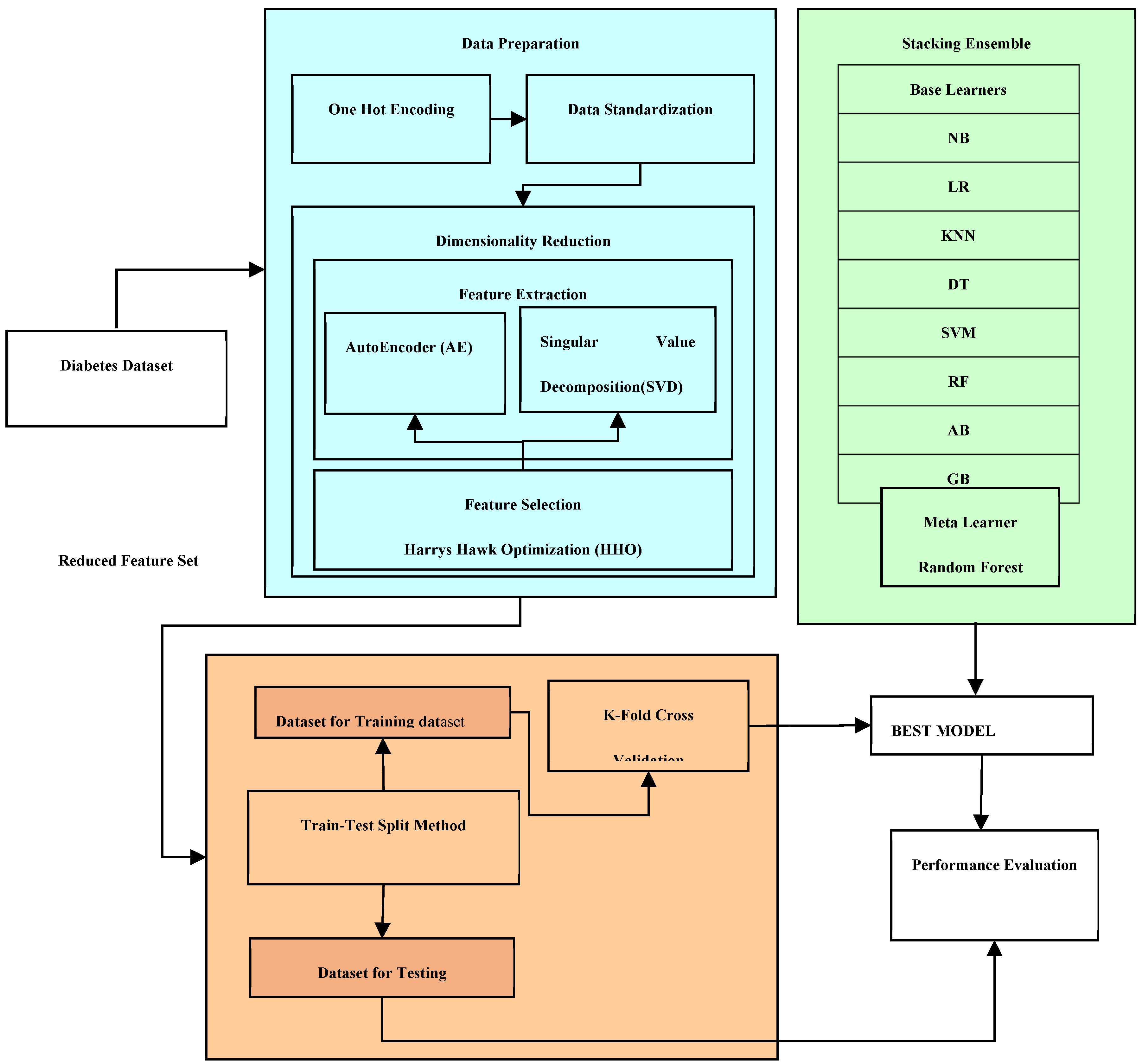
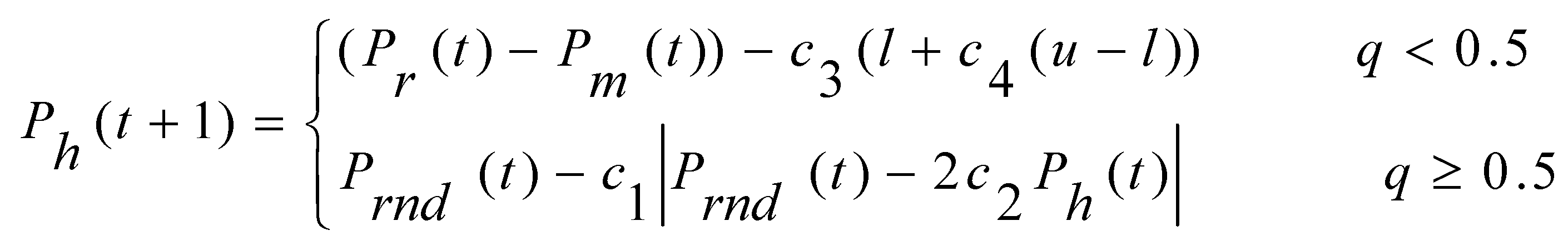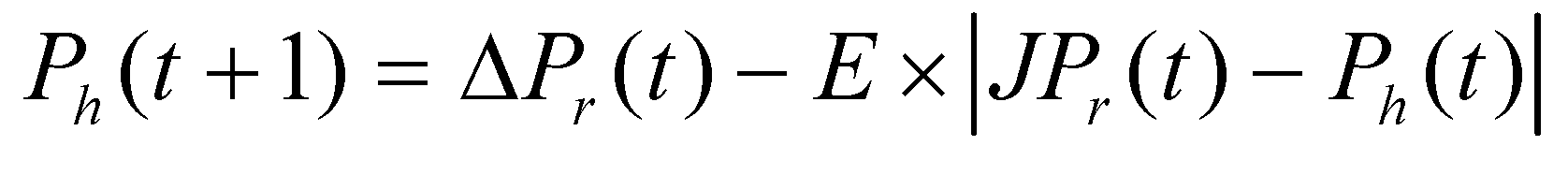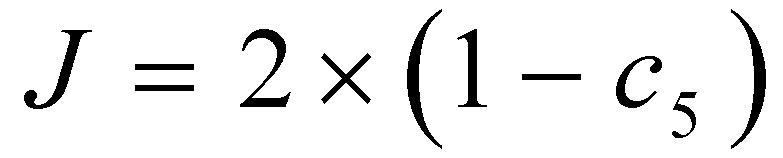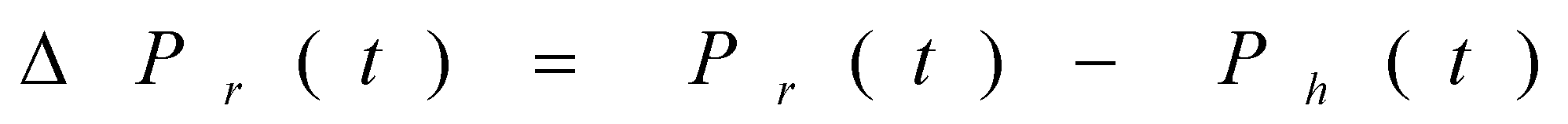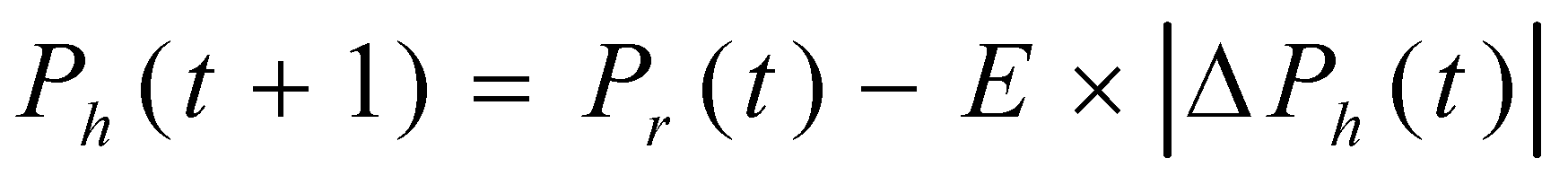This approach involves reducing the number of features by pinpointing the most significant attributes strongly correlated to the target attribute. It not only results in a lesser cost incurred with storage and computation due to smaller feature set but enhanced performance.
Several advantages accompany the reduction of the feature set. For instance, it aids in diminishing data overfitting by eliminating irrelevant features, thereby reducing noise. Additionally, the accuracy of the model improves as misleading input variables are removed. Moreover, a smaller dataset size translates to reduced time required for constructing and training the model.
In supervised learning, traditional feature selection methods fall into three categories: filter, wrapper, and intrinsic methods. In this current work, we employ a wrapper based feature selection employing a bio-inspired metaheuristic approach termed as Harris Hawk Optimization. Wrapper methods involve utilizing various subsets of the feature set to assess the performance of an ML model for each subset. Each subset is assigned a performance evaluation score, and the one demonstrating the highest score is chosen. The quest for the optimal feature subset can follow methodical or stochastic approaches. The former might employ strategies like best-first search, while the latter could involve algorithms like random hill-climbing.
Feature selection, aimed at maximizing the accuracy of an ML model, can be regarded as an optimization problem, particularly when dealing with a large feature set. The optimization function aims to minimize both the size of the feature subset and the classification error. Assuming there are N features, the total possible subsets amount to 2^N. This exponential increase in search space makes a brute-force approach via exhaustive search impractical. Therefore, meta-heuristics are employed to navigate this extensive search space.
In this context, numerous algorithms have been proposed, including Particle Swarm Optimization [
34,
35], Artificial Bee Colony [
36], Differential Evolution [
37], Bat algorithm [
38], Moth Flame Optimization [
39], Grey Wolf Optimizer [
40], Butterfly optimization algorithm [
41], Sine Cosine Algorithm [
42], CSA and HHO. Among these, HHO stands out as a recently introduced algorithm that has garnered considerable attention within the research community due to its simplicity, ease of implementation, efficiency, and fewer parameters.
4.2.1. Harris Hawk Optimization Algorithm
HHO stands as a well-regarded swarm-based optimization algorithm, characterized by its gradient-free approach and a dynamic interplay between exploration and exploitation phases. First introduced in 2019 within the prestigious Journal of Future Generation Computer Systems (FGCS), this algorithm quickly garnered attention within the research community due to its adaptable structure, remarkable performance, and ability to yield high-quality results. Its fundamental concept draws inspiration from the cooperative behaviors and hunting techniques of Harris' hawks in nature, particularly their "surprise pounce" strategy. In the present day, numerous proposals have emerged aiming to enhance the capabilities of HHO, resulting in several improved versions of the algorithm published in esteemed journals by Elsevier and IEEE.
The genesis of the idea behind HHO is both elegant and straightforward. Harris hawks exhibit a diverse range of team hunting patterns, adapting to the dynamic scenarios and evasion tactics employed by their prey, often involving intricate zig-zag maneuvers. They coordinate their efforts, waiting until the opportune moment to launch a synchronized attack from multiple directions.
From an algorithmic perspective, HHO incorporates several key features contributing to its effectiveness. Notably, the "escaping energy" parameter exhibits a dynamic and randomized time-varying nature, enhancing and harmonizing the exploratory and exploitative behaviors of HHO. This dynamic aspect facilitates a seamless transition between exploration and exploitation phases. HHO employs various exploration mechanisms based on the average location of hawks, bolstering its exploratory tendencies during initial iterations. Additionally, it employs diverse LF-based patterns featuring short-length jumps to enrich its exploitative behaviors when conducting local searches.
A progressive selection scheme empowers search agents to incrementally advance their positions and select only superior ones, elevating solution quality and intensification capabilities throughout the optimization process. HHO employs a range of search strategies before selecting the optimal movement step, further strengthening its exploitation tendencies. The randomized jump strength aids candidate solutions in achieving a balance between exploration and exploitation. Lastly, HHO incorporates adaptive and time-varying components to tackle the challenges posed by complex feature spaces, which may contain local optima, multiple modes, and deceptive optima.
Irrespective of the array of algorithms, they share a common characteristic: their search steps encompass two phases—exploration (diversification) and exploitation (intensification). During the exploration phase, the algorithm aims to extensively traverse various regions and facets of the feature space by leveraging and emphasizing its randomized operators. Consequently, a well-designed optimizer's exploratory behaviors should possess a sufficiently enriched random nature, efficiently distributing randomly-generated solutions across diverse areas of the problem's topography during the initial stages of the search process.
Subsequently, the exploitation phase typically follows the exploration phase. Here, the optimizer directs its attention toward the vicinity of higher-quality solutions situated within the feature space. This phase intensifies the search within a local region rather than encompassing the entire landscape. An adept optimizer should strike a reasonable, delicate balance between the tendencies for exploration and exploitation. Otherwise, there's an increased risk of getting trapped in local optima (LO) and encountering drawbacks related to premature convergence.
Exploration Phase
The HHO introduces an exploration mechanism inspired by the behavior of Harris' hawks. These hawks rely on their keen eyes to track and detect prey, yet sometimes, the prey remains elusive. Consequently, the hawks adopt a strategy of waiting, observing, and monitoring the desert landscape, potentially for several hours, in their pursuit to detect prey. Within the HHO framework, the candidate solutions are likened to the Harris' hawks, with the best candidate solution in each step analogous to the intended prey or a near-optimal solution.
In the HHO context, Harris' hawks symbolize candidate solutions perched randomly at various locations, employing two distinct strategies to detect prey. Under an assumption of equal probability (s) for each perching strategy, these hawks decide their perching location based on two considerations: firstly, positioning themselves close enough to other family members to facilitate coordinated attacks (as detailed in Eq. (1)) when s < 0.5; secondly, perching on random tall trees situated within the group's home range (as described in Eq. (1)) when s ≥ 0.5.
In the equation(1) Ph represents the position vector of the hawk, Pr represents the position vector of the rabbit, Pm repreents the average of position vector of all hawks, Prnd represents the random position vector, t represents the time-instant / iteration, t+1 represents the next successive iteration, c1, c2, c3, c4 and q are random numbers in the range (0,1) updated in each iteration, l and b represent the lower and upper bounds of the locations of the hawks.
The HHO algorithm has the ability to transition from exploration to exploitation, and subsequently switch between various exploitative behaviors, all guided by the prey's escaping energy given by the equation (2) below:
In the equation (2), E represents the escaping energy of the prey, T is the total number of iterations, t is the current iteration, and E0 represents the initial state of energy ranging and varying between (-1,1) at each iteration.
Exploitation Phase
During the exploitation phase, the HHO algorithm executes a surprise attack by targeting the identified prey encountered in the exploration phase. Based on the escape patterns of the prey and the hunting approaches of Harris' hawks, the HHO suggests four potential strategies to simulate the attack phase. Prey typically aim to flee when faced with danger. If we assume r represents the likelihood of successful (r < 0.5) or unsuccessful (r ≥ 0.5) escape prior to a surprise attack, the hawks react by employing either a hard or soft encirclement tactic to capture the prey. This approach involves surrounding the prey from various directions with differing force levels based on the prey's remaining energy.
To implement this approach and allow the HHO to alternate between soft and hard besiege procedures, the E parameter comes into play. Specifically, when |E| is equal to or greater than 0.5, the gentle besieging occurs, while if |E| is less than 0.5, the intense besieging takes place.
Soft Besiege
The rabbit retains sufficient energy and attempts to evade through random deceptive leaps, yet ultimately fails to escape when the following condition is satisfied: r ≥ 0.5 and |E| ≥ 0.5. Throughout these evasion attempts, the Harris' hawks gently encircle it, leading the rabbit to fatigue further before executing a sudden pounce. This conduct adheres to the subsequent set of rules:
between the position vector of the rabbit and the current location, J stands for the energy of the rabbit for jumping throughout the escaping process. c5 is the random variable between (0, 1).
Hard Besiege
The prey is extremely fatigued with minimal energy for escape when the following condition is satisfied: r ≥ 0.5 and |E| < 0.5. Furthermore, the Harris' hawks barely surround the targeted prey before executing the surprise pounce. In such a scenario, it is termed as hard besiege and below the equation is used to update the current positions.