1. Introduction
Adeno-Associated Viruses (AAVs, genus
Dependoparvovirus) are the leading vector for delivering gene therapies [
1,
2,
3,
4]. Recombinant AAVs can package foreign genes into their capsid [
1,
5,
6], and until recently, efforts to enhance gene delivery had focused on tailoring and improving capsids. However, efforts are increasingly shifting on improving the viral replicase, encoded by the rep gene [
7,
8,
9].
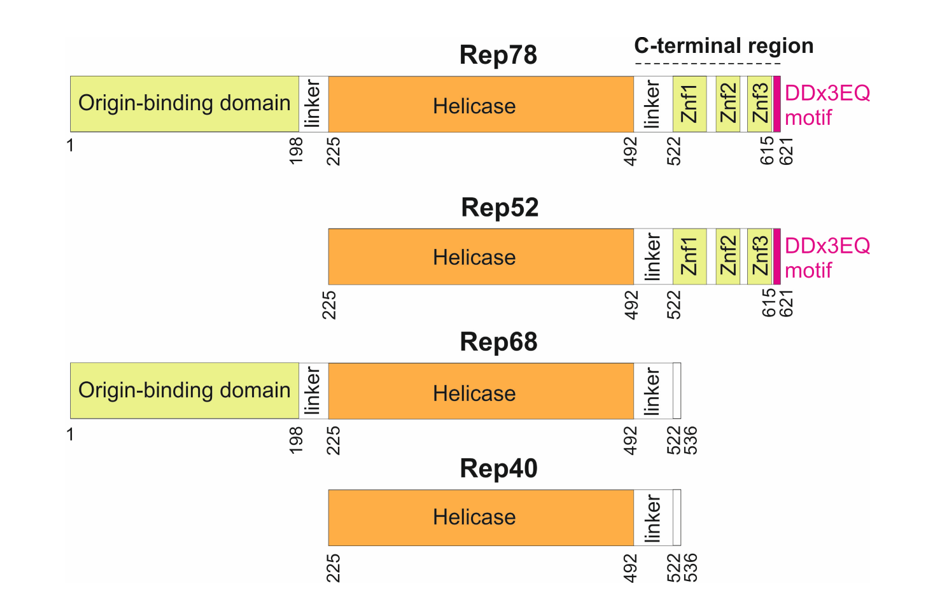
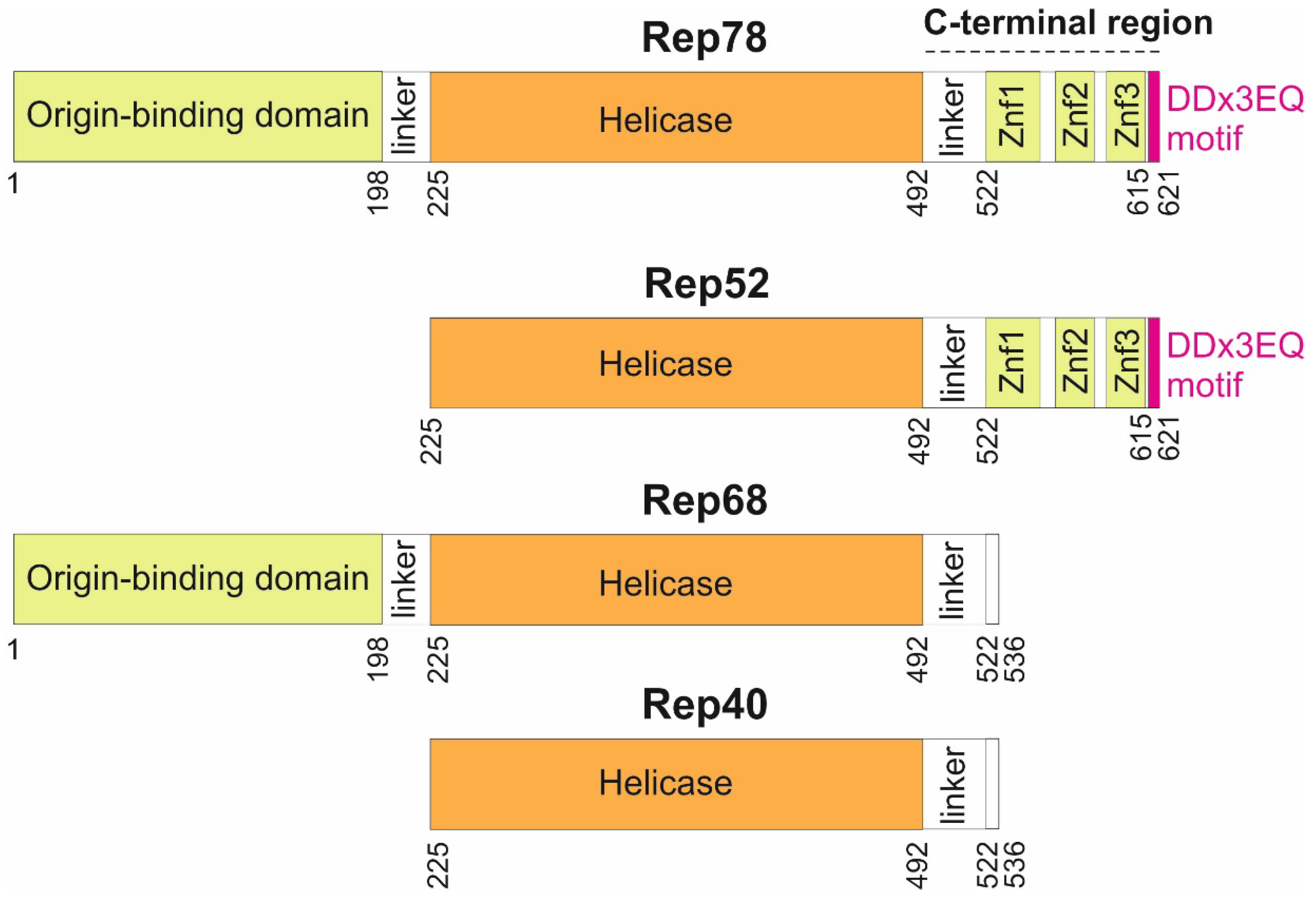
Rep encodes 4 protein isoforms (
Figure 1) thanks to a combination of alternative promoters and alternative splicing sites [
10]: two long isoforms (Rep78 and Rep68), and two short isoforms (Rep52 and Rep40). The larger Rep proteins, Rep78 and Rep68, are required to replicate the genome, while Rep52 and Rep40 facilitate packaging of the genome [
11]. Rep78 and Rep68 are sufficient for recombinant AAV production [
12].
Three main regions have been delineated in Rep78 (
Figure 1): an origin-binding domain [
13]; a helicase domain [
14]; and a C-terminal region, predicted to contain zinc-fingers [
15]. While the origin-binding and helicase domains have been systematically investigated, there has been no in-depth sequence analysis of the C-terminus beyond the putative identification of three zinc fingers [
15]. Here we examined sequences of Rep 78 across all dependoparvoviruses, beyond the usual ones employed in gene therapy (AAV1 to AAV13), and discovered that Rep78 contains a strictly C-terminal motif conserved in most dependoparvoviruses.
The representation is to scale. Znf: Zinc finger. The DDx3EQ motif was discovered in the present study.
2. Materials and Methods
Protein Sequence Analysis
We extracted
Dependoparvovirus sequences from NCBI’s Genbank [
43] on July 1
st 2024. We used Psi-Coffee [
44] for multiple sequence alignment. Alignments are shown with Jalview [
45], using the ClustalX colouring scheme [
46]. We used flDPnn [
47] for predicting disordered regions.
The compositional bias of the DDx3EQ motif was assessed using Composition Profiler [
48] against two datasets: 1) SwissProt (version 51) [
49]; 2) a dataset composed of all dependoparvovirus Rep78 sequences (available in File S2), after having removed their DDx3EQ motif, i.e. the last C-terminal 7 aa in each sequence.
Sequence Motif Searches
We looked for known motifs similar to the DDx3EQ motif using Comparimotif [
22] and TOMtOM [
23].
We also used TOMtOM [
23] to scan the database Prosite (april 2021 release) [
51].
We looked for proteins containing the DDx3EQ motif using Patternsearch, ran from the web-based version of the MPI toolkit [
52] (
https://toolkit.tuebingen.mpg.de/) against three databases that are subsets from Genbank [
43]: 1) the database nr_vir70_12Mar containing viral proteins clustered at 70% sequence identity on 12th March 2024; 2) the database Homo Sapiens_4Jul containing Homo Sapiens proteins on 4th July 2024; 3) the database PDB_nr_12_Mar containing proteins with an experimentally solved 3D structure on 12th March 2024. We used as input the regular expression [DEP]-D-{P}-{P}-{P}-E-Q> that follows the Prosite syntax [
53], in which {P} corresponds to an excluded P aa, and ‘>’ specifies that the motif must be C-terminal.
3D Structure Prediction and Visualization
We predicted the 3D structure of the C-terminal region of Rep78 using Alphafold3 [
16] with 3 zinc atoms. AlphaFold3 outputs a measure of reliability of the 3D structure for each aa, pLDDT. pLDDT ≥ 0.70 corresponds to a reliable prediction, and pLDDT ≥ 0.90 to a highly reliable prediction (expected to be competitive with an experimentally solved 3D structure) [
54]. Structures were visualized using ChimeraX [
55].
3. Results
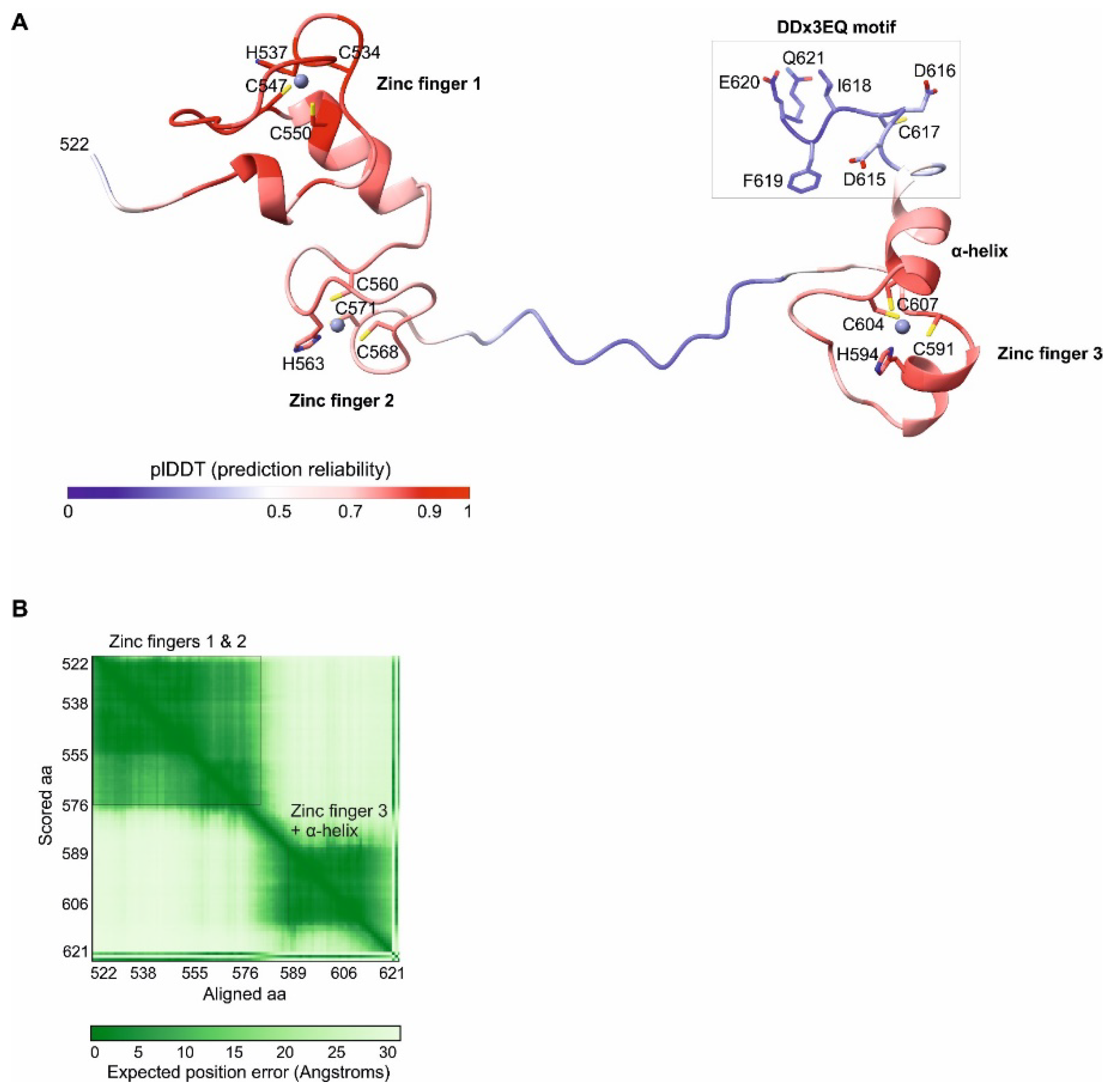
The C-Terminal Region of Rep78 Contains 3 Predicted Zinc Fingers and Flexible Regions
We analyzed the Rep78 protein of AAV2, the
Dependoparvovirus model species (Genbank accession number YP_680423.1, see
Table 1). The C-terminal region of Rep78 starts with a linker predicted to be disordered (aa 493-521 in AAV2, see
Figure 1). We modelled the 3D structure of the remaining C-terminal part (aa 522-621) using Alphafold3 [
16] (the coordinates of the model are in File S1). The model contains two regions reliably predicted to adopt a fixed 3D structure (in red in
Figure 2A; see also
Figure 1):
aa 525-573 are composed of two zinc fingers (named 1 and 2) of the CHCC type (
Figure 2A, left). These zinc fingers are predicted to be in contact and to adopt a fixed conformation relative to each other (
Figure 2B).
aa 587-612 are composed of a third zinc finger, also of the CHCC type, followed by a predicted α-helix (
Figure 2A, right).
The remaining regions are not reliably modelled by Alphafold3, despite being predicted to be ordered, which indicates that they are conformationally flexible; they are visible as blue or white ribbons in
Figure 2.
The C-Terminal Region of Dependoparvoviral Rep78 and Rep52 Contains a Conserved Motif, DDx3EQ, Not Similar to a Known Motif
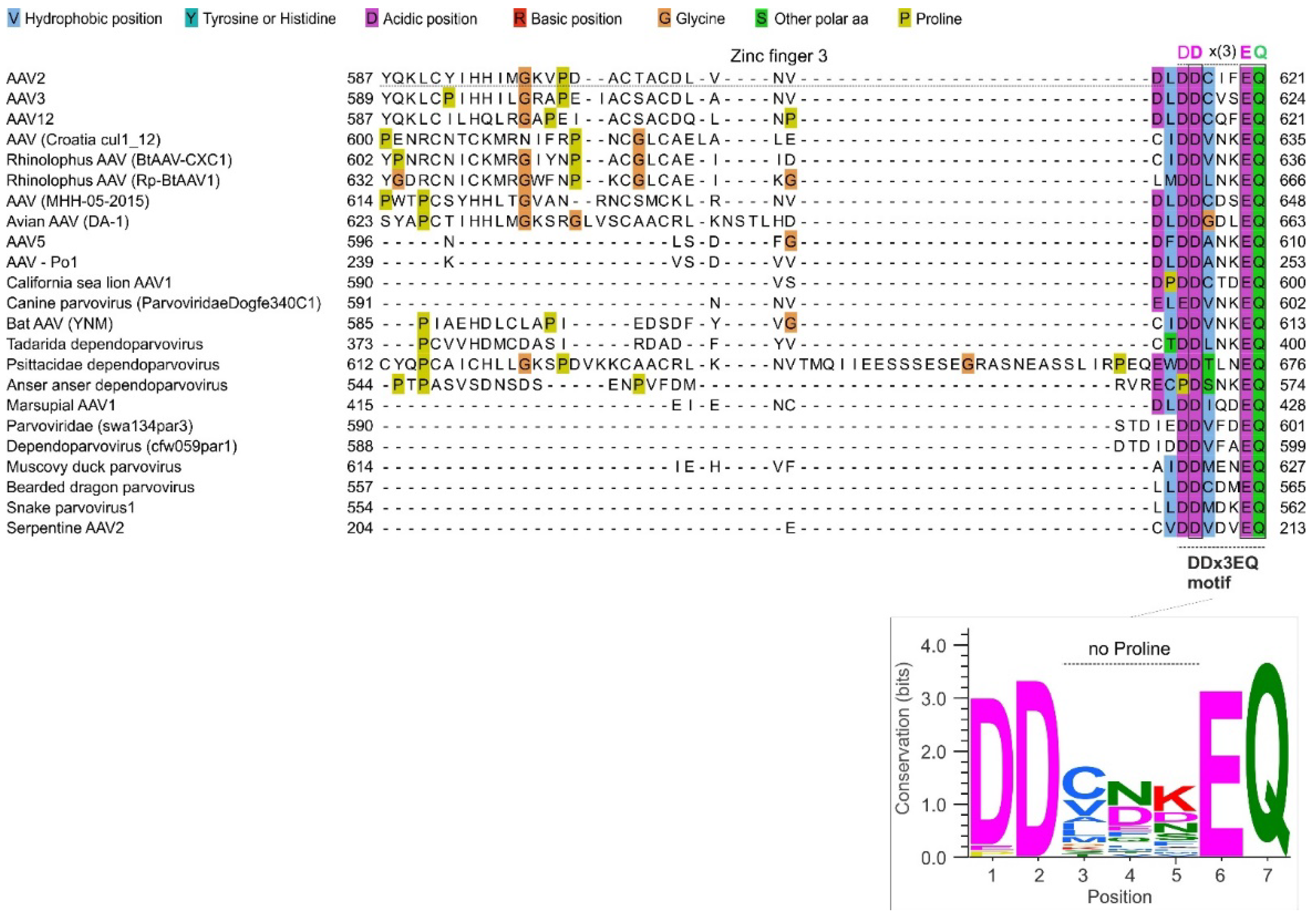
The C-terminal region of Rep78 is highly variable in sequence across dependoparvoviruses, as shown in
Figure 3 (see also File S2). Yet we noticed that in almost all dependoparvoviruses, it contains a D-D-x(3)-E-Q sequence motif (in which x(3) represents a consecutive stretch of any three aa) at the very C-terminus (aa 615-621 in AAV2). The motif is shown in
Figure 3, right, and for simplicity we will refer it as DDx3EQ.
In all dependoparvoviruses the last aa of the motif, Q, is also the last aa of Rep78. This strictly C-terminal position confers a markedly enhanced specificity to the motif [
17] (see Discussion).
Only a handful of dependoparvovirus Rep78 proteins do not have the DDx3EQ motif (File S2): the related viruses desmodus rotundus dependoparvovirus (
Dependoparvovirus chiropteran2) [
18] and feline dependoparvovirus (
Dependoparvovirus carnivoran1) [
19]; canary dependoparvoviruses 1 and 2 [
20]; and five bird dependoparvoviruses [
21]: isolates ltt164par2 (Genbank accession number QLF86430.1), sis142par1 (QKE54964.1), zftwig05par3 (QKN88780.1), wpk049par01 (QKE60686.1), and avian AAV isolate BR_DF12 (YP_010802670.1). The latter presents a striking case. Its rep gene contains a long (1803 nucleotides) reading frame overlapping that of Rep78, which encodes a potential protein of 243 aa ending with a C-terminal DDx3EQ motif. The sequence of that protein and its location within rep are presented in File S3.
Finally, we found that the DDx3EQ motif is not similar to a known motif, according to both Comparimotif [
22] and TOMtOM [
23] (see Methods).
The Motif Contains Three Strictly Conserved aa, Is Highly Negatively Charged and Devoid of Prolines
The frequency of each aa at each position of the motif is shown in
Figure 3, bottom panel. Three positions are strictly conserved (
Figure 3, bottom panel): an aspartate in position 2 (D616 in AAV2), a glutamate in position 6 (E620), and a glutamine in position 7 (Q621). Position 1 almost exclusively contains an aspartate (D615), rarely a glutamate (also negatively charged) or a proline. Position 3 is enriched in hydrophobic aa; position 4 is enriched in negatively charged aa, depleted in hydrophobic aa, and contains no positively charged aa; and position 5 is enriched in polar aa, in particular charged ones.
The motif is significantly (P<0.005) enriched in negative aa compared both to the protein database SwissProt and to the rest of Rep78 (see Methods); its negative charge is expected to be further increased by its C-terminal carboxylate ion (COO–).
Strikingly, the motif is completely devoid of prolines, except at position 1 in anser anser dependoparvovirus (
Figure 3) and a few related species, suggesting that forming an α-helix might be required for its function.
Figure 3.
The variable C-terminal region of dependoparvovirus Rep78 contains a DDx3EQ motif. Top panel: Sequence alignment of the C-terminal region of Rep78 among representative dependoparvoviruses. Note its high variability, and a conserved DDx3EQ motif at the very C-terminus. Bottom panel: sequence logo of the DDx3EQ motif, made using WebLogo [
24].
Figure 3.
The variable C-terminal region of dependoparvovirus Rep78 contains a DDx3EQ motif. Top panel: Sequence alignment of the C-terminal region of Rep78 among representative dependoparvoviruses. Note its high variability, and a conserved DDx3EQ motif at the very C-terminus. Bottom panel: sequence logo of the DDx3EQ motif, made using WebLogo [
24].
A Conserved DDx3EQ Motif Is Found in One Protein from a Eukaryotic Virus and in One Human Protein
To obtain clues regarding the function of the DDx3EQ motif, we searched for other proteins from either eukaryotic viruses or humans that would have the motif conserved in at least another species (see Methods).
In eukaryotic viruses other than dependoparvoviruses, we could only identify one protein with a conserved C-terminal DDx3EQ motif: the protease 2A from the genus
Enterovirus. As an example, the C-terminus of the protease 2A of enterovirus D (NP_740416.1) is EDdamEQ, i.e. with an E in position 1 instead of the D most commonly found in dependoparvovirus Rep78 – see
Figure 3, bottom panel).
The motif is found in the species Enterovirus A-B, D, F and H-J but not in Enterovirus E, G and K nor in the three species Rhinovirus A, B and C. In Enterovirus C, the motif is degenerate, i.e. there is an E in position 2 instead of the strictly conserved D. File S4 presents an alignment of enterovirus proteases 2A that have the DDx3EQ motif.
The enteroviral protease 2A is cleaved immediately after the conserved Q of the motif by another enteroviral protease, 3C [
25]. Apart from this Q, no position of the DDx3EQ motif corresponds to the cleavage specificity of 3C, whose main specificity determinant is an A three aa upstream of the Q at which the cleavage occurs (i.e. in position 4 of the motif) [
26]. Therefore, the presence of the motif in the 2A protease does not stem from a requirement for cleavage by the 3C protease. Interestingly, removing the 5 aa immediately upstream of the C-terminal Q (i.e. most of the motif) from the 2A protease of poliovirus (
Enterovirus C) is lethal without affecting its protease function [
27].
The DDx3EQ motif forms a coil with no regular secondary structure in the 2A protease of coxsackievirus B4 (
Enterovirus B) [
28], similar to the Alphafold3 prediction for the AAV2 Rep78 motif (
Figure 2A). The motif is not visible in the structure of the related coxsackievirus B3 2A protease, suggesting that it is flexible [
29].
Finally, we could only identify a single human protein with a conserved C-terminal DDx3EQ motif: Cep57L1 (Centrosomal protein 57 kDa-like protein 1, Uniprot accession number Q8IYX8). Its C-terminal 7aa are DDimwEQ. The motif is conserved across amniotes (clade
Amniota). File S5 presents an alignment of Cep57L1 orthologs that have the DDx3EQ motif. Cep57L1 contributes to maintaining centriole engagement during interphase [
30]. No functional data is available regarding the role of its C-terminus, to our knowledge, and in a recent study, Cep57L1 was not part of the proteins identified as having mutations of their C-terminus that causes disease in humans [
31].
4. Discussion
The DDx3EQ Motif Should Have a High Specificity Despite Its Short Length, and Is Probably Essential for AAV Replication
Only a handful of strictly C-terminal sequence motifs have been described in eukaryotic viruses [
17,
32]. The C-terminal position confers a high recognition specificity to these motifs, even when relatively short, because only one free carboxy group is found in each protein, at the C-terminus, where it can be recognized by specialized enzymes. For example, the average length of a human protein being ~600aa, a motif containing a glutamine with a free, C-terminal carboxy group is found 600 times less frequently than a glutamine
within a non C-terminal motif [
17].
Given the high rate of evolution of viral proteins, the DDx3EQ motif is most probably essential for dependoparvoviruses, since it is conserved in almost the whole genus. We cannot infer its function from published experimental studies, since to our knowledge no study tested the effect of substitutions or deletions of the very C-terminus of Rep78 (aa 608-621, beyond zinc finger 3) on the replication of wild-type AAVs. The further downstream substitution we are aware is in aa 607, in zinc finger 3 [
15], and the second most downstream substitution we are aware of concerns aa 540 [
33].
Interestingly, we could only identify a single viral protein (the enterovirus protease 2A) and a single human protein (Cep57L1) with a conserved DDx3EQ motif. We note that the presence of the motif in these proteins may be coincidental and does not imply functional similarity to Rep78.
A Hypothesis: The DDx3EQ Motif May Bind the DNA-Binding Interface of the Origin-Binding Domain of Rep78
“No one believes a hypothesis except its originator, but everyone believes an experiment except the experimenter” – William Ian Beardmore Beveridge
A study on human C-terminal motifs found that they typically have either of three functions, in decreasing order of frequency [
34]: 1) directing post-translational modification [
35]; 2) binding another protein(s); 3) directing trafficking through the cell [
17]. We can only provide a meaningful hypothesis regarding the second function, i.e. binding a protein.
Given the considerable rate of sequence evolution of viral proteins, the fact that such a short motif contains three strictly conserved aa suggests that it binds either a cellular protein or a highly conserved region of a viral protein. A prime candidate would be the DNA-binding interface of the origin-binding domain of Rep78, which is well conserved in sequence and positively charged, while the DDx3EQ motif is highly negatively charged. This hypothesis is biologically meaningful since a) the DDx3EQ motif and the origin-binding domain are always in close proximity, being part of the same protein; b) binding of the motif would provide a mechanism for regulating the interaction of this domain with the inverted terminal repeats during the replication cycle [
36]. In this scenario, the DDx3EQ motif of Rep 52, the shorter isoform of Rep78, could not interact in
cis with the origin-binding domain, since Rep52 is devoid of this domain (
Figure 1).
We emphasize that this scenario is merely proposed as a biological hypothesis meant to guide experiments.
The DDx3EQ Motif Might Not Be Necessary for Recombinant AAV Production, But Substitutions Within It Improve Production
The DDx3EQ motif is found in most taxa of AAVs relevant for gene therapy [
37], i.e. AAV 1-4 and 6-13 (
Dependoparvovirus primate 1), AAV5 (
Dependoparvovirus mammalian1), and porcine AAV1 (unclassified) [
38]. A recent study found that in the absence of Rep78, Rep68 was not sufficient for efficient recombinant AAV production [
8], indicating that the C-terminal region of Rep78 is also required. It would be interesting to determine whether the DDx3EQ motif in particular contributes to this requirement.
In that regard, a recent study systematically tested the effect of all single aa substitutions in Rep78 and Rep68 on the production of recombinant AAVs [
9]. Substitutions of conserved positions of the motif or introduction of prolines (normally absent from the motif) did not result in significantly lower production, indicating that the DDx3EQ motif is not necessary for production of recombinant AAVs, at least in the conditions tested.
Intriguingly, in that study, not only were some substitutions neutral but most even had a mildly beneficial effect on recombinant AAV production (i.e. in
Figure 2 of [
9], the last 7aa of Rep78 form a red “patch”). We will detail these briefly below. The study tested two production platforms. In the first one, pCMV-Rep78/68, Rep68 and Rep78 were produced and mutate from one plasmid, and the other AAV proteins (Rep40, Rep52, and the capsid proteins) were produced from other plasmids. In the second platform, wtAAV2, all AAV proteins were produced from a single plasmid and all four Rep proteins were thus mutated simultaneously.
Although numerous substitutions were mildly beneficial, only a few were
significantly beneficial. In the first platform, no substitution had a significant fitness effect. In the wtAAV2 platform, three substitutions significantly improved production: I618T, affecting position 4 (T being observed at this position in some dependoparvoviruses, see
Figure 2, bottom panel); F619N, affecting position 5 (N being seen at this position in some dependoparvoviruses); and E620S, which affects the strictly conserved E in position 6.
In summary, the DDx3EQ motif might not be necessary for efficient production of recombinant AAVs, but substitutions within it have the potential to improve production. Note that
recombinant AAV production, as measured in [
9], and
wild-type AAV replication are not identical processes. As such, it is possible that the DDx3EQ motif may be essential for wild-type AAV replication but dispensable for recombinant AAV production.
Sequence Motifs Can Be Identified Even Within Highly Variable Protein Regions by Examining Alignment of Orthologs
As
Figure 2 makes it clear, the DDx3EQ motif is clearly visible in an alignment of
Dependoparvovirus Rep78, even by a non-expert. Numerous such motifs can be identified in viral proteins using simple visual examination (eg soyuz1 and soyuz2 in
Paramyxovirinae [
39]).
Conversely, these motifs are not detectable even with advanced homology detection software commonly used to ascribe function to viral proteins [
40] (such as PSI-BLAST [
41] or HHpred[
42]), because they are too short (7-20 aa) and “hidden” within a highly variable region. Therefore, we recommend to systematically align variable regions of orthologous proteins across suitable evolutionary distances (i.e. genus or subfamily) and to examine them for conserved sequence motifs.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and supplementary materials.
Acknowledgments
I thank N Davey for his insights regarding motif analysis and N Jain, L Galibert, and J Qiu for feedback on the manuscript.
Conflicts of Interest
The author declares no conflicts of interest.
References
- Pupo A, Fernández A, Low SH, François A, Suárez-Amarán L, Samulski RJ. AAV vectors: The Rubik’s cube of human gene therapy. Molecular Therapy. 2022;30: 3515–3541. [CrossRef]
- Wang D, Tai PWL, Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov. 2019;18: 358–378. [CrossRef]
- Weinmann J, Grimm D. Next-generation AAV vectors for clinical use: an ever-accelerating race. Virus Genes. 2017;53: 707–713. [CrossRef]
- Li C, Samulski RJ. Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet. 2020;21: 255–272. [CrossRef]
- Aponte-Ubillus JJ, Barajas D, Peltier J, Bardliving C, Shamlou P, Gold D. Molecular design for recombinant adeno-associated virus (rAAV) vector production. Appl Microbiol Biotechnol. 2018;102: 1045–1054. [CrossRef]
- Catalán-Tatjer D, Tzimou K, Nielsen LK, Lavado-García J. Unravelling the essential elements for recombinant adeno-associated virus (rAAV) production in animal cell-based platforms. Biotechnology Advances. 2024;73: 108370. [CrossRef]
- Mietzsch M, Eddington C, Jose A, Hsi J, Chipman P, Henley T, et al. Improved Genome Packaging Efficiency of Adeno-associated Virus Vectors Using Rep Hybrids. Dutch RE, editor. J Virol. 2021;95: e00773-21. [CrossRef]
- Johari YB, Pohle TH, Whitehead J, Scarrott JM, Liu P, Mayer A, et al. Molecular design of controllable recombinant adeno-associated virus (AAV) expression systems for enhanced vector production. Biotechnology Journal. 2024;19: 2300685. [CrossRef]
- Jain NK, Ogden PJ, Church GM. Comprehensive mutagenesis maps the effect of all single-codon mutations in the AAV2 rep gene on AAV production. eLife. 2024;12: RP87730. [CrossRef]
- Qiu J, Pintel D. Processing of adeno-associated virus RNA. Front Biosci. 2008;13: 3101–3115. [CrossRef]
- King JA. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. The EMBO Journal. 2001;20: 3282–3291. [CrossRef]
- Hölscher C, Kleinschmidt JA, Bürkle A. High-level expression of adeno-associated virus (AAV) Rep78 or Rep68 protein is sufficient for infectious-particle formation by a rep-negative AAV mutant. J Virol. 1995;69: 6880–6885. [CrossRef]
- Im DS, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61: 447–457. [CrossRef]
- Smith RH, Kotin RM. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3’-to-5’ polarity. J Virol. 1998;72: 4874–4881. [CrossRef]
- Saudan P. Inhibition of S-phase progression by adeno-associated virus Rep78 protein is mediated by hypophosphorylated pRb. The EMBO Journal. 2000;19: 4351–4361. [CrossRef]
- Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024;630: 493–500. [CrossRef]
- Sharma S, Schiller MR. The carboxy-terminus, a key regulator of protein function. Critical Reviews in Biochemistry and Molecular Biology. 2019;54: 85–102. [CrossRef]
- De Souza W, Dennis T, Fumagalli M, Araujo J, Sabino-Santos G, Maia F, et al. Novel Parvoviruses from Wild and Domestic Animals in Brazil Provide New Insights into Parvovirus Distribution and Diversity. Viruses. 2018;10: 143. [CrossRef]
- Li Y, Gordon E, Idle A, Altan E, Seguin MA, Estrada M, et al. Virome of a Feline Outbreak of Diarrhea and Vomiting Includes Bocaviruses and a Novel Chapparvovirus. Viruses. 2020;12: 506. [CrossRef]
- Zhang Y, Talukder S, Bhuiyan MSA, He L, Sarker S. Opportunistic sampling of yellow canary (Crithagra flaviventris) has revealed a high genetic diversity of detected parvoviral sequences. Virology. 2024;595: 110081. [CrossRef]
- Dai Z, Wang H, Wu H, Zhang Q, Ji L, Wang X, et al. Parvovirus dark matter in the cloaca of wild birds. GigaScience. 2022;12: giad001. [CrossRef]
- Edwards RJ, Davey NE, Shields DC. CompariMotif: quick and easy comparisons of sequence motifs. Bioinformatics. 2008;24: 1307–1309. [CrossRef]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble W. Quantifying similarity between motifs. Genome Biol. 2007;8: R24. [CrossRef]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14: 1188–1190. [CrossRef]
- Laitinen OH, Svedin E, Kapell S, Nurminen A, Hytönen VP, Flodström-Tullberg M. Enteroviral proteases: structure, host interactions and pathogenicity: Pathogenicity of enteroviral proteases. Rev Med Virol. 2016;26: 251–267. [CrossRef]
- Blom N, Hansen J, Brunak S, Blaas D. Cleavage site analysis in picornaviral polyproteins: Discovering cellular targets by neural networks. Protein Science. 1996;5: 2203–2216. [CrossRef]
- Li X, Lu H-H, Mueller S, Wimmer E. The C-terminal residues of poliovirus proteinase 2Apro are critical for viral RNA replication but not for cis- or trans-proteolytic cleavage. Journal of General Virology. 2001;82: 397–408. [CrossRef]
- Baxter NJ, Roetzer A, Liebig H-D, Sedelnikova SE, Hounslow AM, Skern T, et al. Structure and Dynamics of Coxsackievirus B4 2A Proteinase, an Enyzme Involved in the Etiology of Heart Disease. J Virol. 2006;80: 1451–1462. [CrossRef]
- Peters CE, Schulze-Gahmen U, Eckhardt M, Jang GM, Xu J, Pulido EH, et al. Structure-function analysis of enterovirus protease 2A in complex with its essential host factor SETD3. Nat Commun. 2022;13: 5282. [CrossRef]
- Ito KK, Watanabe K, Ishida H, Matsuhashi K, Chinen T, Hata S, et al. Cep57 and Cep57L1 maintain centriole engagement in interphase to ensure centriole duplication cycle. Journal of Cell Biology. 2021;220: e202005153. [CrossRef]
- FitzHugh ZT, Schiller MR. Systematic Assessment of Protein C-Termini Mutated in Human Disorders. Biomolecules. 2023;13: 355. [CrossRef]
- Sobhy H. A Review of Functional Motifs Utilized by Viruses. Proteomes. 2016;4: 3. [CrossRef]
- Di Pasquale G, Chiorini JA. PKA/PrKX activity is a modulator of AAV/adenovirus interaction. EMBO J. 2003;22: 1716–1724. [CrossRef]
- Sharma S, Toledo O, Hedden M, Lyon KF, Brooks SB, David RP, et al. The Functional Human C-Terminome. PLoS One. 2016;11: e0152731. [CrossRef]
- Chen L, Kashina A. Post-translational Modifications of the Protein Termini. Front Cell Dev Biol. 2021;9: 719590. [CrossRef]
- Hickman AB, Ronning DR, Perez ZN, Kotin RM, Dyda F. The Nuclease Domain of Adeno-Associated Virus Rep Coordinates Replication Initiation Using Two Distinct DNA Recognition Interfaces. Molecular Cell. 2004;13: 403–414. [CrossRef]
- Issa SS, Shaimardanova AA, Solovyeva VV, Rizvanov AA. Various AAV Serotypes and Their Applications in Gene Therapy: An Overview. Cells. 2023;12: 785. [CrossRef]
- Puppo A, Bello A, Manfredi A, Cesi G, Marrocco E, Corte MD, et al. Recombinant Vectors Based on Porcine Adeno-Associated Viral Serotypes Transduce the Murine and Pig Retina. Qiu J, editor. PLoS ONE. 2013;8: e59025. [CrossRef]
- Karlin D, Belshaw R. Detecting Remote Sequence Homology in Disordered Proteins: Discovery of Conserved Motifs in the N-Termini of Mononegavirales phosphoproteins. Haslam NJ, editor. PLoS ONE. 2012;7: e31719. [CrossRef]
- Kuchibhatla DB, Sherman WA, Chung BYW, Cook S, Schneider G, Eisenhaber B, et al. Powerful sequence similarity search methods and in-depth manual analyses can identify remote homologs in many apparently “orphan” viral proteins. J Virol. 2014;88: 10–20. [CrossRef]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25: 3389–3402. [CrossRef]
- Hildebrand A, Remmert M, Biegert A, Söding J. Fast and accurate automatic structure prediction with HHpred: Structure Prediction with HHpred. Proteins. 2009;77: 128–132. [CrossRef]
- Sayers EW, Bolton EE, Brister JR, Canese K, Chan J, Comeau DC, et al. Database resources of the National Center for Biotechnology Information in 2023. Nucleic Acids Res. 2023;51: D29–D38. [CrossRef]
- Floden EW, Tommaso PD, Chatzou M, Magis C, Notredame C, Chang J-M. PSI/TM-Coffee: a web server for fast and accurate multiple sequence alignments of regular and transmembrane proteins using homology extension on reduced databases. Nucleic Acids Res. 2016;44: W339-343. [CrossRef]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25: 1189–1191. [CrossRef]
- Procter JB, Thompson J, Letunic I, Creevey C, Jossinet F, Barton GJ. Visualization of multiple alignments, phylogenies and gene family evolution. Nat Methods. 2010;7: S16-25. [CrossRef]
- Hu G, Katuwawala A, Wang K, Wu Z, Ghadermarzi S, Gao J, et al. flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat Commun. 2021;12: 4438. [CrossRef]
- Vacic V, Uversky VN, Dunker AK, Lonardi S. Composition Profiler: a tool for discovery and visualization of amino acid composition differences. BMC Bioinformatics. 2007;8: 211. [CrossRef]
- Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot. Methods Mol Biol. 2007;406: 89–112. [CrossRef]
- Kumar M, Michael S, Alvarado-Valverde J, Mészáros B, Sámano-Sánchez H, Zeke A, et al. The Eukaryotic Linear Motif resource: 2022 release. Nucleic Acids Research. 2022;50: D497–D508. [CrossRef]
- Sigrist CJA, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, et al. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41: D344-347. [CrossRef]
- Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, et al. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 2018;430: 2237–2243. [CrossRef]
- de Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34: W362-365. [CrossRef]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596: 583–589. [CrossRef]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis: UCSF ChimeraX Visualization System. Protein Science. 2018;27: 14–25. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).