence of More Than One Chronic Disease): This dataset focuses on the prevalence and management of individuals with multiple chronic diseases, also known as multimorbidity. It highlights the need for comprehensive care strategies to address the complexities of managing multiple health conditions simultaneously. The CDM system tracks individuals diagnosed with Type 2 diabetes, asthma, chronic obstructive pulmonary disease (COPD), and cardiovascular diseases, including stable heart failure, ischaemic heart disease, cerebrovascular disease (stroke/TIA), and atrial fibrillation. This dataset helps identify subsets of individuals with similar underlying risk factors.
- Diabetes, Medication, and Physical Activity: This dataset explores the relationship between diabetes management, medication usage, and the role of physical activity. It emphasizes the impact of lifestyle changes, particularly exercise, on medication requirements and overall diabetes control. Engaging in physical activity fewer than three times per week is identified as a risk factor for developing chronic diseases.
- Age, Physical Activity, and Hospital Admission due to Chronic Disease: This dataset explores the correlation between a patient’s age, level of physical activity, and the frequency of hospital admissions related to chronic diseases. It emphasizes the importance of promoting physical activity, particularly among older adults, to reduce hospitalizations.
- Blood Pressure Control among People with Diabetes: This dataset focuses on the critical need to manage blood pressure in individuals with diabetes. Drug therapy is recommended for those with diastolic blood pressure above 90 and systolic blood pressure above 140. Proper blood pressure control is essential for preventing complications such as stroke, coronary events, and kidney disease.
- Cardiovascular Disease among People with Diabetes: This dataset identifies diabetes patients who are at an increased risk of developing cardiovascular disease. It highlights the importance of regular screenings and preventive measures to mitigate this risk.
- Hospital Admissions among People with Diabetes: This dataset compiles data from patients registered in both the CDM and HIPE systems, focusing on the number of hospital admissions for individuals diagnosed with Type 2 diabetes. Its goal is to identify patterns and causes of hospitalizations, improving diabetes management and reducing healthcare costs.
- Amputations among People with Diabetes: This dataset tracks the incidence of amputations in individuals with diabetes, often resulting from complications like neuropathy, poor circulation, and other risk factors. It highlights the importance of preventive care, such as regular foot screenings and early interventions, to reduce diabetes-related amputations.
- Identifying Patients in a Demographic Location Based on Gender: This dataset categorizes patients within specific demographic locations by gender. The data can be used to tailor healthcare services and design targeted outreach programs.
- Identifying Patients in a Demographic Location Based on Age: This dataset focuses on identifying patients within specific demographic locations, categorized by age. Understanding the age distribution allows healthcare providers to address the specific needs of different age groups more effectively. Individuals aged 45 and older are more prone to developing chronic diseases and other risk factors.
- Medications for People with Both Diabetes and Hypertension: This dataset examines the various medications prescribed to individuals managing both diabetes and hypertension. It underscores the importance of addressing both conditions concurrently to reduce health risks. This data also aids clinicians in developing better care models for these patients
- Identify Patient Subgroups with Shared Conditions: This dataset identifies subgroups of patients with similar health conditions, enabling healthcare providers to develop targeted interventions. Such insights can improve the effectiveness of treatment plans and enhance patient outcomes.
3.1. Technical Methodology
-
Parameters:condition_type: Used to determine which query to run based on the provided value.identification: An identifier (could be MRN, ID, mobile, etc.) to be used in the SQL queries.
- Dynamic Table Creation: The procedure dynamically creates a table named result_value based on the type of data being fetched, which is determined by the condition_type provided.
- CASE Structure: Depending on the value of condition_type, a corresponding query is executed. Each case uses a specific function (e.g., hipe_data, cdm_data, etc.) to retrieve data from various tables.
- ELSE Clause: If none of the provided conditions match, the procedure raises a notice saying “Check selected function”
- Parameters: The function accepts several parameters, including the table names (TABLE1, TABLE2, TABLE3, TABLE4, TABLE5, TABLE6) and filtering conditions (WHERECLAUSE1, WHERECLAUSE2, WHERECLAUSE3).
- Dynamic SQL Query: The query variable is constructed using the format function, which dynamically inserts table names and WHERE conditions into the SQL statement.
- Joins: It performs several JOIN operations between the tables to gather patient data like MRN, IHI, contact details, diagnosis, andor screening details.
- WHERE Clause: The query filters data based on screening date, diagnosis, and chronic diseases.
- Execution: The dynamically generated query is executed, and the result set is returned using RETURN QUERY EXECUTE query.
-
F1_mrn: Example 1 a retrieves data based on the mrn (medical record number) using the hipe_data function. This function integrates all the systems in the local schema and provides details regarding each individual patient from all systems.call dynamiccaseprocedure(’F1mrn’,’10164260’);select * from resultvalue;Sample Query 1.
-
F1_id: Example 2 retrieves data based on the Individual Health Identifiers using the cdm_data function. This function integrates CDM, PCRS and RetinaScreen systems from the local schema and provides details regarding an individual from all systems.call dynamiccaseprocedure(’F1id’,’10043’);select * from resultvalue;Sample Query 2.
-
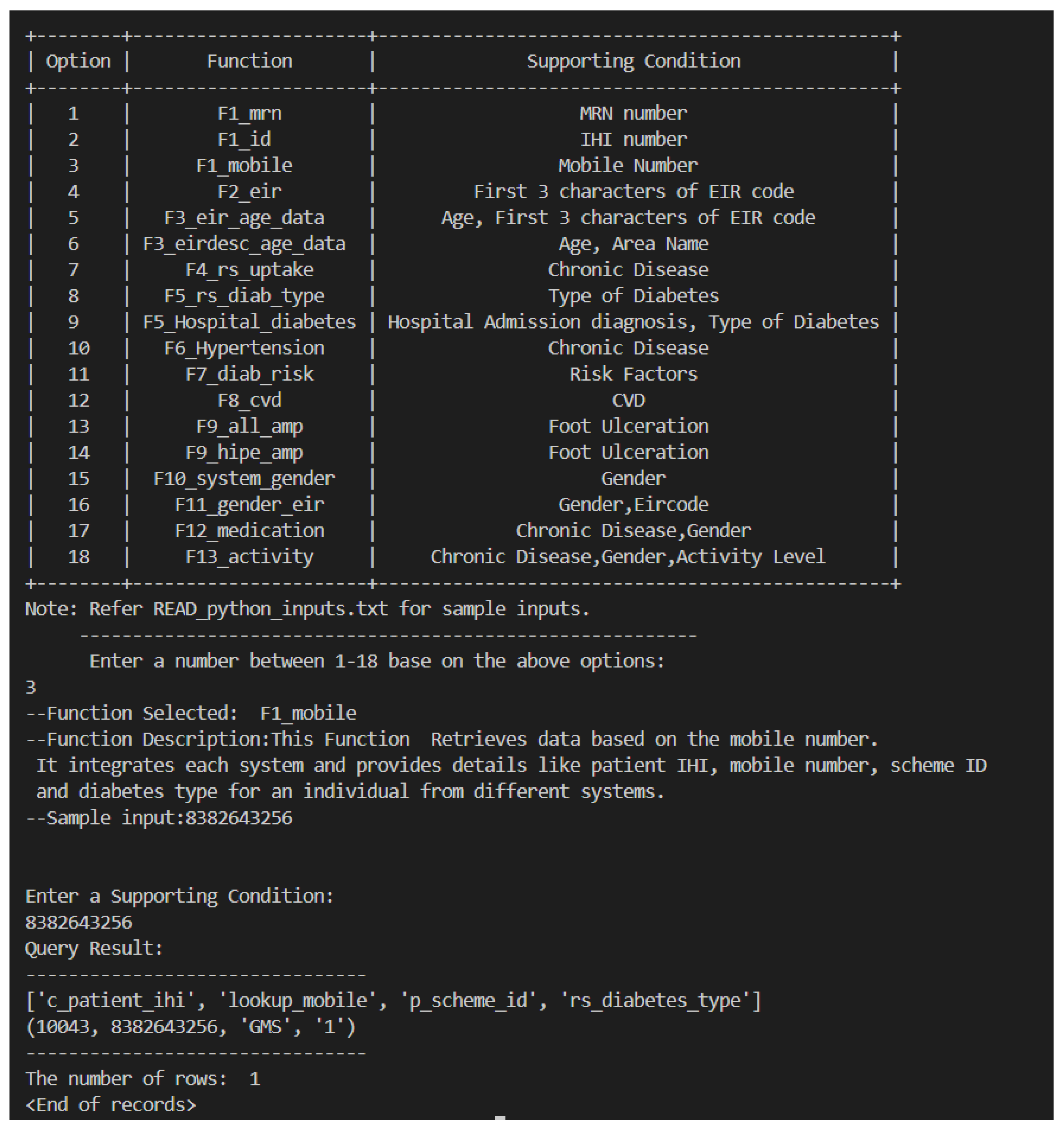
F1_mobile: Example 3 retrieves data based on the mobile number using the rs_data function. It provides a similar functionality as the previous function however the mobile number uniquely identifies the patient.call dynamiccaseprocedure(’F1mobile’,’8382643256’);select * from resultvalue;Sample Query 3.
-
F2_eir: Example 4 retrieves patient data based on the EIR code using the eir_data function. The first three characters of the Eircode that identify the area are stored in the database.call dynamiccaseprocedure(’F2eir’,’F52’);select * from resultvalue;Sample Query 4.
-
F3_eir_age_data: Example 5 retrieves data for patients filtered on both age and Eircode using the eir_age function. It takes a minimum age and the first three characters of an Eircode as parameters and retrieves patient details such as name, sex, address, age, and Eircode. This function is designed to retrieve data for up to 3 Eircodes in a single query.call dynamiccaseprocedure(’F3eirabove45data’,’F52’);select * from resultvalue;Sample Query 5.
-
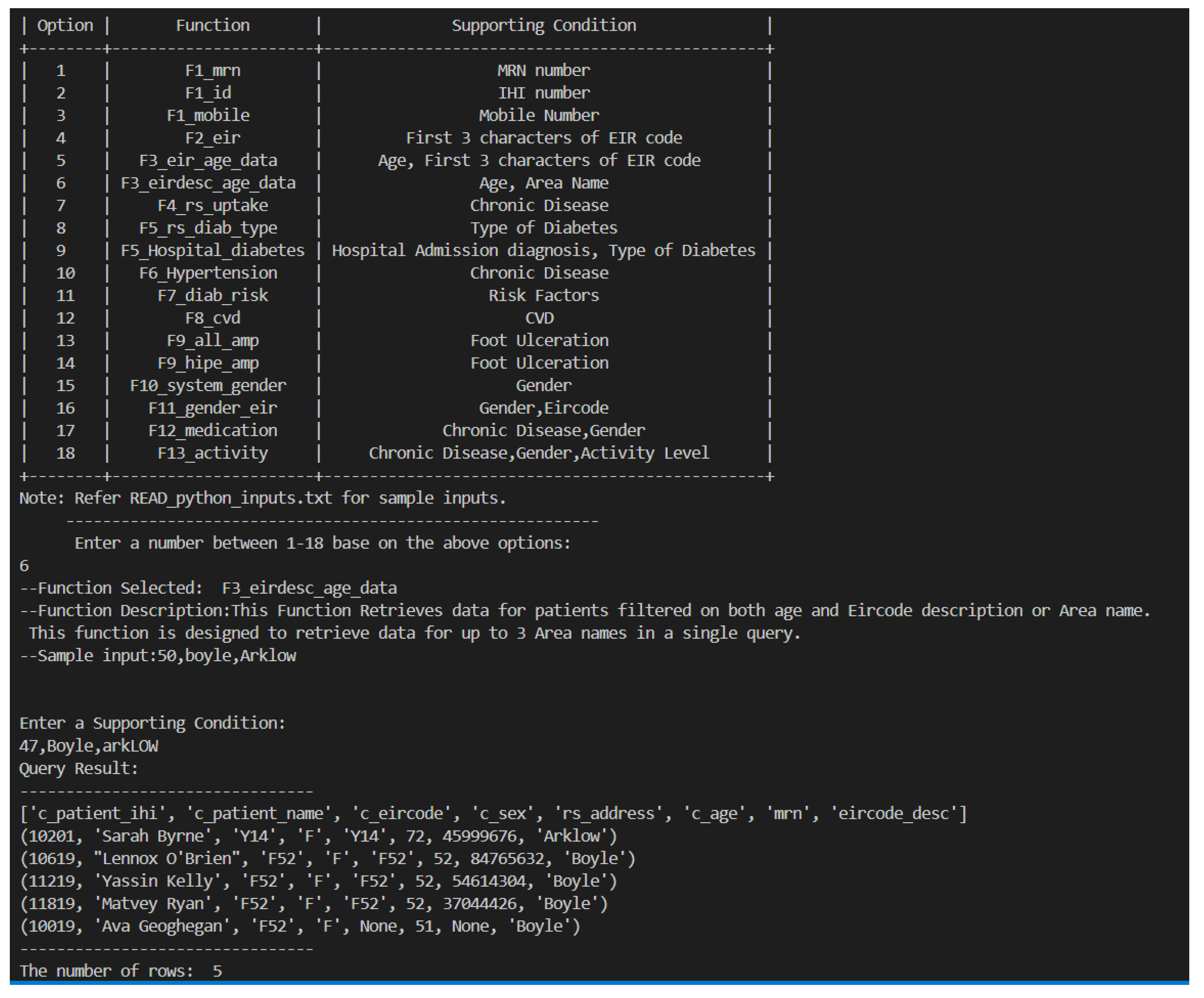
F3_eirdesc_age_data: Example 6 retrieves data for patients filtered on both age and Eircode description using the eirdesc_age_data function. It takes a minimum age and the area name as parameters and retrieves patient details such as name, sex, address, age, and Eircode description. This function is designed to retrieve data for up to 3 Area names in a single query.call dynamiccaseprocedure(’F3eirdescabove45data’,’Boyle’);select * from resultvalue;Sample Query 6.
-
F4_rs_uptake: Example 7 retrieves data related to patients who choose not to enrol in the Retinopathy programme for the prevention of Retinopathy but were admitted to the hospital and diagnosed with Retinopathy.call dynamiccaseprocedure(’F4rsuptake’,’Type 2 diabetes’);select * from resultvalue;Sample Query 7.
-
F5_rs_diab_type: Example 8 retrieves data related to the patient suffering from diabetes but a distinction on the type of diabetes is made using the rs_diab_type function.call dynamiccaseprocedure(’F5rsdiabtype’,’1’);select * from resultvalue;Sample Query 8.
-
F5_Hospital_diabetes: Example 9 retrieves data based on hospitalization due to any condition and type of diabetes.call dynamiccaseprocedure(’F5rsdiabtype’,’2’);select * from resultvalue;Sample Query 9.
-
F6_Hypertension: Example 10 retrieves data related to patients who are diagnosed with both diabetes and hypertension using the diab_hyp function.call dynamiccaseprocedure(’F6Hypertension’,’Type 2 diabetes’);select * from resultvalue;Sample Query 10.
-
F7_diab_risk: Example 11 retrieves data related to diabetes risk factors like physical activity, age, chronic diseases or any other risk factors using the diab_risk function. Other risk factors that have been included are: overweight or obesity, age 45 or older, parent or sibling with type 2 diabetes, being physically active less than 3 times a week, have non-alcoholic fatty liver disease (NAFLD).call dynamiccaseprocedure(’F7diabrisk’, ’sibling with type 2 diabetes,non-alcholic fatty liver disease,parent with type 2 diabetes, ethnicity, overweight’);select * from resultvalue;Sample Query 11.
-
F8_cvd: Example l2 retrieves data related to diabetic patients who are also diagnosed with one or more cardiovascular diseases using the diab_cvd function. Different Cardiovascular diseases mentioned in the CDM booklet are Stable Heart Failure, Ischaemic Heart Disease, Cerebrovascular Disease (Stroke / TIA) and/or Atrial Fibrillation.call dynamiccaseprocedure(’F7diabrisk’, ’sibling with type 2 diabetes, ethnicity’);select * from resultvalue;Sample Query 12.
-
F9_all_amp: Example 13 retrieves data related to diabetic amputations using the amputation function.call dynamiccaseprocedure(’F8cvd’, ’Atrial fibrillation,Ischaemic Heart Disease, Stroke,Stable Heart Failure’);select * from resultvalue;Sample Query 13.
-
F9_hipe_amp: Example 14 retrieves data related to diabetic patients who have had amputations and are registered in the hospital system using the amputation_hipe function.call dynamiccaseprocedure(’F8cvd’, ’Stable Heart Failure’);select * from resultvalue;Sample Query 14.
-
F10_system_gender: Example 15 retrieves data based on gender (used as an identifier here) using the gender_data function.call dynamiccaseprocedure(’F9allamp’, ’Foot Ulceration’);select * from resultvalue;Sample Query 15.
-
F11_gender_eir: Example 16 retrieves gender-related data based on Eircode using the gender_eir_data function.call dynamiccaseprocedure(’F9hipeamp’, ’Foot Ulceration’);select * from resultvalue;Sample Query 16.
-
F12_medication: Example 17 retrieves data regarding the diabetic and hypertension patients’ medication data using the diab_hyp_med function to understand treatment provided in different parts of the country.call dynamiccaseprocedure(’F10systemgender’, ’F’);select * from resultvalue;Sample Query 17.
-
F13_activity: Example 18 retrieves activity-related data using the diab_hyp_act function. The data can be retrieved using different chronic diseases, gender and physical activity frequency.call dynamiccaseprocedure(’F11gendereir’, ’F,F93’);select * from resultvalue;Sample Query 18.
4. Conclusions
Acknowledgments
References
- C. Batini, M. Lenzerini and S. Navathe A comparative analysis of methodologies for database schema integration. ACM Computing Surveys (CSUR) 1986, 18, 323–362. [Google Scholar] [CrossRef]
- D. Brizan and A. Tansel. A Survey of entity resolution and record linkage methodologies. Communications of the IIMA 2006, 6. [Google Scholar]
- J. Dahmen and D. Cook. SynSys: A synthetic data generation system for healthcare applications, Sensors 2019, 19.
- The FHIR Specification, 2019, available at: https://www.hl7.org/fhir/overview.html.
- B. Franz, A. Schuler and O. Krauss. Applying FHIR in an integrated health monitoring system. EJBI 2015, 11, 51–56. [Google Scholar]
- A. Hermann, “Federated data systems: Balancing innovation and trust in the use of sensitive data,” in World Economic Forum, 2019.
- V. M. Ngo, G. Sood, F. Donohue, P. Kearney, C. Buckley, and M. Roantree, “Using HL7-FHIR as an Integration Platform for Chronic Disease Services Management and Planning in the Irish Healthcare Sector,” in Joint International Conference of ISEH, ICEPH & ISEG on Environment and Health 2024, 2024, pp. 1–8.
- Junqiao C. et., al. The validity of synthetic clinical data: a validation study of a leading synthetic data generator (Synthea) using clinical quality measures. BMC medical informatics and decision making, 19:1, pp.1-9, BioMed Central, 2019.
- D. Nie and M. Roantree, “Detecting multi-relationship links in sparse datasets,” in ICEIS, 2019.
- Y. Park, M. Shankar, B. Park and J. Ghosh. Graph databases for large-scale healthcare systems: A framework for efficient data management and data services. 2014 IEEE 30th International Conference on Data Engineering Workshops, pp. 12-19, IEEE Press, 2014.
- R. Saripalle, C. Runyan and M. Russell. Using HL7 FHIR to achieve interoperability in patient health record. Journal of biomedical informatics 2019, 94. [Google Scholar]
- M Scriney, S McCarthy, A McCarren, P Cappellari, M Roantree. Automating data mart construction from semi-structured data sources. The Computer Journal 2019, 62, 394–413. [Google Scholar] [CrossRef]
- Engineering Data Assets for Public Health Applications: A Covid-19 Case Study. M. Scriney, M. Timilsina, E. Curry, L. Porwol, D. Nie, D. Dahley, J. Fernandez, M. D’Aquin and M. Roantree. 2023 IEEE International Conference on Big Data, pp. 1853-1862, IEEE, 2023.
- A. Sheth and J. Larson. Federated database systems for managing distributed, heterogeneous, and autonomous databases. ACM Computing Surveys 1990, 22, 186–236. [Google Scholar]
- Walinjkar and J. .Woods. FHIR tools for healthcare interoperability. Biomedical Journal of Scientific and Technical Research 2018, 9. Biomedical Research Network.
- M. Frisby. Operationalizing Neo4j for use within Healthcare by leveraging HL7 FHIR Standards, 2021. available at: https://github.com/Optum/CyFHIR.

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



