1. Introduction
Nematodes are microscopic organisms under the large phylum
Nematoda and are often found in microbial communities in soil and several other environments [
1]. In soil environments, different types of nematodes are present, including free-living nematodes (FLNs) and plant-parasitic nematodes (PPNs). FLNs consist of several different feeding groups including bacterivores, fungivores, omnivores, and predators [
2]. Conversely, PPNs feed on plant roots to gain nutrients for survival. To pierce the root tissue of the plant, PPNs use their stylet and may also secrete enzymes that aid in the degradation of the plant cell wall [
2]. PPNs cause severe damage to agricultural crops both directly as well as indirectly, costing
$125 billion in crop losses worldwide [
1], and are thereby considered the world’s most damaging plant pathogen [
3,
4]. In Canada, PPNs cause great yield losses for several different crops, up to 15% every year [
5].
A healthy agricultural soil consists of an environment with a good balance of beneficial FLNs as they contribute to nutrient cycling and the release of nitrogen in the soil [
6,
7]. Nematodes are also part of several trophic levels in the soil food web and play a crucial role in balancing the environment by feeding on various soil microorganisms and being a food source to other predators [
6,
8]. Moreover, nematodes are sensitive to disturbances and changes in their population dynamics also reflect changes in the soil microbial communities, making nematodes great bio-indicators of soil health and fertility [
6,
9]. To improve soil quality and crop yield in a sustainable manner, farmers have started implementing the use of cover crops during the off-season [
10]. This allows the enrichment and protection of soil as well as suppression of weeds, disease, or pests, resulting in increased soil microbial communities and improved soil health [
11]. Research has shown that different cover crops not only suppress PPN populations [
12,
13], but also increase FLN populations when incorporated into the soil [
14,
15]. This is because certain cover crops are non-hosts to the damaging PPNs [
16]. Moreover,
Brassica cover crops have been found to suppress PPNs infection by hydrolysis of glucosinolate compounds found within the plant, a process termed biofumigation [
17,
18,
19]. Studies have also shown that certain cover crops may serve as potential hosts to PPNs, allowing these populations to grow and cause further damage to the cash crop [
20,
21]. It is therefore important that the growers select proper cover crops that are poor or non-hosts to the PPNs of concern in their agricultural systems to promote a beneficial nematode community and a healthy soil environment.
Although research has been conducted to look at specific cover crops and their effect on microbial communities in the soil, how different types of cover crops affect both FLN and PPN populations is largely unknown and in demand. In the current investigation, we aimed at gaining a better understanding of the impact of cover crops on nematode communities, and how the growers can mitigate crop yield losses by implementing the most beneficial cover crops to their agricultural systems. The main objectives of this study were to determine the effect of legume (Cowpea, Vigna unguiculata) and non-legume (Pearl Millet, Pennisetum glaucum) cover crops as well as their mixture on the abundance, structure and diversity of both FLN and PPN communities in a spinach cultivation system.
2. Results
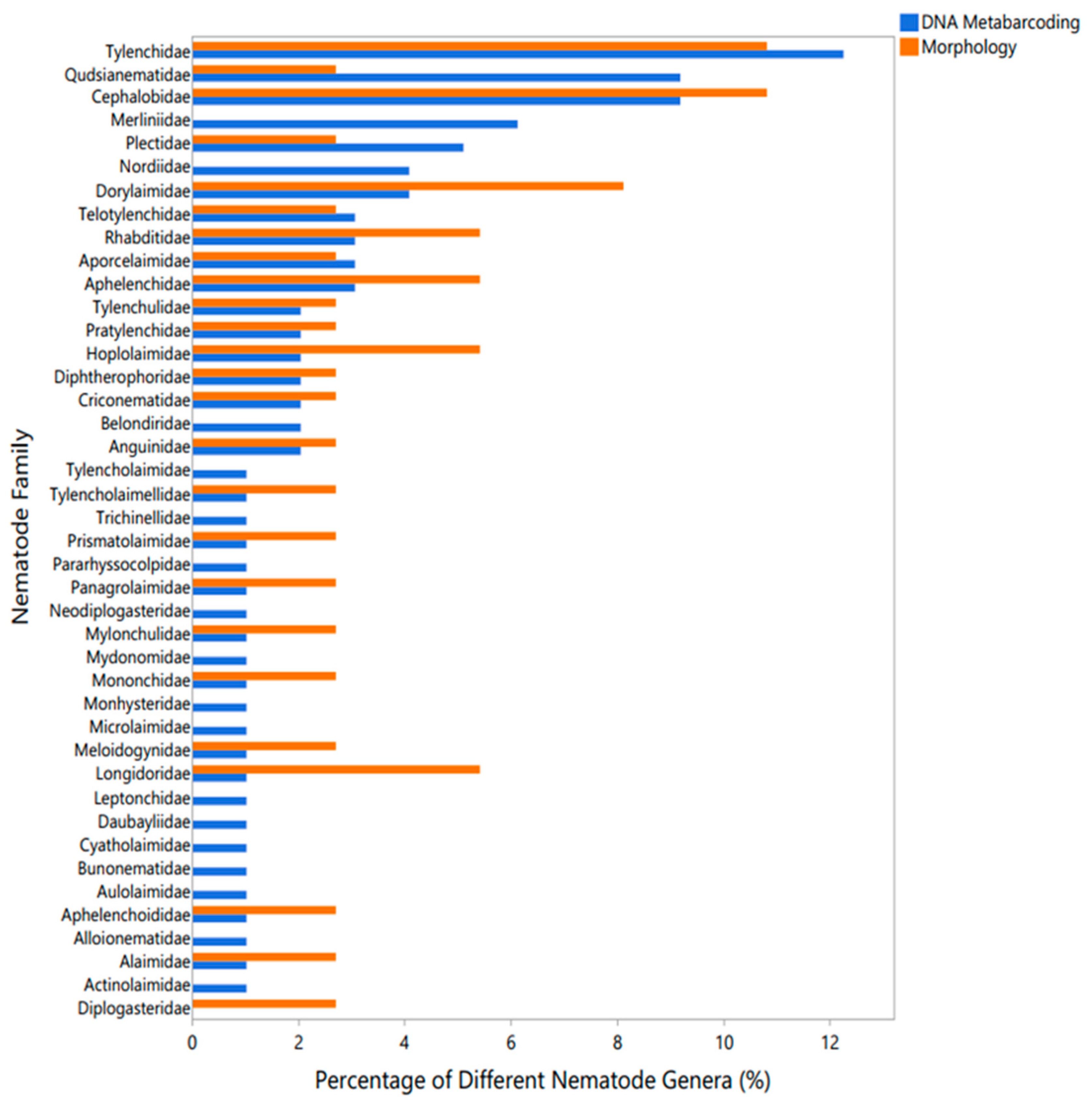
Using the morphological and DNA metabarcoding analyses, 42 nematode families were identified in all study soil samples (
Figure 1). Of these 42 families, 41 were identified using the DNA metabarcoding method and 25 were identified using morphological analysis, with 24 families found by both methods. Most nematode families identified only using DNA metabarcoding were generally found to represent the rare taxa, with 1% of different nematode genera detected within the families of Tylencholaimidae, Tylencholaimellidae, Trichinellidae, Prismatolaimidae, Pararhyssocolpidae, Panagrolaimidae, Neodiplogasteridae, Mylonchulidae, Mydonomidae, Mononchidae, Monhysteridae, Microlaimidae, Meloidogynidae, Longidoridae, Leptonchidae, Daubayliidae, Cyatholaimidae, Bunonematidae, Aulolaimidae, Aphelenchoididae, Alloionematidae, Alaimidae, and Actinolaimidae (
Figure 1).
The most diverse nematode families that were well-represented by both DNA metabarcoding and morphological analysis included Tylenchidae (12.3% of different nematode genera for DNA metabarcoding and 10.8% for morphology) and Cephalobidae (9.2% and 10.8%, respectively). Certain nematode families were over-represented in one identification method (Qudsianematidae, 9.2% for DNA metabarcoding versus 2.7% for morphology; Dorylaimidae, 4.1% versus 8.1%, respectively), while other families showed similar representation using both DNA metabarcoding and morphological analysis (Tylenchulidae, 2% versus 2.7%; Pratylenchidae, 2% versus 2.7%) (
Figure 1).
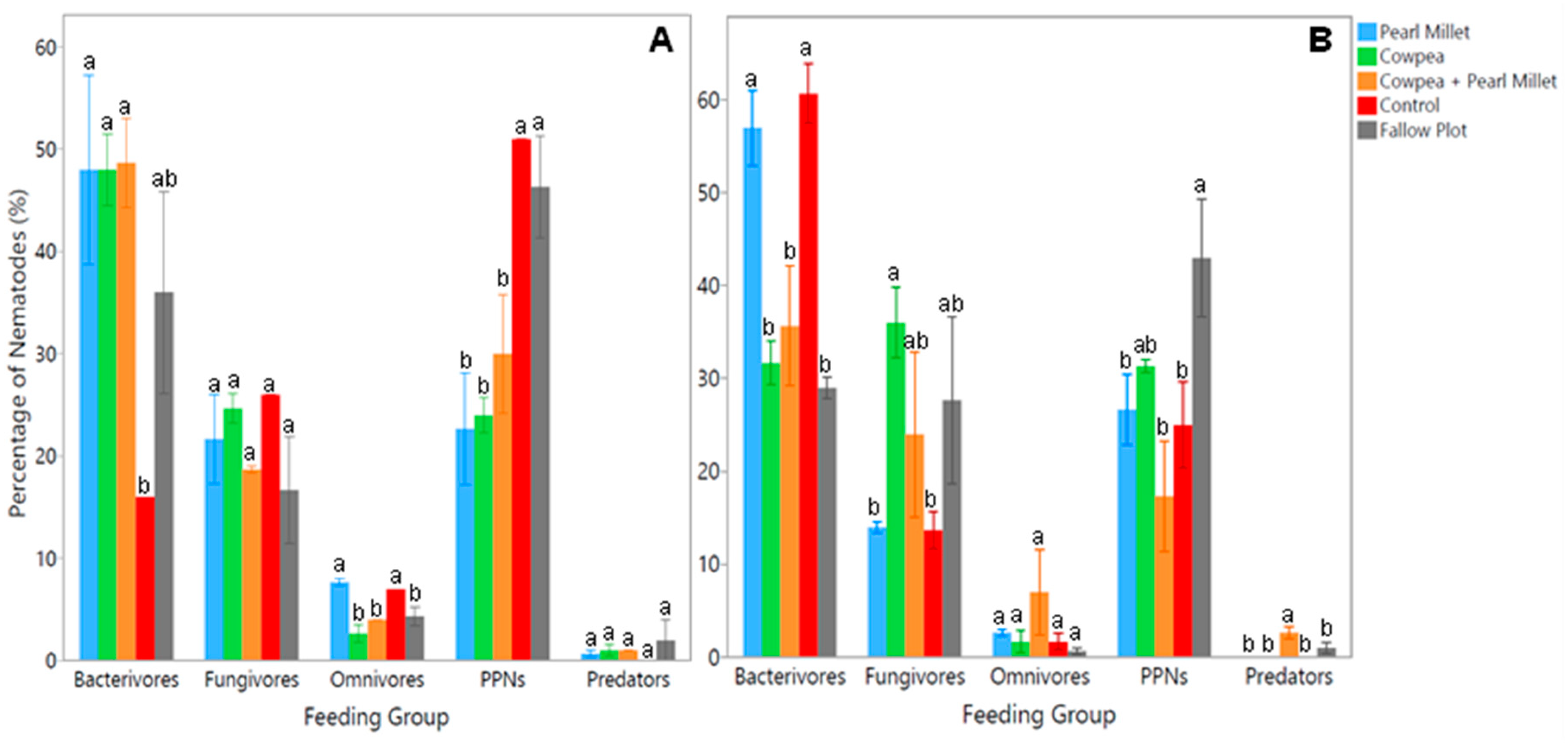
Extracted nematodes from soil samples were examined under the microscope to identify species present based on morphological characteristics and classify them into respective feeding groups. Results from the time of cover crop maturity (Time 1) showed significant differences (
p < 0.05) between cover crop treatments for the bacterivores, herbivores (PPNs), and omnivores (
Figure 2A). The average number of bacterivores was found to be significantly higher (
p < 0.05) in the Cowpea, Pearl Millet, and mixture of both cover crop treatments, having 100% more bacterivores than the Control. Conversely, the average number of PPNs was observed to be significantly higher (
p < 0.05) in the Control and Fallow Plot, with 53% more PPNs than the mixture of Cowpea and Pearl Millet and 67% more PPNs than the Pearl Millet and Cowpea treatments (
Figure 2A). In terms of omnivores, the average number was significantly higher (
p < 0.05) in the Pearl Millet and Control treatments compared to all other experimental treatments, both having 67% more omnivores. No significant differences were observed between treatments for the fungivore and predator feeding groups at the time of cover crop maturity.
Results from the spinach harvest time (Time 2) showed significant differences (
p < 0.05) between cover crop treatments for the bacterivores, herbivores (PPNs), and predators (
Figure 2B). The average number of bacterivores was found to be significantly higher (
p < 0.05) in the Control, with 53% more bacterivores than the mixture of Cowpea and Pearl Millet and 67% more than Cowpea and the Fallow Plot. Pearl Millet also had significantly more (
p < 0.05) bacterivores than the mixture of covers with a difference of 49% and the same for Cowpea and Fallow Plot with a difference of 64%. The Fallow Plot had significantly higher (
p < 0.05) PPNs than Pearl Millet and the Control by 44% and the mixture of Cowpea and Pearl Millet by 84% (
Figure 2B). The average number of predators showed the opposite trend, as the mixture of cover crops had significantly more (
p < 0.05) predators than Pearl Millet, Cowpea, the Control and Fallow Plot by at least 100%. The average number of fungivores was significantly higher (
p < 0.05) in the Cowpea treatment than Pearl Millet and the Control by at least 80%. No significant differences were observed between cover crop treatments for the omnivore feeding group at the spinach harvest time. Thus, the implementation of cover crops significantly influenced soil nematode communities with Cowpea, Pearl Millet and their mixture largely increasing FLNs such as bacterivores, and the Control and Fallow Plot generally increasing PPNs.
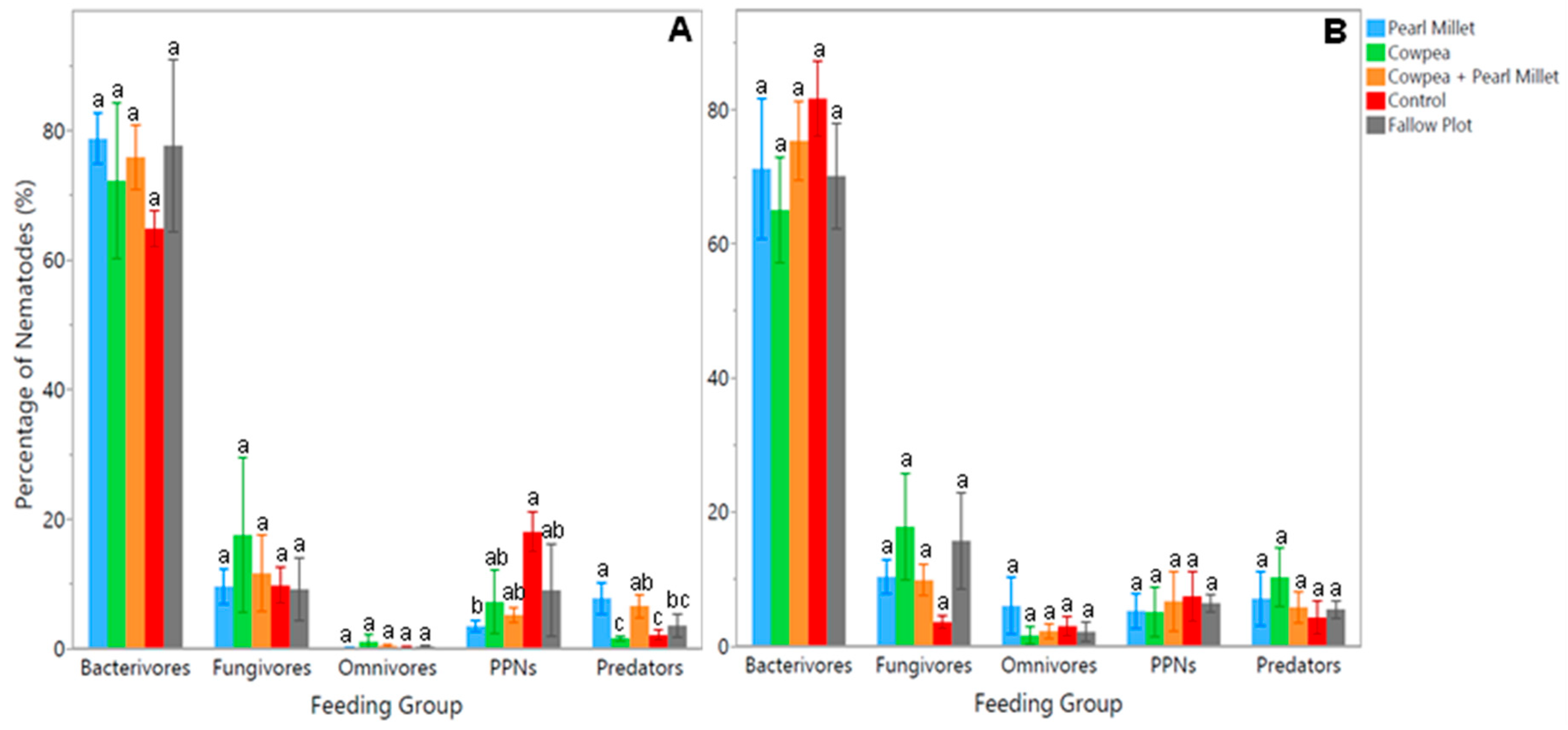
Soil samples were also analyzed using DNA metabarcoding analysis in order to have a broader understanding of nematode populations and soil health for each of the cover crop treatments. Results from the time of cover crop maturity (Time 1) exhibited significant differences (
p < 0.05) between treatments for the herbivores (PPNs) and predators (
Figure 3A). The average number of PPNs was noticed to be significantly higher (
p < 0.05) in the Control treatment, with 120% more PPNs than Pearl Millet. On the other hand, the average number of predators was found to be significantly higher (
p < 0.05) in Pearl Millet, with 107% more predators than the Fallow Plot and 133% more than Cowpea and the Control (
Figure 3A). No significant differences were noted between treatments for the bacterivore, fungivore, and omnivore feeding groups at the time of cover crop maturity. Results from the time of spinach harvest (Time 2) showed no significant differences between cover crop treatments for any of the feeding groups (
Figure 3B).
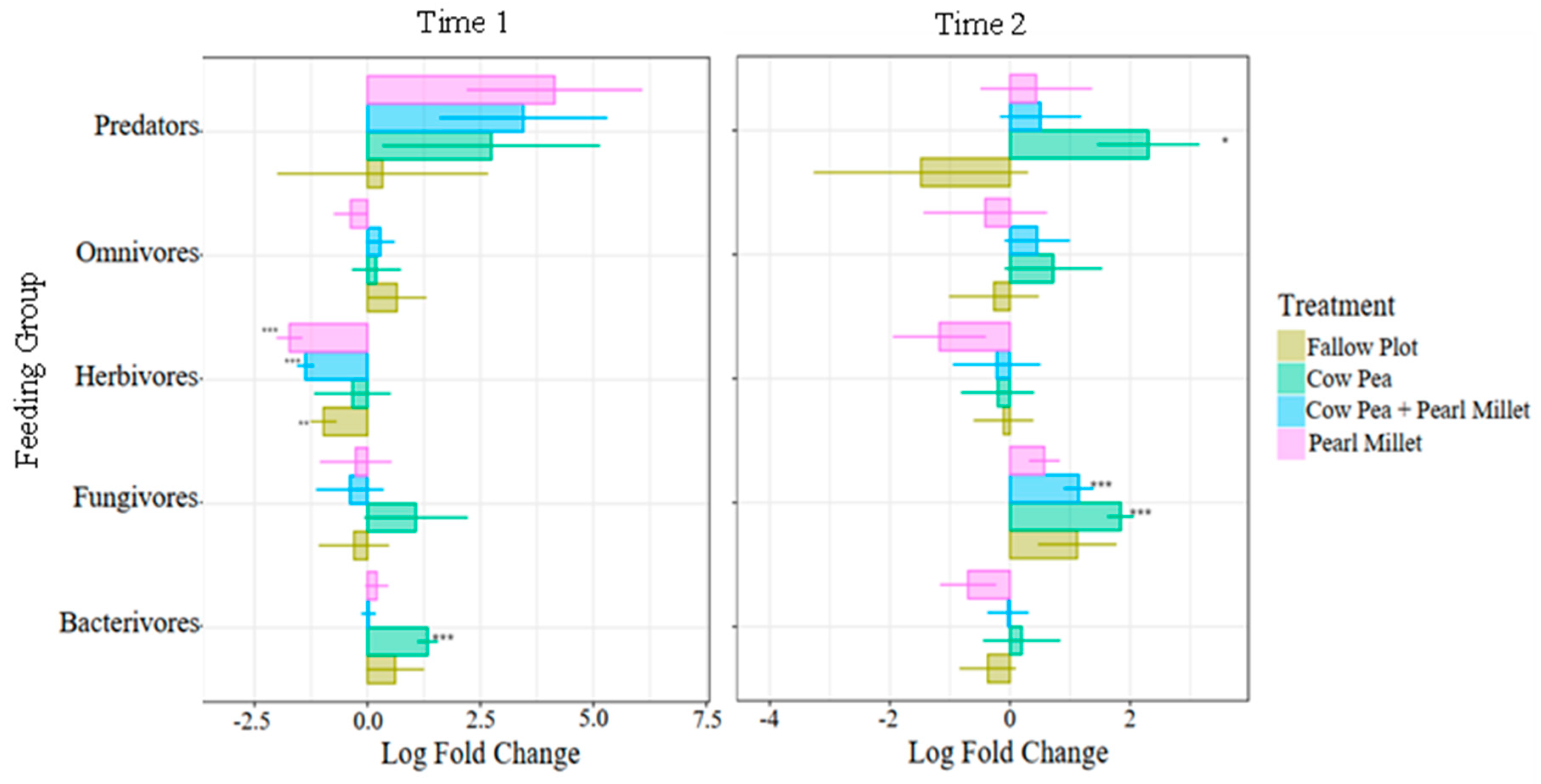
In this study, the log fold differences of feeding groups in the different cover crop treatments were compared to the Control using ANCOM-BC (
Figure 4). With mature cover crops present (Time 1), we found that the relative abundance of herbivores (PPNs) decreased significantly in the Pearl Millet (-1.7 log
e), mixture of Cowpea and Pearl Millet (-1.4 log
e), and the Fallow (-1.0 log
e) treatments. Bacterivores were only found to increase in Cowpea (1.3 log
e) with mature cover crops (Time 1) and were not differentially abundant by spinach harvest (Time 2). At the spinach harvest time, there was a significant increase in predators for Cowpea (2.3 log
e) and a significant increase in fungivores for Cowpea (1.8 log
e) and the Cowpea and Pearl Millet mixture (1.1 log
e) (
Figure 4).
In the present work, ecological indices were calculated for all sets of soil samples, which included richness, representing the number of species present; evenness, representing the equality of abundance of different species; the Shannon-Wiener Index (SWI) value, a measure of species diversity; and the Bonger’s Maturity Index (MI), a measure of environmental disturbance based on nematode species composition. Ecological indices values for Time 1 described the soil environment for treatments after cover crops were seeded and given the appropriate amount of time to mature (
Table 1).
Results at cover crop maturity (Time 1) revealed that there were no significant differences between richness, evenness, or SWI values for all cover crop treatments (
Table 1). The MI value of the Control treatment (2.14) was significantly higher (
p < 0.05) by 22% in comparison with the mixture of Cowpea and Pearl Millet as well as the Cowpea treatments (1.71). These results indicated that the soil in the Control treatment was less disturbed. There were no significant differences between the richness, evenness, SWI, or MI values for all cover crop treatments at the time of spinach harvest (Time 2).
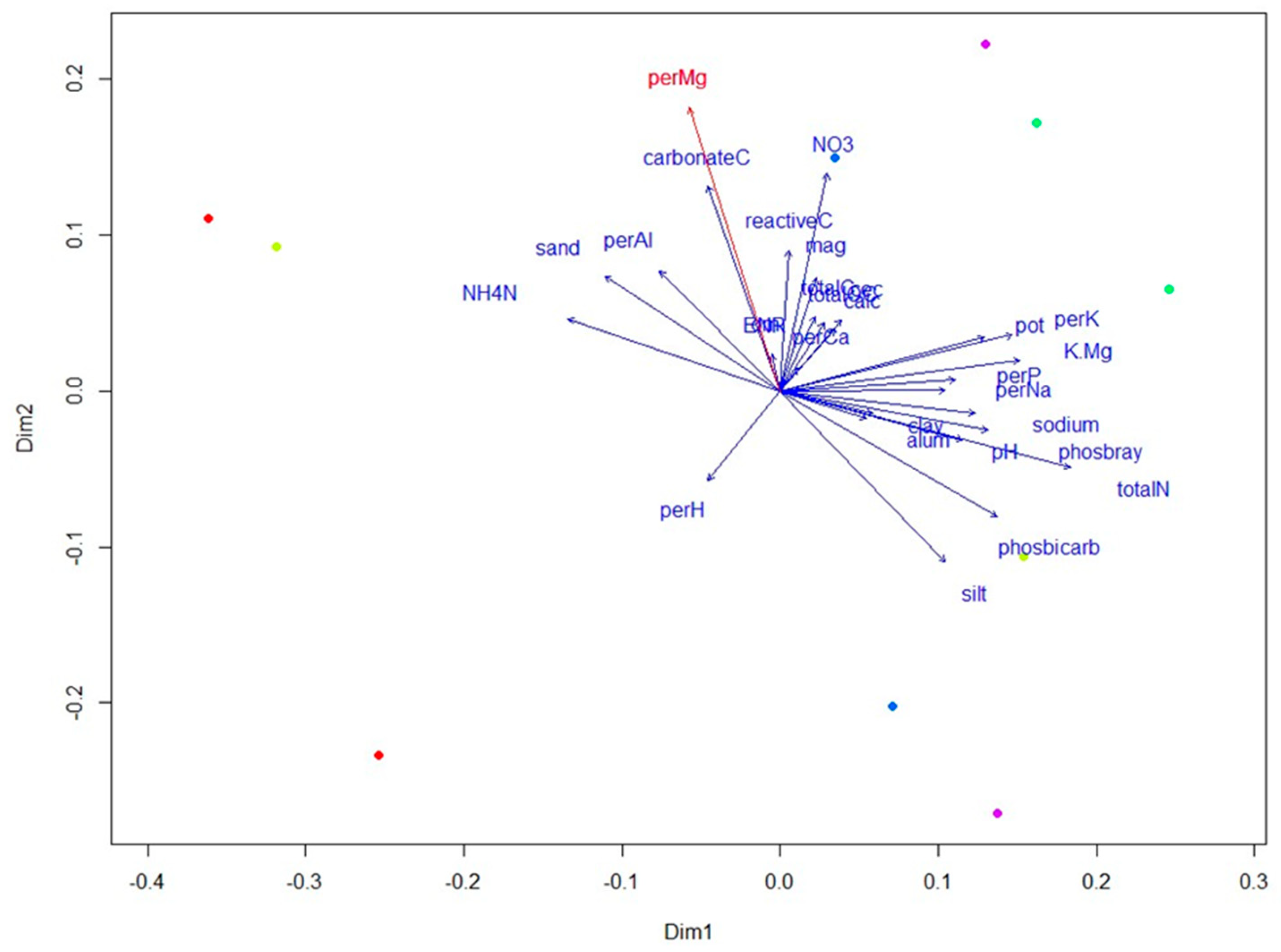
Analysis of soil properties was also performed on all samples to get a broader understanding of how they are correlated with the nematode community composition in cover crop treatments. Additionally, the envfit function was used to fit soil properties onto the Principal Coordinates Analysis (PCoA) ordination from DNA metabarcoding data. Of all soil properties, results showed that the percentage of magnesium (Mg) was significantly correlated to the ordination axes (r
2 = 0.59,
p = 0.04) and was generally higher in samples taken during cover crop maturity (Time 1) (
Figure 5). Total nitrogen (totalN) showed the second highest correlation coefficient with the ordination axes (r
2 = 0.58,
p = 0.07) and was also found to be significantly correlated to the community composition by Mantel test (
p = 0.02). Moreover, magnesium and total nitrogen were significantly correlated with community composition, especially at cover crop maturity.
3. Discussion
In the present investigation, we identified 42 nematode families across all studied soil samples using both morphological and DNA metabarcoding analyses, which revealed key insights into the efficacy and scope of each method. The DNA metabarcoding approach detected a broader range of nematode families, identifying 41 families compared to the 25 found through morphological analysis, with 24 families common to both methods. This discrepancy highlights the greater sensitivity of DNA metabarcoding method, particularly in detecting the rare taxa. Families such as Tylencholaimidae, Tylencholaimellidae and a few others were only detected by DNA metabarcoding, likely due to their lower abundance, making them less visible to traditional morphological approaches. These rare taxa, though not abundant, can play crucial roles in soil ecosystems, and their detection underscores the importance of integrating molecular techniques in nematode community studies [
22,
23].
Among the families well-represented by both methods, Tylenchidae and Cephalobidae stood out, comprising significant portions of the nematode communities (12.3% and 9.2% for DNA metabarcoding, and 10.8% each for morphology). The consistency in detecting these families across methods validates the reliability of the morphological approach for more common and morphologically distinctive nematodes. However, discrepancies such as the over-representation of Qudsianematidae in DNA metabarcoding (9.2% versus 2.7% in morphology) and the higher detection of Dorylaimidae through morphological analysis (8.1% versus 4.1% in DNA metabarcoding) suggest that each method has biases, potentially linked to the differing sensitivities to DNA extraction efficiency and morphological identification challenges [
24,
25].
The temporal dynamics of nematode communities were also explored by comparing cover crop treatments at two key time points: cover crop maturity and spinach harvest. Morphological analysis at cover crop maturity revealed significant variations in the abundance of different feeding groups. Bacterivores were notably more abundant in Cowpea, Pearl Millet, and their mixture, showing a 100% increase compared to the Control. This suggests that these cover crops may enhance bacterial populations, which in turn support larger bacterivore communities. Conversely, PPNs were significantly more abundant in the Control and Fallow plots, with 53% more PPNs than in the mixture of Cowpea and Pearl Millet, and 67% more than in the individual Cowpea and Pearl Millet treatments. This indicates that cover crops can suppress PPN populations, potentially through root exudates that deter these pests or through fostering beneficial microbes that outcompete them. These results are consistent with past studies finding that legume and non-legume cover crop mixes often provide better PPN suppression compared to single-species [
26,
27]. However, the non-legume cover crops included in most studies consist of
Brassica species, as they contain glucosinolate compounds that suppress PPN infection [
28,
29]. Omnivores, on the other hand, were more prevalent in the Pearl Millet and Control treatments, which may reflect the broader trophic flexibility of this group in these particular soil environments.
By the time of spinach harvest, the nematode community composition had found to be shifted. Bacterivores were now the most abundant in the Control, suggesting a decline in bacterial activity in cover-cropped soils as the season progressed. Interestingly, Pearl Millet still supported more bacterivores than the cover crop mixture, possibly due to differences in root exudate composition or microbial community structure between these treatments. PPNs were most numerous in the Fallow Plot, which lacked the protective influence of cover crops, leading to higher pest pressure. Predators, crucial for natural nematode pest control, were significantly more abundant in the cover crop mixture, highlighting the potential of diverse cover cropping to enhance predatory nematode populations and thereby contribute to biological control. Research has also shown that combining more than one cover crop can lead to increased multi-functionality of covers and improved outcomes compared to implementing a monoculture in an agricultural system [
27,
30]. This concept is supported by findings from [napp,Swinton[
29], who suggested that legume and grass cover crop mixtures are among the most effective combinations for producing high-quality residues in agricultural soil due to the complementarity of both crop types.
DNA metabarcoding corroborated these findings with additional insights. At cover crop maturity, PPNs were significantly more abundant in the Control treatment, with a 120% increase compared to Pearl Millet. This further emphasizes the suppressive effect of Pearl Millet on nematode pests. Predators, on the other hand, were 107% more abundant in Pearl Millet compared to the Fallow Plot and 133% more than in Cowpea and the Control, reinforcing the idea that certain cover crops can enhance beneficial nematode populations. The higher abundance of predatory nematodes in the Pearl Millet treatment could also explain the lower abundance of PPNs in this same treatment, as predatory nematodes provide top-down control of these damaging populations [
31]. By spinach harvest, no significant differences were observed among treatments for any feeding groups, suggesting a convergence in nematode community composition as the cropping cycle progressed.
The differential abundance analysis provided further depth to these observations. With mature cover crops present, the relative abundance of PPNs decreased significantly in Pearl Millet, the Cowpea and Pearl Millet mixture, and Fallow treatments, confirming the suppressive effect of these cover crops. On the contrary, bacterivores increased significantly in Cowpea at cover crop maturity, reflecting the promotion of bacterial populations under this cover crop [
27]. By spinach harvest, predators increased significantly in Cowpea, and fungivores were more abundant in both Cowpea and the cover crop mixture, highlighting the dynamic shifts in soil food webs influenced by different cover cropping strategies. The mixture of both cover crops also revealed significantly higher fungivores than the Control at this time-point. Fungivores are an important nematode feeding group as they feed on plant-pathogenic fungi, reducing the risk of crop damage by these microorganisms [
32].
Ecological indices, including richness, evenness, SWI and the MI, were calculated to assess broader soil ecosystem health. At cover crop maturity, there were no significant differences in richness, evenness, or SWI among treatments, demonstrating that overall species diversity remained stable across different cover crop treatments. However, the MI value was significantly higher in the Control treatment, suggesting that the soil in this treatment was less disturbed, potentially due to the absence of cover crops, which can disrupt nematode communities through changes in root structure and soil microenvironment [
33,
34,
35]. By the time of spinach harvest, no significant differences were observed in any of the ecological indices, suggesting that the initial effects of cover cropping on nematode communities had diminished over time, possibly due to the uniformity imposed by subsequent agricultural practices. Soil property analysis revealed that magnesium levels were significantly correlated with nematode community composition, particularly at cover crop maturity, where they were generally higher. This finding, along with the significant correlation of total nitrogen with community composition, underscores the complex interactions between soil chemistry and biological communities, influenced by cover crop selection and management practices.
Author Contributions
Conceptualization, E.A. and T.S.; Methodology, E.A., J.A., P.L. and T.S.; Validation, E.A., J.A. O.E. and T.S.; Formal analysis, E.A., J.A., P.L., O.E. J.G. and T.S.; Investigation, E.A., P.L. and T.S.; Data curation, E.A. and J.A.; Writing—original draft, E.A.; Writing—review & editing, E.A., J.A., P.L., O.E., J.G. and T.S.; Supervision, T.S. and P.L.; Project administration, T.S.; Funding acquisition, T.S. All authors have read and agreed to the published version of the manuscript.