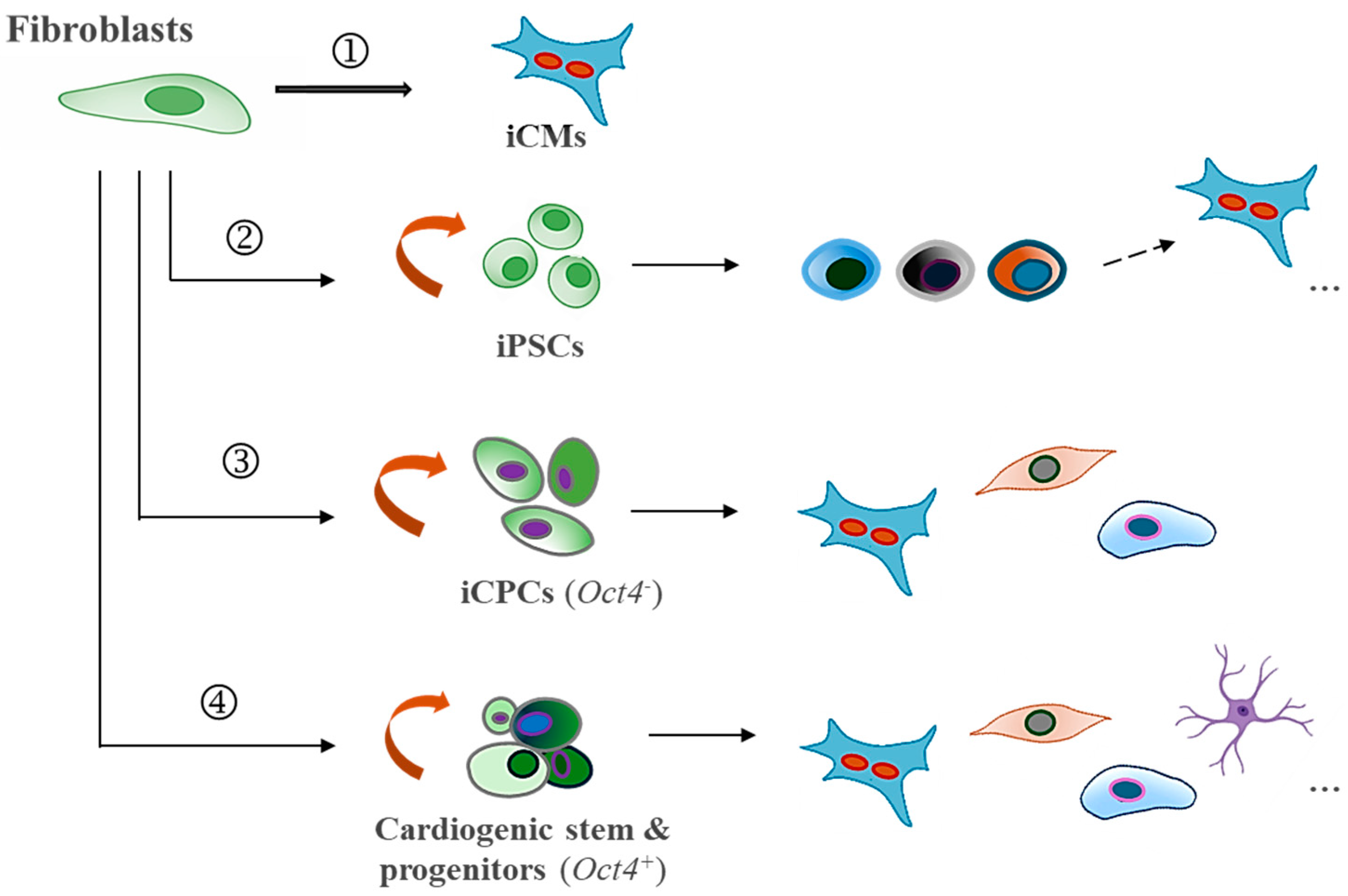
1. Clinical Challenges in Myocardial Regeneration
Heart diseases continue to be a major cause of global morbidity and mortality, with heart failure and myocardial infarction (MI) being the most significant contributors. Myocardial infarction, commonly known as a heart attack, occurs due to a blockage in the coronary arteries, leading to ischemia and the death of cardiomyocytes. If left untreated, the damaged myocardium is progressively replaced by fibrotic scar tissue, eventually leading to congestive heart failure (CHF), characterized by decreased contractile function and further deterioration of overall heart performance. [
1,
2,
3,
4,
5] Despite advancements in medical management, the prognosis for CHF remains poor, with a 5-year survival rate of only 50%. Current treatments primarily focus on improving heart function through medications that reduce cardiac workload and prevent further damage, such as angiotensin-converting enzyme (ACE) inhibitors, beta-adrenergic blockers, mechanical devices (e.g., ventricular assist devices, pacemakers), and surgical interventions (e.g., heart transplantation). However, these approaches mainly manage symptoms rather than restore the lost function of damaged cardiac tissue. They are also limited by low recovery efficacy, serious complications, and the global scarcity of donor hearts for transplantation. [
6,
7,
8,
9] A significant challenge is that the adult human heart has very limited regenerative capacity after cell loss, hampering the recovery of myocardial function after injury.
To reverse the loss of myocytes and promote myocardial regeneration, cell transplantation has been a major area of research over the past two decades. Strategies have been developed to administer exogenous stem or reserve cells, such as bone marrow stem cells, skeletal muscle cells, cardiac progenitors, or pluripotent stem cells, into the infarct zone to enhance cardiac repair and functional recovery. Unfortunately, clinical trials involving stem cell transplantation have largely been disappointing, often due to inadequate implant phenotypes, poor survival, limited engraftment of transplanted cells into the host myocardium, or the risk of cardiac arrhythmias. [
10,
11,
12,
13,
14] These challenges emphasize the need for a substantial paradigm shift in the field of regenerative medicine.
2. The Promise of Cardiac Cellular Reprogramming
The Nobel Prize-winning discovery of induced pluripotent stem cell (iPSC) technology in 2006, along with subsequent findings that iPSCs could be re-differentiated into cardiomyocyte-like cells, soon led to the discovery that specialized reprogramming factors (
e.g., Gata4, Mef2C, and Tbx5) could be administered to cardiac fibroblasts to directly transdifferentiate them into “induced cardiomyocyte-like” (iCM) cells without going through a pluripotency stage. [
15,
16,
17] Cardiac cellular transdifferentiation thus offers the potential to regenerate functional iCMs from cardiac fibroblasts
in situ, turning scar tissue into functional myocardium while overcoming the challenges posed by exogenous cell delivery strategies. Studies, including our own, have shown that administering reprogramming factors can increase ventricular ejection fraction by up to 30% in both acute and chronic animal models of myocardial infarction, while reducing infarct size by nearly half. [
18,
19,
20,
21,
22] These findings support the premise that direct cellular reprogramming is a promising new treatment for cardiomyopathy. Despite these encouraging results, however, it was soon revealed that cardiac fibroblasts from higher-order species (e.g., human, pig) are more resistant to trans-differentiation than rodent cells, likely due to more complex epigenetic barriers to reprogramming. [
23,
24,
25,
26,
27,
28,
29,
30] Expanded reprogramming cocktails have been explored to overcome this hurdle in human systems, incorporating additional transcription factors, microRNAs, small molecules, chemicals, growth factors, or p63 pathway inhibition. [
31,
32,
33,
34,
35] While these approaches have improved outcomes to varying degrees, further in-depth exploration of innovative cocktails and mechanistic investigations is still necessary to fully realize their potential for clinical applications.
Another reprogramming approach involves the generation of induced cardiac progenitor cells (iCPCs). iCPCs are engineered from cardiac fibroblasts to acquire unlimited self-renewal capacity and cardiac potency, enabling them to differentiate into all three cardiac lineages: cardiomyocytes, endothelial cells, and smooth muscle cells. iCPCs do not typically express key pluripotency genes such as Oct4 and Nanog, but are characterized by their expression of heart-specific multipotent progenitor markers such as Nkx2.5, Isl1, Flk1, and Gata4. As a result, iCPCs provide several benefits compared to iCMs or iPSCs, including their capacity to regenerate key cardiac lineages required for myocardial repair, a reduced risk of tumorigenesis, and the potential for autologous transplantation with minimized immune rejection.[
36,
37,
38,
39,
40] Despite these benefits, the iCPC approaches often involve complex reprogramming factors and induction procedures that can hinder their clinical application. Current methods utilize up to 11 or 5 cardiac factors along with JAK/STAT signaling mediators,[
41] multiple pluripotency regulators with refined chemical conditions or small molecules,[
42,
43] Ets2 and Mesp1 with TGF-β pathway members,[
44] or CRISPR activation of endogenous genes including
Gata4, Nkx2.5 and
Tbx5. [
45,
46,
47] These studies collectively support the applicability and regenerative capacity of iCPCs both in vitro and
in vivo. However, despite the theoretical feasibility of producing iCPCs in situ through the local delivery of the required reprogramming factors, few successful attempts have been documented to date. This is likely due to challenges in delivering multiple factors and managing iCPC induction and progression within the myocardial microenvironment. Thus, there is a pressing need for refined reprogramming cocktails and improved induction strategies to advance this approach. [
48,
49,
50]
3. Induced Partial Cellular Fate Transitions and Recent Advances
As an additional strategy, the induction of cell fate transitions into a partially reprogrammed state has gained increasing attention. In fact, partial cell fate transitions represent one of the most natural processes occurring in the body. In response to injury or stress, certain tissue cells can partially revert to a more primitive state, regaining characteristics associated with stem cells, such as self-renewal and multipotency. These features promote reparative regeneration, rejuvenation, and healing while minimizing the risks of tumor formation. This adaptive response is essential for maintaining homeostasis and ensuring organism survival.
In induced partial reprogramming, many protocols have utilized time-controlled transient expression of Yamanaka factors (i.e., Oct4, Sox2, Klf4, and c-Myc; OSKM) to trigger this process without reaching full pluripotency. Additionally, several non-OSKM approaches are being explored. In comparison to the iCM and iCPC approaches, partial reprogramming presents theoretical advantages for cardiac regeneration. By reverting to a plastic state, partially reprogrammed cells can dynamically adapt to the specific needs of the injured heart tissue, promoting the generation of damaged cell types for comprehensive repair. This method also mitigates the risk of uncontrolled cell proliferation or differentiation, ensuring a safer, more refined pathway to restoring heart function after injury. This balance of flexibility and inherent precision positions partial reprogramming as a particularly promising strategy in cardiac regenerative medicine.
Nevertheless, the pluripotency factor-induced mixed intermediate stem and progenitor states, along with the broad spectrum of acquired differentiation potentials, underscore that partial reprogramming remains a promising yet intricate and less well-defined concept. This complexity spans investigations involving diverse starting cell types, various pluripotency or multipotency induction protocols, growth signaling mediators, and different in vitro and in vivo models, making it challenging to precisely delineate the molecular and epigenetic modifications involved. To date, many OSKM-induced cardiac cell induction models have been explored, often in combination with small molecules, chemicals, or supportive culturing materials, as summarized below.
3.1. Partial Reprogramming of Cardiomyocytes In Vitro
The earliest efforts in partial (or ‘shortcut’) reprogramming emerged as researchers sought to adapt the principles of iPSC technology, directing the process toward cardiogenesis without reverting to full pluripotency. In 2011, Efe et al. transiently overexpressed OSKM in mouse embryonic fibroblasts (MEFs) and tail-tip fibroblasts (TTFs) to induce partially reprogrammed non-pluripotent colonies. These colonies were then exposed to defined media containing BMP4 and the JAK inhibitor JI1, which induced the cardiac progenitor cell (CPC) markers Nkx2.5, Gata4, and Flk1, and directed differentiation into spontaneously contracting patches of cardiomyocytes. [
51] In another study, Wang et al. transiently overexpressed Oct4 alone, along with small molecules SB431542 (a TGFβ inhibitor), CHIR99021 (a GSK3β inhibitor), Parnate (an LSD1 inhibitor), and Forskolin (a cAMP activator) in MEFs and TTFs. This combination similarly induced contracting iCM clusters through an intermediate proliferative CPC stage, with CPCs also differentiating into endothelial cells and smooth muscle cells under specific conditions.[
52] Furthermore, Fu et al. used an entirely chemical combination of CHIR99021, Forskolin, RepSox (an ALK5 inhibitor), valproic acid (an HDAC inhibitor), Parnate, and TTNPB (a retinoid activator) to reprogram these two fibroblast types, generating cardiomyocyte-like cells through a CPC stage without reaching full pluripotency.[
53] In human foreskin fibroblasts (HFFs), Cao et al. identified a combination of nine small molecules that epigenetically activated cardiogenic gene programs, resulting in sequential expression of gene markers for mesoderm (
KDR, MESP1, BRACHYURY), second heart field progenitors (
ISL1, HAND2, MEF2C, GATA4), and cardiomyocytes (
TNNT2, MYH6, NPPA), ultimately producing chemically induced beating cardiomyocytes (ciCMs).[
23] Additionally, various supporting materials such as Matrigel, engineered polyethylene glycol (PEG) hydrogels, and 3D fibrin gels have been employed in these models to enhance reprogramming efficiency. [
54,
55] Studies have also been conducted in bone marrow-derived mesenchymal stromal cells (MSCs), where co-culture with embryonic cardiomyocytes induced their expression of cardiac markers such as Nkx2.5 and ANF, while the MSCs retained their stromal cell phenotype. This process involves a dedifferentiation intermediate, characterized by a 2.6±0.7-fold increase in Oct4 expression. Silencing Oct4 hindered this MSC-to-CM transition, suggesting that Oct4 upregulation plays a crucial role in this partial reprogramming process. [
56]
In another study targeting cultured postnatal cardiomyocytes extracted from both rats and mice, temporary expression of adenoviral OSKM induced a partially dedifferentiated state characterized by enhanced cell proliferation. This state was marked by the temporary downregulation of cardiomyocyte-specific markers (
cTNT, Myh6, Myh7), disassembly or absence of sarcomere structures, increased expression of dedifferentiation markers (
Dppa4, E-cadherin), and the presence of proliferative Ki67+ cardiomyocytes. Notably, cardiomyocyte-specific morphology, gene expression, and contractile activity were spontaneously recovered by day 15 following viral transduction. [
57]
Supporting the partial reprogramming and rejuvenation concept, Chuang et al. demonstrated the feasibility of partially converting iPSC-derived cardiomyocytes into neurons by introducing the neurogenic transcription factors Brn2, Ascl1, Myt1l, and NeuroD. This approach generated a significant number of cells expressing markers of both cardiomyocytes and neurons, suggesting the presence of cells in an intermediate, partially-reprogrammed state. The observation that only three or four neuronal transcriptional factors are needed to convert mesoderm-derived, electrophysiologically-active CMs into ectoderm-derived, electrophysiologically-active neuronal-like cells suggests that a small number of key transcription factors are sufficient to regulate distinct cell fates. [
58] Taken together, these in vitro studies highlight a partially cardiogenic fate transition process stimulated by various factors in both human and rodent models. They provide valuable insights for mechanistic exploration, innovative protocols, and applications such as cardiac tissue engineering, drug screening, and organoid development. However, since the starting fibroblasts are not necessarily of cardiac origin, further investigations are needed to assess their in situ cardiac regenerative relevance and enhance their therapeutic potential.
3.2. In Vivo Models of Partial Reprogramming
In animal models, most in vivo partial reprogramming studies have centered on longevity research and rejuvenation, with the aim of epigenetically reversing markers of cellular aging and restoring youthful functionality. Many significant studies on this topic have been thoroughly reviewed elsewhere. [
59,
60,
61,
62,
63,
64,
65,
66,
67,
68] Additionally, tissue-targeted partial reprogramming strategies have been explored. For instance, Wang et al. reported that transient myofiber-specific OSKM expression using the
Acta1-Cre system activated muscle stem cells (satellite cells, or SCs) in mice, which accelerated muscle regeneration. Mechanistically, OSKM expression in myofibers regulates genes crucial for the SC microenvironment, such as p21, which in turn downregulates the myofiber-secreted niche factor Wnt4. [
69] Hishida et al. developed regulatable hepatocyte-specific OSKM expression in
Alb-Cre mice, which rapidly and transiently induces a proliferative, plastic progenitor state, resulting in enhanced liver regeneration. [
70] Similarly, Kim et al. induced in vivo partial dedifferentiation of intestinal epithelial cells using OSKM, revealing molecular changes akin to those observed during the intestinal regeneration process, which facilitated tissue regeneration following damage. [
71] OSKM-induced partial reprogramming also reduced fibrosis and improved tissue healing in mouse excisional and incisional wound models by inhibiting fibroblast transdifferentiation to myofibroblasts. [
72] In the neural context, transient reprogramming has demonstrated promise in several animal models of central nervous system (CNS) diseases, promoting the generation of new neurons, improving functional outcomes, and reducing scar formation. [
73,
74,
75,
76,
77,
78,
79] Additionally, non-OSKM approaches have also been documented. Chandrakanthan et al. transiently applied a combination of 5-azacytidine and the growth factor PDGF-AB to mature bone and fat cells, inducing the formation of induced multipotent stem (iMS) cells that expressed low levels of pluripotency genes, including Oct4, Myc, Sox2, Klf4, and Nanog. These iMS cells demonstrated long-term self-renewal, serial clonogenicity, and contributed to tissue regeneration in a context-dependent manner when introduced in vivo, without developing teratomas. [
80]
In the cardiac field, however, studies investigating in vivo partial reprogramming approaches remain very limited. Two notable studies have explored this area. Kisby et al. intramyocardially injected short-term adenoviral vectors encoding OSKM factors into both healthy and MI-injured mouse hearts. The expression of these factors induced transient upregulation of several endogenous pluripotency genes (
endo-Oct3/4,
Gdf3) and reprogramming-related genes (
Cdh1, Fut4), while markers of fully dedifferentiated cells, including
Nanog, remained silenced, with no teratoma formation observed, consistent with a partially reprogrammed process. However, this strategy was unable to elicit a significant regenerative response in the injured myocardium, possibly due in part to the relatively low rate of vector transduction (an average of 11.8%) in the myocardium. [
81] In comparison, in a separate study, Chen et al. employed temporally controlled, heart-specific OSKM expression in mice, which led to the dedifferentiation of adult cardiomyocytes and reentry into the cell cycle, with a gene expression profile resembling that of fetal cardiomyocytes. In MI-injured hearts, this approach notably decreased fibrotic scar size and improved ventricular ejection fraction, highlighting its potential therapeutic relevance. However, it is important to note that with long-term OSKM treatments, cardiomyocytes either entered an irreversible neonatal state that could not sustain function or progressed to a pluripotent state that led to teratoma formation, resulting in premature death. [
82] This dose- and time-controlled pattern aligns with the partial reprogramming process and trajectory in somatic cells, further underscoring its significance in therapeutic settings.
4. Advancing the Cardiac Partial Reprogramming Strategies
Building on the outcomes of previous studies, there is a growing imperative to develop innovative methodologies that can enhance current reprogramming paradigm in cardiac cells. In a recent study, our team explored the
Sall gene family member, Sall4, due to its unique roles in pluripotent stem cells, cardiac progenitors, and heart morphogenesis. [
83] Our work expands upon the promising findings that overexpressing Sall4 alongside Gata4, two transcription factors known for their distinct and overlapping roles in resident CPCs, iCM transdifferentiation, iPSC and iCPC reprogramming, and heart development, can potently induce partial cellular fate transitions in cardiac fibroblasts. Specifically, the data demonstrated that overexpression of these two factors drove the emergence of a stem/progenitor-like cell population in rodent cardiac fibroblasts, with approximately 32 ± 6.4% expressing Nkx2.5 and 13 ± 3.6% expressing Oct4. Additionally, 29.2 ± 15.0% were detected as Flk1
+, while the percentages of Nanog
+ or SSEA1
+ cells remained below 5%. These cells exhibited remarkable clonogenicity and extended ex vivo expandability, showing a high susceptibility to differentiate into various cardiac and non-cardiac cell types, including contractile cardiomyocytes, endothelial cells, smooth muscle cells, and neuron-like cells. Molecular analysis revealed that the SALL4-GATA4 complex activates crucial stemness-related signaling pathways such as PI3K/Akt, Hippo, and Wnt, while synergistically stimulating pluripotency and cardiac gene promoters and repressing fibrogenic genes. This synergistic action suggests a primitive transition process capable of driving cardiac regeneration. Furthermore, significant stem/progenitor-like transitions and robust cardiogenic differentiation were observed in both human and rodent cardiac fibroblasts but not in skin fibroblasts. This distinction potentially highlights the cardiac specificity and therapeutic applications of this approach. Together, these findings open new avenues for advancing cardiac regenerative therapies and tissue engineering.
In comparison to existing cardiac reprogramming strategies (see
Figure 1), this approach presents several unique strengths: (1) the use of only two factors, Sall4 and Gata4, both of which naturally contribute to cardiogenic regeneration; (2) a simplified, cardiac fibroblast-targeted procedure that does not require specialized induction media to drive the process; (3) a high primitive fate (
e.g., OCT4+/Nkx2.5+) conversion rate and proliferative cellular production in both rodent and human models, along with increased sensitivity to generate multiple heart cell fractions necessary for cardiac repair; (4) anticipated improved myocardial regeneration and integration following localized delivery of these factors; (5) minimal tumorigenic risks in the adult heart, partly supported by in vivo reports utilizing xenotransplantation models or intramyocardial gene delivery in myocardial infarction animal models; [
84,
85] and (6) given the simplicity of using just two genes, practical non-integrating delivery approaches, such as modified RNA, refined proteins, or CRISPR/Cas9-mediated gene activation technology could be readily explored for enhanced safety and efficiency. These strengths make this
non-OSKM approach highly promising for streamlined therapeutic applications. Nevertheless, despite the compelling advantages, significant limitations need to be addressed in the next steps of the studies. These include: (1) a limited understanding of the cellular and epigenetic mechanisms regulating Sall4 and Gata4 interactions in cardiac fibroblasts and fibroblasts from various tissue origins and species, necessitating further investigation to optimize safety, efficacy, and specificity; (2) uncertainty regarding the exact responsive subpopulations within the phenotypically heterogeneous resident cardiac fibroblasts and specific molecular pathways involved, which requires further characterization; (3) challenges in controlling the compositions and fates of reprogrammed and spontaneously differentiated cells, which may affect the consistency and effectiveness of the therapy; (4) the need for systematic in vivo mechanistic and reparative validations to confirm therapeutic benefits in diseased models; and (5) uncertainty surrounding the long-term effects of Sall4/Gata4-induced reprogramming, including safety and sustained functionality in vivo. Taken together, while the potential of Sall4 and Gata4 in cardiac reprogramming is evident, ongoing research is required to address these aspects and fully realize their therapeutic potential.
5. Prospective Considerations for Enhancing Therapeutic Cardiac Reprogramming
As the field of cardiac reprogramming progresses, several prospective enhancements warrant consideration to optimize outcomes and broaden therapeutic applicability. Firstly, the choice of starting cells should ideally focus on those of cardiac origin, such as cardiac fibroblasts, which are more applicable for in situ conversion. Utilizing these cells leverages their inherent properties and profiles, as they possess unique epigenetic landscapes and signaling pathways tailored to the heart’s microenvironment. These cells also naturally express many cardiogenic genes that are critical for reprogramming, such as Gata4, Tbx20, Mef2c, and Nkx2-5, which are often absent in non-cardiac cells. [
86,
87,
88] Additionally, cardiac fibroblasts critically engage in scar formation and pathological remodeling following myocardial injury.
Importantly, off-target reprogramming effects can lead to significant undesired consequences. In this regard, distinguishing cell heterogeneity and responsive subfractions through techniques such as single-cell RNA sequencing, phenotypic profiling, and epigenomic analysis will be beneficial for significantly improving efficacy, guiding targeted therapies, and optimizing reprogramming methods. For example, previous studies have identified adult cardiac-resident colony forming units-fibroblasts (CFU-Fs) expressing Oct4, but not Sox2 or Nkx2.5. [
89] Diverse progenitor-like fractions have also been characterized based on markers such as Sca1, Isl1, Pdgfr, Flk1, and Thy1. [
36,
89,
90,
91,
92] Such cell subtypes maybe more susceptible to specific reprogramming factors. As an example, exogenous Oct4 with either Sox2 or Klf4 partially reprogramed Thy1
+/Sca1
+ fibroblasts to a primitive stage concurrently expressing mixed lineage markers. [
93] Therefore, targeting cardiac-specific cell types and subfractions ensures relevant and more effective therapeutic outcomes, including the regeneration of lost cardiomyocytes and essential supporting cells through pathways such as paracrine signaling, extracellular matrix remodeling, neurovascular coupling, and angiogenesis—all of which can seamlessly integrate with the heart’s structural and functional demands. [
94,
95,
96,
97,
98]
Secondly, it will be essential to optimize the reprogramming factors and treatment duration, as these elements significantly influence reprogramming efficiency and the resultant cell fate transitions. The Sall4/Gata4 transcriptome study identifies several pluripotency genes and a range of potential transcriptional candidates such as
Sox17,
Gata6,
Sall1,
Tbx1,
Srf, along with other relevant genes like
Hcn4, which play critical roles in various aspects of the cellular reprogramming process. Briefly, Sox17 and Gata6 are key regulators of endodermal and mesodermal lineage specification, with the interaction between SOX17 and OCT4 being particularly important in cardiac progenitor cell development. [
99] Gata6 shares overlapping roles with Gata4 in regulating the expression of Sall4 and inducing pluripotency. [
100] Sall1, similar to Sall4, is involved in maintaining pluripotency and dedifferentiation, supporting the transition of differentiated cells back to a more stem-like state while sustaining the cardiac precursor cell-like features. [
101] Tbx1 contributes to cellular proliferation, differentiation, and early cardiac development, playing a role in both myocardial growth and angiogenesis. [
102] Likewise, serum response factor (SRF) is crucial in regulating the plasticity and adaptability of cardiac fibroblasts. [
101,
103] Meanwhile, Hcn4 is crucial for regulating the electrophysiological properties of cardiac precursor cells, especially in maintaining pacemaker activity. [
104,
105] Together, these factors may differentially enhance the potential of partial reprogramming strategies by driving the dedifferentiation and specification necessary for efficient cardiac regeneration. Additionally, a detailed time-dependent investigation of reprogramming will be beneficial for understanding the precise dedifferentiation and cardiac specification phases, ultimately improving the efficacy of partial cardiac regenerative strategies.
Thirdly, the utilization of non-integrating approaches, such as small molecules, chemicals, and microRNAs, offers a transient and reversible reprogramming process with diminished risks of insertional mutagenesis or long-term genomic alterations. [
106,
107] Leveraging available datasets for these factors, in conjunction with in-depth Sall4/Gata4 epigenetic and microRNA studies, may aid in predicting and establishing new essential effectors. Small molecules can modulate key DNA methylation and histone modification states, as well as signaling pathways such as PI3K/Akt, Hippo, TGF-β, Wnt, and MAPK, all identified in Sall4/Gata4-stimulated pathways in cardiac fibroblasts. Similarly, microRNAs can fine-tune the balance between cellular processes, such as proliferation, survival, and cardiac fate induction, as mediated by Sall4/Gata4. For example, miR-1, miR-133, miR-208, miR-200 family, and miR-590 have been shown to promote cardiomyocyte conversion and phenotypes, as associated to Gata4
or Sall4. [
30,
108,
109,
110] All of these could be harnessed to enhance partial cardiac reprogramming. Additionally, combining reprogramming with tissue engineering strategies, such as 3D bioprinting of cardiac patches or biomaterials, [
111,
112] could offer synergistic benefits by providing structural support and facilitating the integration of newly generated cardiomyocytes into heart tissue.
Lastly, new strategies should emphasize the development of human-specific reprogramming factors and techniques, ensuring that approaches are tailored to the unique characteristics of human cardiac cells. This focus can enhance the efficacy and safety of reprogramming methodologies, ultimately facilitating their translation into clinical practice. Overall, these prospective enhancements—from optimizing cell selection and reprogramming factors to refining non-integrating reprogramming methods and advancing the development of human-specific protocols—represent pivotal steps in advancing cardiac reprogramming therapies toward future clinical applications.
6. Conclusions
In conclusion, cardiac partial cell fate transition strategies offer significant potential for addressing the challenges of myocardial regeneration. These approaches leverage the plasticity of iPSCs, promoting broad tissue regeneration and rejuvenation in the diseased heart, while maintaining the lineage-specific safety and reduced tumor risk associated with iCPCs. By facilitating versatile in situ myocardial regeneration, partial reprogramming strikes a balance between regenerative potential and therapeutic safety. Although significant limitations remain, such as the need for a deeper understanding of intrinsic molecular and epigenetic mechanisms, cellular interactions, and optimization of targeted reprogramming, future advancements are anticipated to bring us closer to achieving safe, effective, and clinically relevant cardiac reprogramming therapies for regenerative medicine.
Funding
This work was supported by Michael E. DeBakey Department of Surgery Faculty Award and in part by NIH grant R01HL121294-01A1.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Savarese, G. & Lund, L.H. Global Public Health Burden of Heart Failure. Card Fail Rev 3, 7-11 (2017). [CrossRef]
- Benjamin, E.J. et al. Heart Disease and Stroke Statistics-2018 Update: A Report From the American Heart Association. Circulation 137, e67-e492 (2018). [CrossRef]
- Aronow, W.S. Heart-failure-complicating acute myocardial infarction. Clin Geriatr Med 23, 123-139 (2007). [CrossRef]
- Jackson, S.L. et al. National Burden of Heart Failure Events in the United States, 2006 to 2014. Circ Heart Fail 11, e004873 (2018). [CrossRef]
- Akhtar, K.H. et al. The spectrum of post-myocardial infarction care: From acute ischemia to heart failure. Prog Cardiovasc Dis 82, 15-25 (2024). [CrossRef]
- Shah, D. & Sen, J. Mechanical Circulatory Support in Cardiogenic Shock: A Narrative Review. Cureus 16, e69379 (2024). [CrossRef]
- Heidenreich, P.A. et al. 2022 AHA/ACC/HFSA Guideline for the Management of Heart Failure: Executive Summary: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 145, e876-e894 (2022). [CrossRef]
- Carberry, J., Marquis-Gravel, G., O'Meara, E. & Docherty, K.F. Where Are We With Treatment and Prevention of Heart Failure in Patients Post-Myocardial Infarction? JACC Heart Fail 12, 1157-1165 (2024). [CrossRef]
- Dar, J.A. & Jacob, J.R. Beta Blockers in Contemporary Cardiology: Is It Better to Cast Them Out? Korean Circ J 54, 165-171 (2024). [CrossRef]
- Rosenzweig, A. Cardiac cell therapy--mixed results from mixed cells. N Engl J Med 355, 1274-1277 (2006). [CrossRef]
- Nguyen, P.K., Rhee, J.W. & Wu, J.C. Adult Stem Cell Therapy and Heart Failure, 2000 to 2016: A Systematic Review. JAMA Cardiol 1, 831-841 (2016). [CrossRef]
- Rosengart, T.K., Patel, V. & Sellke, F.W. Cardiac stem cell trials and the new world of cellular reprogramming: Time to move on. J Thorac Cardiovasc Surg 155, 1642-1646 (2018). [CrossRef]
- Abouzid, M.R. et al. Stem Cell Therapy for Myocardial Infarction and Heart Failure: A Comprehensive Systematic Review and Critical Analysis. Cureus 16, e59474 (2024). [CrossRef]
- Tsai, I.T. & Sun, C.K. Stem Cell Therapy against Ischemic Heart Disease. Int J Mol Sci 25 (2024). [CrossRef]
- Qian, L. et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485, 593-598 (2012). [CrossRef]
- Song, K. et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485, 599-604 (2012). [CrossRef]
- Ieda, M. et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375-386 (2010). [CrossRef]
- Mathison, M. et al. In vivo cardiac cellular reprogramming efficacy is enhanced by angiogenic preconditioning of the infarcted myocardium with vascular endothelial growth factor. J Am Heart Assoc 1, e005652 (2012). [CrossRef]
- Mathison, M. et al. "Triplet" polycistronic vectors encoding Gata4, Mef2c, and Tbx5 enhances postinfarct ventricular functional improvement compared with singlet vectors. J Thorac Cardiovasc Surg 148, 1656-1664 e1652 (2014). [CrossRef]
- Mathison, M. et al. In situ reprogramming to transdifferentiate fibroblasts into cardiomyocytes using adenoviral vectors: Implications for clinical myocardial regeneration. J Thorac Cardiovasc Surg 153, 329-339 e323 (2017). [CrossRef]
- Tani, H. et al. Direct Reprogramming Improves Cardiac Function and Reverses Fibrosis in Chronic Myocardial Infarction. Circulation 147, 223-238 (2023). [CrossRef]
- Perveen, S., Vanni, R., Lo Iacono, M., Rastaldo, R. & Giachino, C. Direct Reprogramming of Resident Non-Myocyte Cells and Its Potential for In Vivo Cardiac Regeneration. Cells 12 (2023). [CrossRef]
- Cao, N. et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 352, 1216-1220 (2016). [CrossRef]
- Christoforou, N. et al. Core Transcription Factors, MicroRNAs, and Small Molecules Drive Transdifferentiation of Human Fibroblasts Towards The Cardiac Cell Lineage. Sci Rep 7, 40285 (2017). [CrossRef]
- Nam, Y.J. et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci U S A 110, 5588-5593 (2013). [CrossRef]
- Wada, R. et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc Natl Acad Sci U S A 110, 12667-12672 (2013). [CrossRef]
- Singh, V.P. et al. Enhanced Generation of Induced Cardiomyocytes Using a Small-Molecule Cocktail to Overcome Barriers to Cardiac Cellular Reprogramming. J Am Heart Assoc 9, e015686 (2020). [CrossRef]
- Garry, G.A. & Olson, E.N. Cardiac Reprogramming: Toward a Total Eclipse of the Failing Heart. Circulation 147, 239-241 (2023). [CrossRef]
- Talkhabi, M., Zonooz, E.R. & Baharvand, H. Boosters and barriers for direct cardiac reprogramming. Life Sci 178, 70-86 (2017). [CrossRef]
- Singh, V.P. et al. MiR-590 Promotes Transdifferentiation of Porcine and Human Fibroblasts Toward a Cardiomyocyte-Like Fate by Directly Repressing Specificity Protein 1. J Am Heart Assoc 5 (2016). [CrossRef]
- Vaseghi, H., Liu, J. & Qian, L. Molecular barriers to direct cardiac reprogramming. Protein Cell 8, 724-734 (2017). [CrossRef]
- Addis, R.C. et al. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J Mol Cell Cardiol 60, 97-106 (2013). [CrossRef]
- Zhou, H., Dickson, M.E., Kim, M.S., Bassel-Duby, R. & Olson, E.N. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proceedings of the National Academy of Sciences of the United States of America 112, 11864-11869 (2015). [CrossRef]
- Patel, V. et al. p63 Silencing induces reprogramming of cardiac fibroblasts into cardiomyocyte-like cells. J Thorac Cardiovasc Surg 156, 556-565 e551 (2018). [CrossRef]
- Pinnamaneni, J.P. et al. p63 silencing induces epigenetic modulation to enhance human cardiac fibroblast to cardiomyocyte-like differentiation. Sci Rep 12, 11416 (2022). [CrossRef]
- Barreto, S., Hamel, L., Schiatti, T., Yang, Y. & George, V. Cardiac Progenitor Cells from Stem Cells: Learning from Genetics and Biomaterials. Cells 8 (2019). [CrossRef]
- Schwach, V. et al. Expandable human cardiovascular progenitors from stem cells for regenerating mouse heart after myocardial infarction. Cardiovasc Res 116, 545-553 (2020). [CrossRef]
- Xu, J., Lian, W., Li, L. & Huang, Z. Generation of induced cardiac progenitor cells via somatic reprogramming. Oncotarget 8, 29442-29457 (2017). [CrossRef]
- Moretti, A. et al. Mouse and human induced pluripotent stem cells as a source for multipotent Isl1+ cardiovascular progenitors. FASEB J 24, 700-711 (2010). [CrossRef]
- Parmacek, M.S. & Epstein, J.A. Pursuing cardiac progenitors: Regeneration redux. Cell 120, 295-298 (2005). [CrossRef]
- Lalit, P.A. et al. Lineage Reprogramming of Fibroblasts into Proliferative Induced Cardiac Progenitor Cells by Defined Factors. Cell Stem Cell 18, 354-367 (2016). [CrossRef]
- Zhang, Y. et al. Expandable Cardiovascular Progenitor Cells Reprogrammed from Fibroblasts. Cell Stem Cell 18, 368-381 (2016). [CrossRef]
- Wang, J. et al. Reprogramming of fibroblasts into expandable cardiovascular progenitor cells via small molecules in xeno-free conditions. Nat Biomed Eng 6, 403-420 (2022). [CrossRef]
- Islas, J.F. et al. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci U S A 109, 13016-13021 (2012). [CrossRef]
- Li, X.H. et al. Generation of Functional Human Cardiac Progenitor Cells by High-Efficiency Protein Transduction. Stem Cells Transl Med 4, 1415-1424 (2015). [CrossRef]
- Jiang, L. et al. CRISPR activation of endogenous genes reprograms fibroblasts into cardiovascular progenitor cells for myocardial infarction therapy. Mol Ther 30, 54-74 (2022). [CrossRef]
- Jiang, L., Liang, J.L., Huang, W., Paul, C. & Wang, Y.G. Reprogramming of Fibroblasts Into Cardiac Progenitor Cells Using Crispr Activation System. Circulation 138 (2018).
- Liu, L., Guo, Y., Li, Z. & Wang, Z. Improving Cardiac Reprogramming for Heart Regeneration in Translational Medicine. Cells 10 (2021). [CrossRef]
- He, X. et al. Advances in Cellular Reprogramming-Based Approaches for Heart Regenerative Repair. Cells 11 (2022). [CrossRef]
- Zhao, H.Y. & Huang, C.X. Conversion of human cardiac progenitor cells using reprogramming factors into heterogeneous cardiac pacemaker-like cells. Journal of Molecular and Cellular Cardiology 141, 53-53 (2020). [CrossRef]
- Efe, J.A. et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol 13, 215-222 (2011). [CrossRef]
- Wang, H. et al. Small molecules enable cardiac reprogramming of mouse fibroblasts with a single factor, Oct4. Cell Rep 6, 951-960 (2014). [CrossRef]
- Fu, Y. et al. Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails. Cell Res 25, 1013-1024 (2015). [CrossRef]
- Kong, Y.P., Carrion, B., Singh, R.K. & Putnam, A.J. Matrix identity and tractional forces influence indirect cardiac reprogramming. Sci Rep 3, 3474 (2013). [CrossRef]
- Smith, A.W. et al. Direct reprogramming of mouse fibroblasts to cardiomyocyte-like cells using Yamanaka factors on engineered poly(ethylene glycol) (PEG) hydrogels. Biomaterials 34, 6559-6571 (2013). [CrossRef]
- Yannarelli, G. et al. OCT4 expression mediates partial cardiomyocyte reprogramming of mesenchymal stromal cells. PLoS ONE 12, e0189131 (2017). [CrossRef]
- Kisby, T., de Lazaro, I., Stylianou, M., Cossu, G. & Kostarelos, K. Transient reprogramming of postnatal cardiomyocytes to a dedifferentiated state. PLoS ONE 16, e0251054 (2021). [CrossRef]
- Chuang, W. et al. Partial Reprogramming of Pluripotent Stem Cell-Derived Cardiomyocytes into Neurons. Sci Rep 7, 44840 (2017). [CrossRef]
- Puri, D. & Wagner, W. Epigenetic rejuvenation by partial reprogramming. Bioessays 45, e2200208 (2023). [CrossRef]
- Gill, D. et al. Multi-omic rejuvenation of human cells by maturation phase transient reprogramming. Elife 11 (2022). [CrossRef]
- Mendelsohn, A.R., Larrick, J.W. & Lei, J.L. Rejuvenation by Partial Reprogramming of the Epigenome. Rejuvenation Res 20, 146-150 (2017). [CrossRef]
- Olova, N., Simpson, D.J., Marioni, R.E. & Chandra, T. Partial reprogramming induces a steady decline in epigenetic age before loss of somatic identity. Aging Cell 18, e12877 (2019). [CrossRef]
- Ocampo, A. et al. In Vivo Amelioration of Age-Associated Hallmarks by Partial Reprogramming. Cell 167, 1719-1733 e1712 (2016). [CrossRef]
- Paine, P.T., Nguyen, A. & Ocampo, A. Partial cellular reprogramming: A deep dive into an emerging rejuvenation technology. Aging Cell 23, e14039 (2024). [CrossRef]
- Singh, P.B. & Zhakupova, A. Age reprogramming: Cell rejuvenation by partial reprogramming. Development 149 (2022). [CrossRef]
- Chondronasiou, D. et al. Multi-omic rejuvenation of naturally aged tissues by a single cycle of transient reprogramming. Aging Cell 21, e13578 (2022). [CrossRef]
- Lu, Y. et al. Reprogramming to recover youthful epigenetic information and restore vision. Nature 588, 124-129 (2020). [CrossRef]
- de Lazaro, I., Cossu, G. & Kostarelos, K. Transient transcription factor (OSKM) expression is key towards clinical translation of in vivo cell reprogramming. EMBO Mol Med 9, 733-736 (2017). [CrossRef]
- Wang, C. et al. In vivo partial reprogramming of myofibers promotes muscle regeneration by remodeling the stem cell niche. Nat Commun 12, 3094 (2021). [CrossRef]
- Hishida, T. et al. In vivo partial cellular reprogramming enhances liver plasticity and regeneration. Cell Rep 39, 110730 (2022). [CrossRef]
- Kim, J. et al. Partial in vivo reprogramming enables injury-free intestinal regeneration via autonomous Ptgs1 induction. Sci Adv 9, eadi8454 (2023). [CrossRef]
- Doeser, M.C., Scholer, H.R. & Wu, G. Reduction of Fibrosis and Scar Formation by Partial Reprogramming In Vivo. Stem Cells 36, 1216-1225 (2018). [CrossRef]
- Cho, H.E. et al. In Vivo Reprogramming Using Yamanaka Factors in the CNS: A Scoping Review. Cells 13 (2024). [CrossRef]
- Gao, X., Wang, X., Xiong, W. & Chen, J. In vivo reprogramming reactive glia into iPSCs to produce new neurons in the cortex following traumatic brain injury. Sci Rep 6, 22490 (2016). [CrossRef]
- Dehghan, S. et al. Oct4 transcription factor in conjunction with valproic acid accelerates myelin repair in demyelinated optic chiasm in mice. Neuroscience 318, 178-189 (2016). [CrossRef]
- Su, Z., Niu, W., Liu, M.L., Zou, Y. & Zhang, C.L. In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat Commun 5, 3338 (2014). [CrossRef]
- Tai, W. et al. In vivo reprogramming of NG2 glia enables adult neurogenesis and functional recovery following spinal cord injury. Cell Stem Cell 28, 923-937 e924 (2021). [CrossRef]
- Seo, J.H. et al. In Situ Pluripotency Factor Expression Promotes Functional Recovery From Cerebral Ischemia. Mol Ther 24, 1538-1549 (2016). [CrossRef]
- Niu, W. et al. In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat Cell Biol 15, 1164-1175 (2013). [CrossRef]
- Chandrakanthan, V. et al. PDGF-AB and 5-Azacytidine induce conversion of somatic cells into tissue-regenerative multipotent stem cells. Proc Natl Acad Sci U S A 113, E2306-2315 (2016). [CrossRef]
- Kisby, T. et al. Adenoviral Mediated Delivery of OSKM Factors Induces Partial Reprogramming of Mouse Cardiac Cells In Vivo. Adv Ther-Germany 4 (2021). [CrossRef]
- Chen, Y. et al. Reversible reprogramming of cardiomyocytes to a fetal state drives heart regeneration in mice. Science 373, 1537-1540 (2021). [CrossRef]
- Gao, H. et al. Sall4 and Gata4 induce cardiac fibroblast transition towards a partially multipotent state with cardiogenic potential. Sci Rep 14, 24182 (2024). [CrossRef]
- Aguila, J.R. et al. SALL4 is a robust stimulator for the expansion of hematopoietic stem cells. Blood 118, 576-585 (2011). [CrossRef]
- Wu, J. et al. Improved Factor Combination for In Vivo Reprogramming of Cardiac Myofibroblast to Cardiomyocyte-Like Cell With Dual Recombinase Tracing. Circulation 148, 1728-1731 (2023). [CrossRef]
- Tao, Y. et al. Robust small molecule-aided cardiac reprogramming systems selective to cardiac fibroblasts. iScience 26, 108466 (2023). [CrossRef]
- Furtado, M.B. et al. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ Res 114, 1422-1434 (2014). [CrossRef]
- Tang, Y. et al. TBX20 Improves Contractility and Mitochondrial Function During Direct Human Cardiac Reprogramming. Circulation 146, 1518-1536 (2022). [CrossRef]
- Chong, J.J. et al. Adult cardiac-resident MSC-like stem cells with a proepicardial origin. Cell Stem Cell 9, 527-540 (2011). [CrossRef]
- Yu, J. et al. Topological Arrangement of Cardiac Fibroblasts Regulates Cellular Plasticity. Circ Res 123, 73-85 (2018). [CrossRef]
- Moretti, A. et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127, 1151-1165 (2006). [CrossRef]
- Mehanna, R.A. et al. Cardiac stem cells: Current knowledge and future prospects. World J Stem Cells 14, 1-40 (2022). [CrossRef]
- Nemajerova, A., Kim, S.Y., Petrenko, O. & Moll, U.M. Two-factor reprogramming of somatic cells to pluripotent stem cells reveals partial functional redundancy of Sox2 and Klf4. Cell Death Differ 19, 1268-1276 (2012). [CrossRef]
- Aires, A. et al. Neurovascular Coupling Impairment in Heart Failure with Reduction Ejection Fraction. Brain Sci 10 (2020). [CrossRef]
- Moore, M., Ryzhov, S., Sawyer, D.B., Gartner, C. & Vary, C.P.H. ALK1 Signaling in Human Cardiac Progenitor Cells Promotes a Pro-angiogenic Secretome. J Cell Signal 5, 122-142 (2024). [CrossRef]
- Khan, A. et al. Biomimetic Approaches in Cardiac Tissue Engineering: Replicating the Native Heart Microenvironment. Cureus 15, e43431 (2023). [CrossRef]
- Yuko, A.E. et al. LIN28a induced metabolic and redox regulation promotes cardiac cell survival in the heart after ischemic injury. Redox Biol 47, 102162 (2021). [CrossRef]
- Tachibana, A. et al. Paracrine Effects of the Pluripotent Stem Cell-Derived Cardiac Myocytes Salvage the Injured Myocardium. Circ Res 121, e22-e36 (2017). [CrossRef]
- Stefanovic, S. et al. Interplay of Oct4 with Sox2 and Sox17: A molecular switch from stem cell pluripotency to specifying a cardiac fate. J Cell Biol 186, 665-673 (2009). [CrossRef]
- Shu, J. et al. GATA family members as inducers for cellular reprogramming to pluripotency. Cell Res 25, 169-180 (2015). [CrossRef]
- Katano, W. et al. Sall1 and Sall4 cooperatively interact with Myocd and SRF to promote cardiomyocyte proliferation by regulating CDK and cyclin genes. Development 150 (2023). [CrossRef]
- Chen, L., Fulcoli, F.G., Tang, S. & Baldini, A. Tbx1 regulates proliferation and differentiation of multipotent heart progenitors. Circ Res 105, 842-851 (2009). [CrossRef]
- Lighthouse, J.K. & Small, E.M. Transcriptional control of cardiac fibroblast plasticity. J Mol Cell Cardiol 91, 52-60 (2016). [CrossRef]
- Raghunathan, S. et al. Conversion of human cardiac progenitor cells into cardiac pacemaker-like cells. J Mol Cell Cardiol 138, 12-22 (2020). [CrossRef]
- Liang, X. et al. HCN4 dynamically marks the first heart field and conduction system precursors. Circ Res 113, 399-407 (2013). [CrossRef]
- Alibhai, F.J. & Li, R.K. Rejuvenation of the Aging Heart: Molecular Determinants and Applications. Can J Cardiol 40, 1394-1411 (2024). [CrossRef]
- He, X. et al. Direct cellular reprogramming techniques for cardiovascular regenerative therapeutics. Can J Physiol Pharmacol 102, 1-13 (2024). [CrossRef]
- Kablak-Ziembicka, A. et al. Cardiac microRNAs: Diagnostic and therapeutic potential. Arch Med Sci 19, 1360-1381 (2023). [CrossRef]
- Abouelnazar, F.A. et al. The new advance of SALL4 in cancer: Function, regulation, and implication. J Clin Lab Anal 37, e24927 (2023). [CrossRef]
- Medlej, A., Mohammad Soltani, B., Javad Mowla, S., Hosseini, S. & Baharvand, H. A novel miRNA located in the GATA4 gene regulates the expression of IGF-1R and AKT1/2 genes and controls cell proliferation. J Cell Biochem 121, 3438-3450 (2020). [CrossRef]
- Liu, T. et al. Advanced Cardiac Patches for the Treatment of Myocardial Infarction. Circulation 149, 2002-2020 (2024). [CrossRef]
- Razavi, Z.S. et al. Advancements in tissue engineering for cardiovascular health: A biomedical engineering perspective. Front Bioeng Biotechnol 12, 1385124 (2024). [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).