Submitted:
25 October 2024
Posted:
28 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Sfriso, R.; Egert, M.; Gempeler, M.; Voegeli, R.; Campiche, R. Revealing the secret life of skin - with the microbiome you never walk alone. Int. J. Cosmet. Sci. 2020, 42(2), 116–126. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2020, 16(3), 143–155. [Google Scholar] [CrossRef] [PubMed]
- Townsend, E.C.; Kalan, L.R. The dynamic balance of the skin microbiome across the lifespan. Biochem. Soc. Trans. 2023, 51(1), 71–86. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Murphy, B.; Grimshaw, S.; Hoptroff, M.; Paterson, S.; Arnold, D.; Cawley, A.; Adams, S.E.; Falciani, F.; Dadd, T.; Eccles, R.; Mitchell, A.; Lathrop, W.F.; Marrero, D.; Yarova, G.; Villa, A.; Bajor, J.S.; Feng, L.; Mihalov, D.; Mayes, A.E. Alteration of barrier properties, stratum corneum ceramides and microbiome composition in response to lotion application on cosmetic dry skin. Sci. Rep. 2022, 12, 5223. [Google Scholar] [CrossRef] [PubMed]
- Ogonowska, P.; Szymczak. K.; Empel, J.; Urbaś, M.; Woźniak-Pawlikowska, A.; Barańska-Rybak, W.; Świetlik, D.; Nakonieczna, J. Staphylococcus aureus from Atopic Dermatitis Patients: Its Genetic Structure and Susceptibility to Phototreatment. Microbiol. Spectr. 2023, 11, e04598–22. [Google Scholar] [CrossRef]
- Wrześniewska, M.; Wołoszczak, J.; Świrkosz, G.; Szyller, H.; Gomułka, K. The Role of the Microbiota in the Pathogenesis and Treatment of Atopic Dermatitis-A Literature Review. Int. J. Mol. Sci. 2024, 25(12), 6539. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Korneev, A.; Peshkova, M.; Koteneva, P.; Gundogdu, A.; Timashev, P. Modulation of the skin and gut microbiome by psoriasis treatment: a comprehensive systematic review. Arch. Dermatol. Res. 2024, 316(7), 374. [Google Scholar] [CrossRef] [PubMed]
- Thio, H.B. The Microbiome in Psoriasis and Psoriatic Arthritis: The Skin Perspective. J. Rheumatol. Suppl. 2018, 94, 30–31. [Google Scholar] [CrossRef] [PubMed]
- Stettler, H.; Crowther, J.M.; Brandt, M.; Boxshall, A.; Lu, B.; de Salvo, R.; Laing, S.; Hennighausen, N.; Bielfeldt, S.; Blenkiron, P. Multi parametric biophysical assessment of treatment effects on xerotic skin. Skin Health Dis. 2021, 1(2), e21. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Qaizar, Z.; Sun, C.; Unbereit, K.; Stettler, H. Skin microbiome effects of long term dexpanthenol based gentle wash and emollient products formulated for dry skin. European Academy of Dermatology and Venereology conference, Berlin, Germany, 11-14 October 2023. [Google Scholar]
- Kligman, A.M. A.M. A brief history of how the dead stratum corneum became alive. In Skin Barrier; Elias, P.M., Feingold, K.R., Eds.; Informa Healthcare Inc.: New York, 2010; pp. 15–24. [Google Scholar]
- Pagac, M.P.; Stalder, M.; Campiche, R. Menopause and facial skin microbiomes: a pilot study revealing novel insights into their relationship. Front. Aging. 2024, 5, 1353082. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Crowther, J.M. Stratum corneum or stratum ecologica? Int. J. Cosmet. Sci. 2019, 41(2), 200–201. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Losada, M.; Crandall, K.A. Spatial diversity of the skin bacteriome. Front. Microbiol. 2023, 14, 1257276. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Brandwein, M.; Fuks, G.; Israel, A.; Hodak, E.; Sabbah, F.; Steinberg, D.; Bentwich, Z.; Shental, N.; Meshner, S. Biogeographical Landscape of the Human Face Skin Microbiome Viewed in High Definition. Acta Derm. Venereol. 2021, 101(11), adv00603. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- 16. Kim, J.; Park, T.; Kim, H.J.; An, S.; Sul, W.J. Inferences in microbial structural signatures of acne microbiome and mycobiome. J. Microbiol. 2021, 59(4), 369–375. [Google Scholar] [CrossRef] [PubMed]
- McCall, L.I.; Callewaert, C.; Zhu, Q.; Song, S.J.; Bouslimani, A.; Minich, J.J.; Ernst, M.; Ruiz-Calderon, J.F.; Cavallin, H.; Pereira, H.S.; Novoselac, A.; Hernandez, J.; Rios, R.; Branch, O.H.; Blaser, M.J.; Paulino, L.C.; Dorrestein, P.C.; Knight, R.; Dominguez-Bello, M.G. Home chemical and microbial transitions across urbanization. Nat. Microbiol. 2020, 5(1), 108–115. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gu, X.; Li, Z.; Su, J. Air pollution and skin diseases: A comprehensive evaluation of the associated mechanism. Ecotoxicol. Environ. Saf. 2024, 278, 116429. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.Q.; Li, X.; Zhang, R.Y.; Yuan, C.; Yan, B.; Humbert, P.; Quan, Z.X. Effects of Investigational Moisturizers on the Skin Barrier and Microbiome following Exposure to Environmental Aggressors: A Randomized Clinical Trial and Ex Vivo Analysis. J. Clin. Med. 2023, 12(18), 6078. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Palaniraj, A.; Jayaraman, V. Production, recovery and applications of xanthan gum by Xanthomonas campestris. J. Food Eng. 2011, 106(1), 1–12. [Google Scholar] [CrossRef]
- Dai, X.; Gao, G.; Wu, M.; Wei, W.; Qu, J.; Li, G.; Ma, T. Construction and application of a Xanthomonas campestris CGMCC15155 strain that produces white xanthan gum. Microbiologyopen 2019, 8, e631. [Google Scholar] [CrossRef]
- De Toni, C.H.; Richter, M.F.; Chagas, J.R.; Henriques, J.A.; Termignoni, C. Purification and characterization of an alkaline serine endopeptidase from a feather-degrading Xanthomonas maltophilia strain. Can. J. Microbiol. 2002, 48(4), 342–348. [Google Scholar] [CrossRef] [PubMed]
- Han, J.H.; Kim, H.S. Skin Deep: The Potential of Microbiome Cosmetics. J. Microbiol. 2024, 62(3), 181–199. [Google Scholar] [CrossRef] [PubMed]
- Woolery-Lloyd, H.; Andriessen, A.; Day, D.; Gonzalez, N.; Green, L.; Grice, E.; Henry, M. Review of the microbiome in skin aging and the effect of a topical prebiotic containing thermal spring water. J. Cosmet. Dermatol. 2023, 22(1), 96–102. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Mao, J.; Diaz, I.; Kopylova, E.; Melnik, A.V.; Aksenov, A.A.; Tipton, C.D.; Soliman, N.; Morgan, A.M.; Boyd, T. Multi-omic approach to decipher the impact of skincare products with pre/postbiotics on skin microbiome and metabolome. Front. Med. (Lausanne) 2023, 10, 1165980. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- De Almeida, C.V.; Antiga, E.; Lulli, M. Oral and Topical Probiotics and Postbiotics in Skincare and Dermatological Therapy: A Concise Review. Microorganisms 2023, 11(6), 1420. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Staudinger, T.; Pipal, A.; Redl, B. Molecular analysis of the prevalent microbiota of human male and female forehead skin compared to forearm skin and the influence of make-up. J. Appl. Microbiol. 2011, 110(6), 1381–1389. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, E.; Åkerström, U.; Berger-Picard, N.; Lataste, S.; Gillbro, J.M. Randomized comparative double-blind study assessing the difference between topically applied microbiome supporting skincare versus conventional skincare on the facial microbiome in correlation to biophysical skin parameters. Int. J. Cosmet. Sci. 2023, 45(1), 83–94. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Jeong, S.E.; Lee, S.; Kim, S.; Han, H.; Jeon, C.O. Effects of cosmetics on the skin microbiome of facial cheeks with different hydration levels. Microbiologyopen 2018, 7(2), e00557. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cui, S.; Pan, M.; Tang, X.; Liu, G.; Mao, B.; Zhao, J.; Yang, K. Metagenomic insights into the effects of cosmetics containing complex polysaccharides on the composition of skin microbiota in females. Front. Cell. Infect. Microbiol. 2023, 13, 1210724. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Iglesia, S.; Kononov, T.; Zahr, A.S. A multi-functional anti-aging moisturizer maintains a diverse and balanced facial skin microbiome. J. Appl. Microbiol. 2022, 133(3), 1791–1799. [Google Scholar] [CrossRef] [PubMed]
- Hwang, B.K.; Lee, S.; Myoung, J.; Hwang, S.J.; Lim, J.M.; Jeong, E.T.; Park, S.G.; Youn, S.H. Effect of the skincare product on facial skin microbial structure and biophysical parameters: A pilot study. Microbiologyopen 2021, 10(5), e1236. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Callejon, S.; Giraud, F.; Larue, F.; Buisson, A.; Mateos, L.; Grare, L.; Guyoux, A.; Perrier, E.; Ardiet, N.; Trompezinski, S. Impact of Leave-on Skin Care Products on the Preservation of Skin Microbiome: An Exploration of Ecobiological Approach. Clin. Cosmet. Investig. Dermatol. 2023, 16, 2727–2735. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, H.; Yu, F.; Wang, C.; Han, Z.; Liu, S.; Chen, D.; Liu, D.; Meng, X.; He, X.; Huang, Z. The impacts of sodium lauroyl sarcosinate in facial cleanser on facial skin microbiome and lipidome. J. Cosmet. Dermatol. 2024, 23(4), 1351–1359. [Google Scholar] [CrossRef] [PubMed]
- van Belkum, A.; Lisotto, P.; Pirovano, W.; Mongiat, S.; Zorgani, A.; Gempeler, M.; Bongoni, R.; Klaassens, E. Being friendly to the skin microbiome: Experimental assessment. Front. Microbiomes 2023, 1, 1077151. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.L.; Park, M. NISC Comparative Sequencing Program; Kong HH, Segre JA. Temporal Stability of the Human Skin Microbiome. Cell 2016, 165(4), 854–866. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Park, J.; Schwardt, N.H.; Jo, J.H.; Zhang, Z.; Pillai, V.; Phang, S.; Brady, S.M.; Portillo, J.A.; MacGibeny, M.A.; Liang, H.; Pensler, M.; Soldin, S.J.; Yanovski, J.A.; Segre, J.A.; Kong, H.H. Shifts in the Skin Bacterial and Fungal Communities of Healthy Children Transitioning through Puberty. J. Invest. Dermatol. 2022, 142(1), 212–219. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Leem, S.; Keum, H.L.; Song, H.J.; Gu, K.N.; Kim, Y.; Seo, J.Y.; Shin, J.G.; Lee, S.G.; Lee, S.M.; Sul, W.J.; Kang, N.G. Skin aging-related microbial types separated by Cutibacterium and α-diversity. J. Cosmet. Dermatol. 2024, 23(3), 1066–1074. [Google Scholar] [CrossRef] [PubMed]
- Radocchia, G.; Brunetti, F.; Marazzato, M.; Totino, V.; Neroni, B.; Bonfiglio, G.; Conte, A.L.; Pantanella, F.; Ciolli, P.; Schippa, S. Women Skin Microbiota Modifications during Pregnancy. Microorganisms 2024, 12(4), 808. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Somerville, D.A. The normal flora of the skin in different age groups. Br. J. Dermatol. 1969, 81(4), 248–258. [Google Scholar] [CrossRef] [PubMed]
- Luna, P.C. Skin Microbiome as Years Go By. Am. J. Clin. Dermatol. 2020, 21 (Suppl 1), 12–17. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Myers, T.; Bouslimani, A.; Huang, S.; Hansen, S.T.; Clavaud, C.; Azouaoui, A.; Ott, A.; Gueniche, A.; Bouez, C.; Zheng, Q.; Aguilar, L.; Knight, R.; Moreau, M.; Song, S.J. A multi-study analysis enables identification of potential microbial features associated with skin aging signs. Front. Aging. 2024, 4, 1304705. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ying, S.; Zeng, D.N.; Chi, L.; Tan, Y.; Galzote, C.; Cardona, C.; Lax, S.; Gilbert, J.; Quan, Z.X. The Influence of Age and Gender on Skin-Associated Microbial Communities in Urban and Rural Human Populations. PLoS One 2015, 10(10), e0141842. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shibagaki, N.; Suda, W.; Clavaud, C.; Bastien, P.; Takayasu, L.; Iioka, E.; Kurokawa, R.; Yamashita, N.; Hattori, Y.; Shindo, C.; Breton, L.; Hattori, M. Aging-related changes in the diversity of women's skin microbiomes associated with oral bacteria. Sci. Rep. 2017, 7(1), 10567. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Murphy, B.; Hoptroff, M.; Arnold, D.; Cawley, A.; Smith, E.; Adams, S.E.; Mitchell, A.; Horsburgh, M.J.; Hunt, J.; Dasgupta, B.; Ghatlia, N.; Samaras, S.; MacGuire-Flanagan, A.; Sharma, K. Compositional Variations between Adult and Infant Skin Microbiome: An Update. Microorganisms 2023, 11(6), 1484. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Leyden, J.J.; McGinley, K.J.; Mills, O.H.; Kligman, A.M. Age-related changes in the resident bacterial flora of the human face. J. Invest. Dermatol. 1975, 65(4), 379–381. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Son, S.M.; Park, H.; Kim, B.K.; Choi, I.S.; Kim, H.; Huh, C.S. Taxonomic profiling of skin microbiome and correlation with clinical skin parameters in healthy Koreans. Sci. Rep. 2021, 11(1), 16269. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Luebberding, S.; Krueger, N.; Kerscher, M. Skin physiology in men and women: in vivo evaluation of 300 people including TEWL, SC hydration, sebum content and skin surface pH. Int. J. Cosmet. Sci. 2013, 35(5), 477–483. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Tang, M.; Dong, K.; Zhou, J.; Wang, D.; Song, L. Changes in the skin microbiome during male maturation from 0 to 25 years of age. Skin Res. Technol. 2023, 29(9), e13432. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Russo, E.; Di Gloria, L.; Cerboneschi, M.; Smeazzetto, S.; Baruzzi, G.P.; Romano, F.; Ramazzotti, M.; Amedei, A. Facial Skin Microbiome: Aging-Related Changes and Exploratory Functional Associations with Host Genetic Factors, a Pilot Study. Biomedicines 2023, 11(3), 684. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ottinger, S.; Robertson, C.M.; Branthoover, H.; Patras, K.A. The human vaginal microbiota: from clinical medicine to models to mechanisms. Curr. Opin. Microbiol. 2024, 77, 102422. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489(7415), 220–230. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chaudhary, P.P.; Kaur, M.; Myles, I.A. Does "all disease begin in the gut"? The gut-organ cross talk in the microbiome. Appl. Microbiol. Biotechnol. 2024, 108(1), 339. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhou, Z.; Feng, Y.; Xie, L.; Ma, S.; Cai, Z.; Ma, Y. Alterations in gut and genital microbiota associated with gynecological diseases: a systematic review and meta-analysis. Reprod. Biol. Endocrinol. 2024, 22(1), 13. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wallen-Russell, C.; Wallen-Russell, S. ; Meta Analysis of Skin Microbiome: New Link between Skin Microbiota Diversity and Skin Health with Proposal to Use This as a Future Mechanism to Determine Whether Cosmetic Products Damage the Skin. Cosmetics 2017, 4(2), 14. [Google Scholar] [CrossRef]
- Carvalho, M.J.; S Oliveira, A.L.; Santos Pedrosa, S.; Pintado, M.; Pinto-Ribeiro, I.; Madureira, A.R. Skin Microbiota and the Cosmetic Industry. Microb. Ecol. 2023, 86(1), 86–96. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Thompson, A.; Jaros, J.; Blackcloud, P.; Hsiao, J.; Shi, V.Y. Updated understanding of Staphylococcus aureus in atopic dermatitis: From virulence factors to commensals and clonal complexes. Exp. Dermatol. 2021, 30(10), 1532–1545. [Google Scholar] [CrossRef] [PubMed]
- Blicharz, L.; Rudnicka, L.; Samochocki, Z. Staphylococcus aureus: an underestimated factor in the pathogenesis of atopic dermatitis? Postepy. Dermatol. Alergol. 2019, 36(1), 11–17. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tauber, M.; Balica, S.; Hsu, C.Y.; Jean-Decoster, C.; Lauze, C.; Redoules, D.; Viodé, C.; Schmitt, A.M.; Serre, G.; Simon, M.; Paul, C.F. Staphylococcus aureus density on lesional and nonlesional skin is strongly associated with disease severity in atopic dermatitis. J. Allergy. Clin. Immunol. 2016, 137, 1272–1274.e3. [Google Scholar] [CrossRef] [PubMed]
- Cleary, J.M.; Lipsky, Z.W.; Kim, M.; Marques, C.N.H.; German, G.K. Heterogeneous ceramide distributions alter spatially resolved growth of Staphylococcus aureus on human stratum corneum. J. R. Soc. Interface. 2018, 15(141), 20170848. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Imokawa, G.; Abe, A.; Jin, K.; Higaki, Y.; Kawashima, M.; Hidano, A. Decreased level of ceramides in stratum corneum of atopic dermatitis: an etiologic factor in atopic dry skin? J. Investig. Dermatol. 1991, 96(4), 523–526. [Google Scholar] [CrossRef]
- Watanabe, M.; Tagami, H.; Horii, I.; Takahashi, M.; Kligman, A.M. Functional analyses of the superficial stratum corneum in atopic xerosis. Arch. Dermatol. 1991, 127(11), 1689–1692. [Google Scholar] [CrossRef]
- Deng, L.; Costa, F.; Blake, K.J.; Choi, S.; Chandrabalan, A.; Yousuf, M.S.; Shiers, S.; Dubreuil, D.; Vega-Mendoza, D.; Rolland, C.; et al. S. aureus drives itch and scratch-induced skin damage through a V8 protease-PAR1 axis. Cell 2023, 186, 5375–5393.e25. [Google Scholar] [CrossRef] [PubMed]
- Rahrovan, S.; Fanian, F.; Mehryan, P.; Humbert, P.; Firooz, A. Male versus female skin: What dermatologists and cosmeticians should know. Int. J. Womens Dermatol. 2018, 4(3), 122–130. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jemec, G.B.; Serup, J. Scaling, dry skin and gender. A bioengineering study of dry skin. Acta Derm. Venereol. Suppl. (Stockh.) 1992, 177, 26–28. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Su, Y.; Zheng, B.; Wen, S.; Liu, D.; Ye, L.; Yan, Y.; Elias, P.M.; Yang, B.; Man, M.Q. Gender-related characterization of sensitive skin in normal young Chinese. J. Cosmet. Dermatol. 2020, 19(5), 1137–1142. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Farage, M.A. The Prevalence of Sensitive Skin. Front. Med. 2019, 6, 98. [Google Scholar] [CrossRef]
- Callewaert, C.; Ravard Helffer, K.; Lebaron, P. Skin Microbiome and its Interplay with the Environment. Am. J. Clin. Dermatol. 2020, 21 (Suppl 1), 4–11. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Skowron, K.; Bauza-Kaszewska, J.; Kraszewska, Z.; Wiktorczyk-Kapischke, N.; Grudlewska-Buda, K.; Kwiecińska-Piróg, J.; Wałecka-Zacharska, E.; Radtke, L.; Gospodarek-Komkowska, E. Human Skin Microbiome: Impact of Intrinsic and Extrinsic Factors on Skin Microbiota. Microorganisms 2021, 9(3), 543. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nguyen, U.T.; Kalan, L.R. Forgotten fungi: the importance of the skin mycobiome. Curr. Opin. Microbiol. 2022, 70, 102235. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jo, J.H.; Kennedy, E.A.; Kong, H.H. Topographical and physiological differences of the skin mycobiome in health and disease. Virulence 2017, 8(3), 324–333. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Meray, Y.; Gençalp, D.; Güran, M. Putting It All Together to Understand the Role of Malassezia spp. in Dandruff Etiology. Mycopathologia 2018, 183(6), 893–903. [Google Scholar] [CrossRef] [PubMed]
- Leyden, J.J.; Kligman, A.M. Interdigital athlete's foot: new concepts in pathogenesis. Postgrad. Med. 1977, 61(6), 113–116. [Google Scholar] [CrossRef] [PubMed]
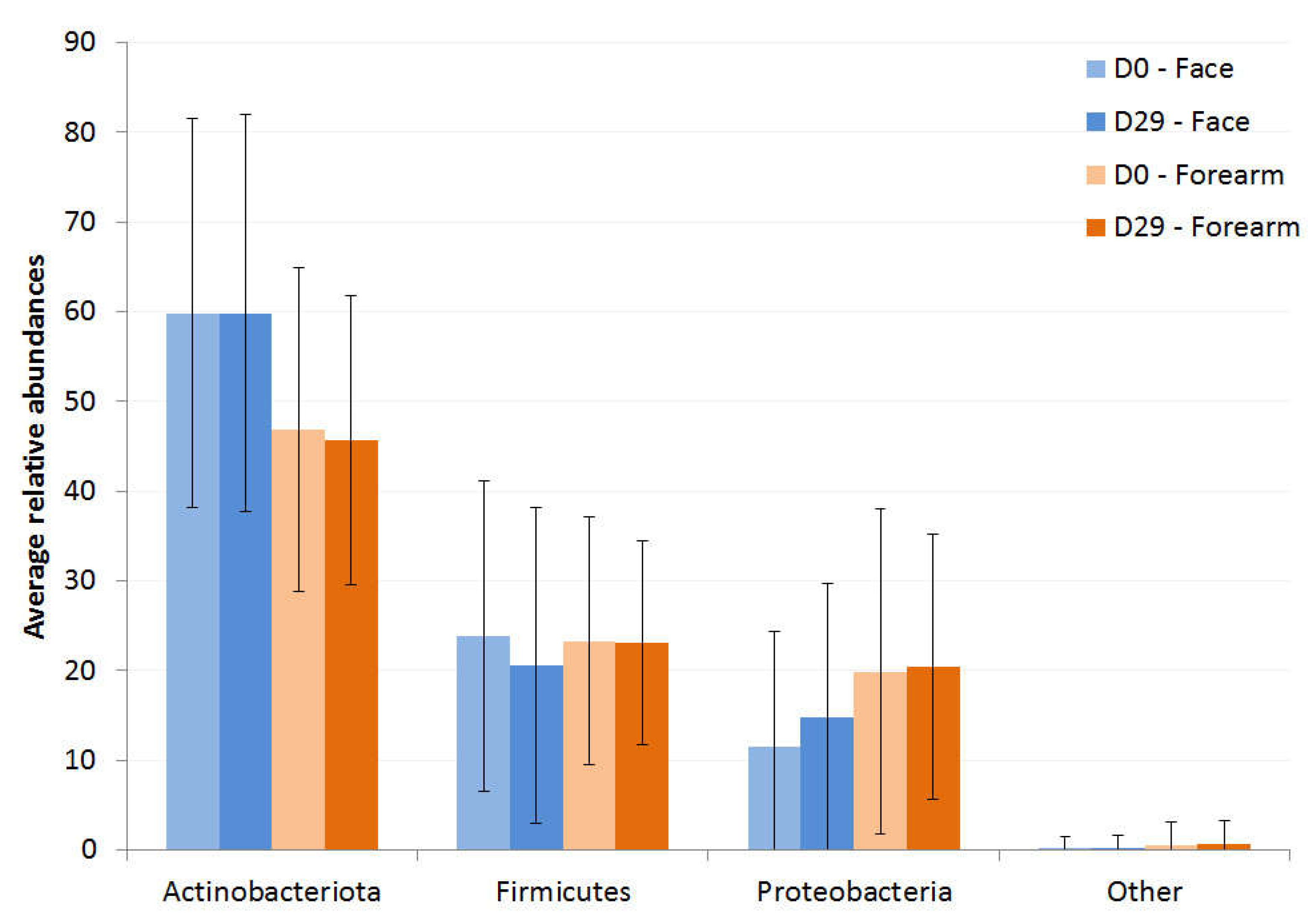
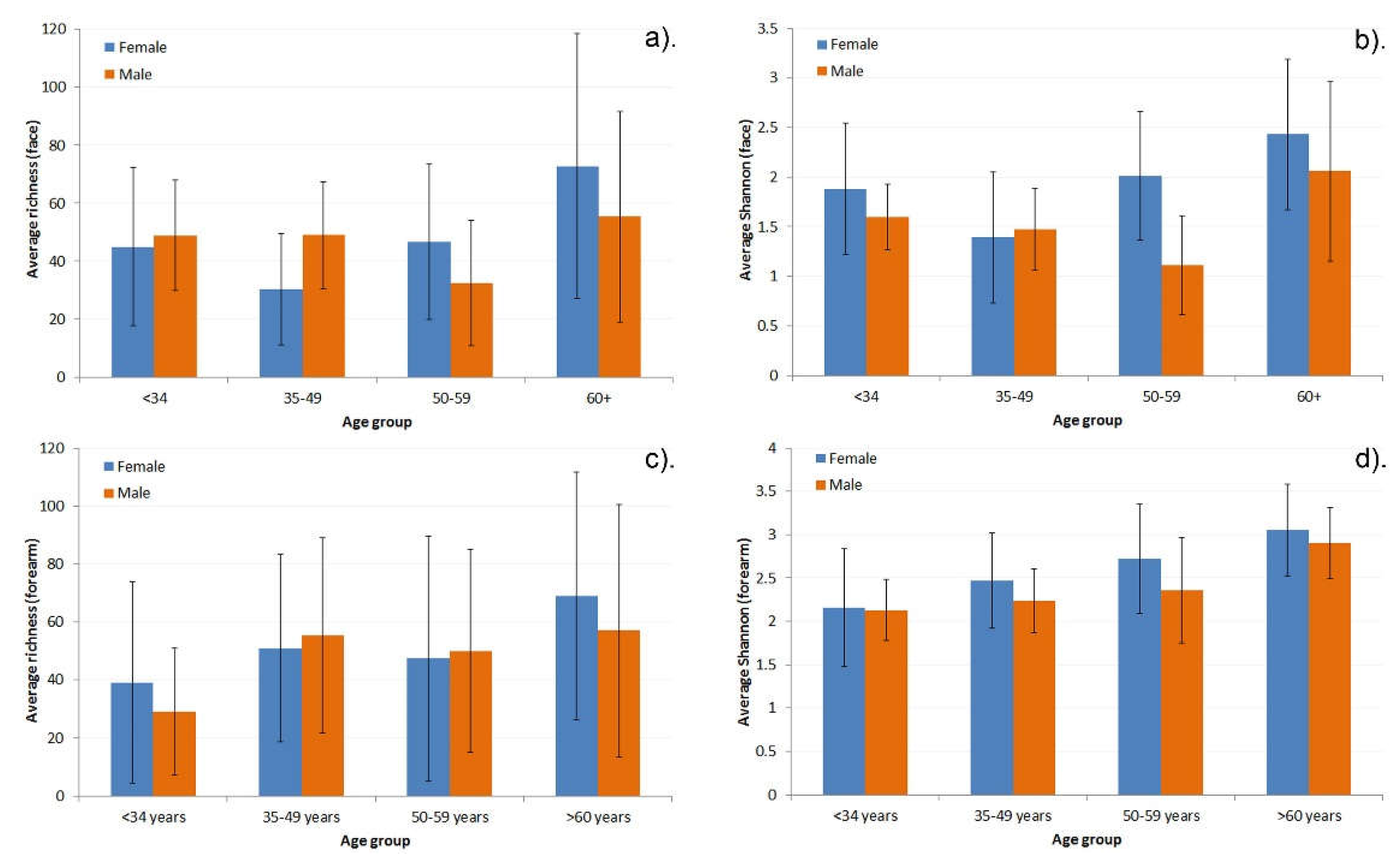
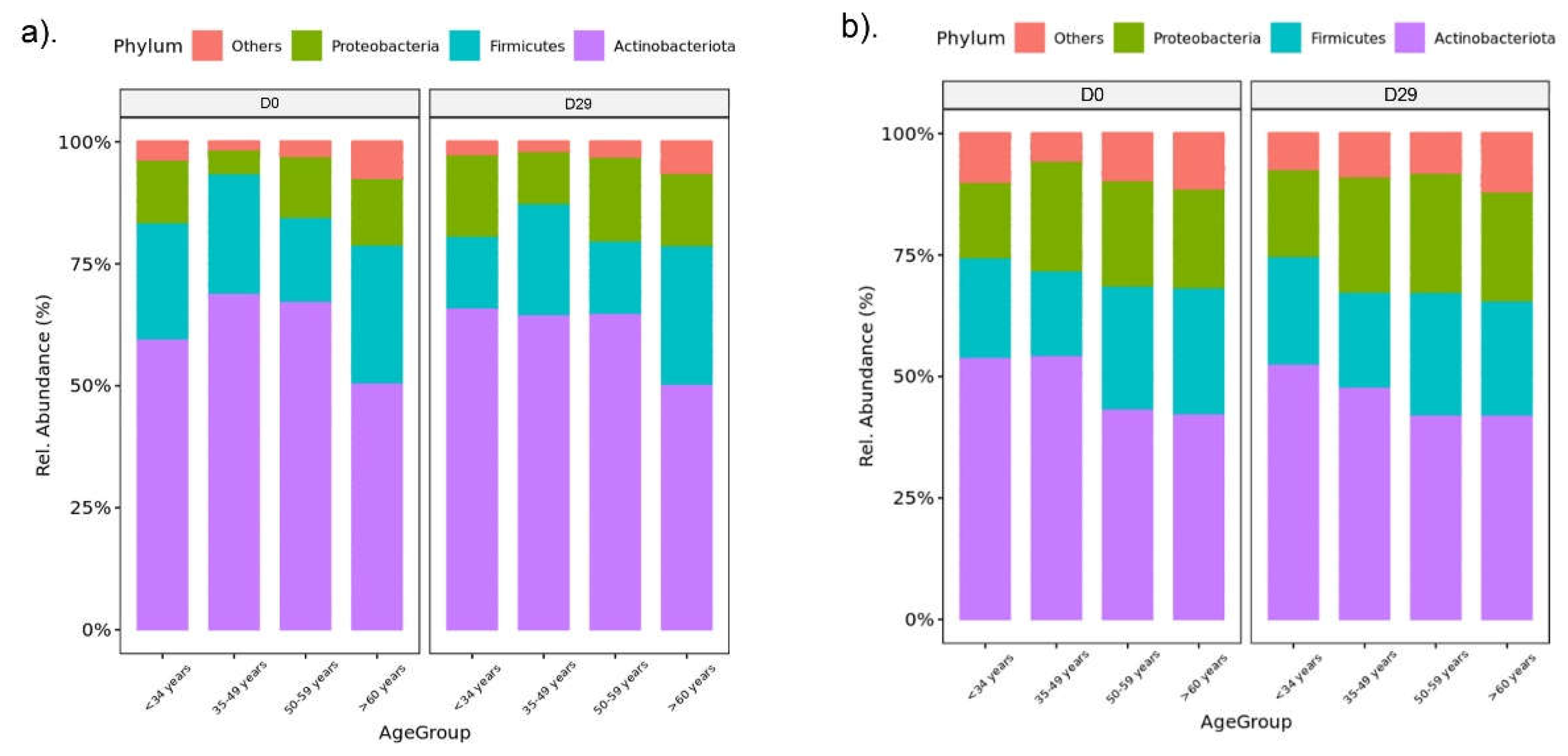
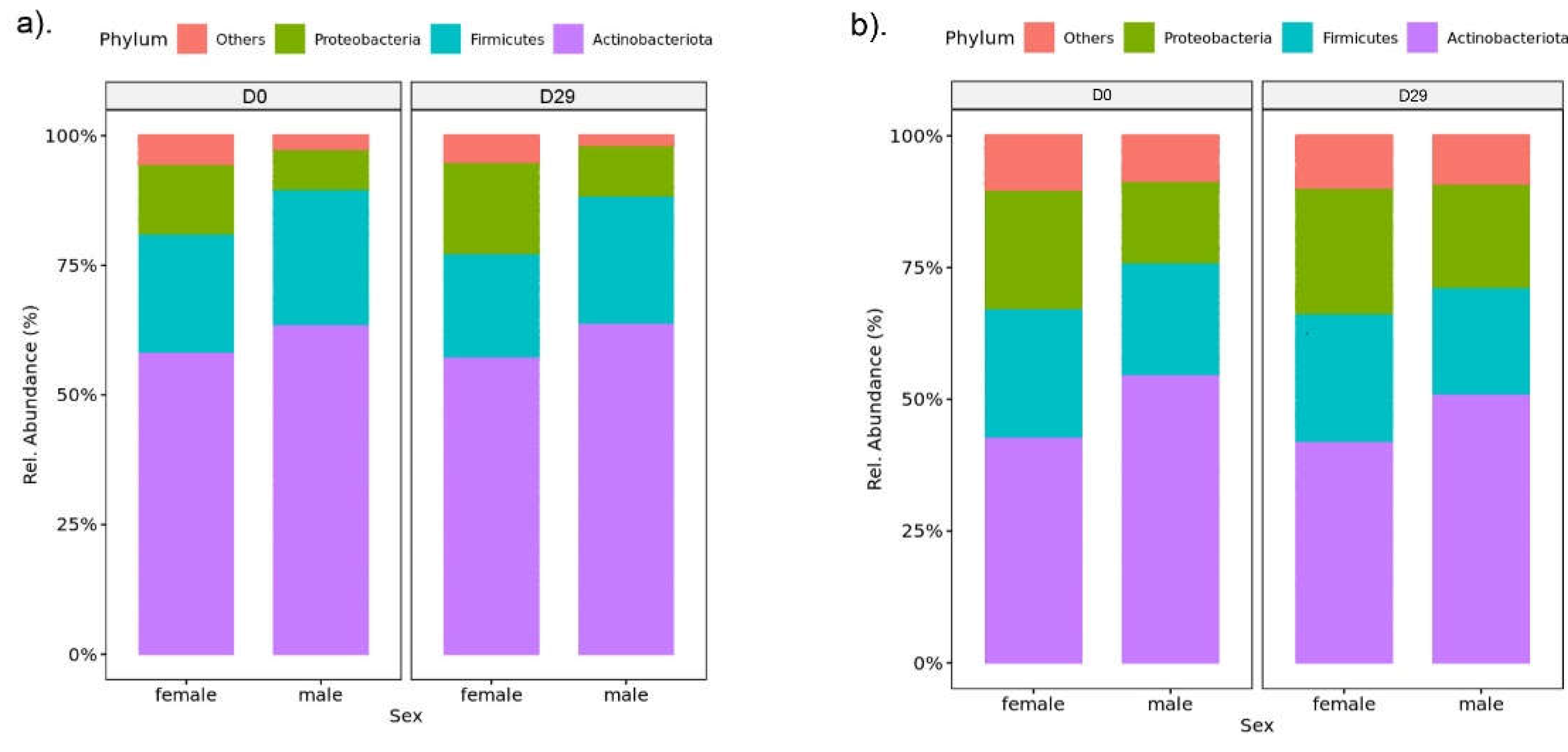
| Variable | Male | Female | ||||
| N | % | Mean (SD) | N | % | Mean (SD) | |
| Age (range 22-65) | 52.1 (12.4) | 51.3 (13.1) | ||||
| Fitzpatrick skin type I II III IV V |
0 15 10 2 1 |
0 53.6 35.7 7.1 3.6 |
8 34 19 1 1 |
12.6 54.0 30.2 1.6 1.6 |
||
| Ethnicity White Black Asian |
23 3 2 |
82.1 10.7 7.2 |
61 2 0 |
96.8 3.2 0 |
| Product Code | Product Name | |
| DFCD | Dexpanthenol repair complex (DRC) Face cream day | |
| DFCN | DRC Face cream night | |
| DFC-SPF | DRC Face cream (SPF 25) | |
| DFW | Dexpanthenol Face wash | |
| DBL | DRC Body lotion (low lipid level) | |
| DBH | DRC Body balm (high lipid level) | |
| DHC | DRC Hand cream | |
| DBW | Dexpanthenol Body wash | |
| Product | Body site | D0 (Shannon Index) | D29 (Shannon Index) | Difference | |
| DFCD | Face | 1.297 (1.120, 1.598) | 1.263 (1.083, 1.559) | -0.034 (NS) | |
| DFCN | Face | 2.352 (1.525, 2.224) | 2.149 (1.529, 2.135) | -0.203 (NS) | |
| DFC-SPF | Face | 1.601 (1.293, 1.792) | 1.658 (1.326, 1,872) | +0.057 (NS) | |
| DFW | Face | 1.831 (1.476, 2.027) | 1.741 (1.287, 1.902) | -0.090 (NS) | |
| DBL | Forearm | 2.808 (2.250, 2.637) | 2.900 (2.439, 2.864) | +0.092 (NS) | |
| DBH | Forearm | 2.738 (2.442, 2,714) | 3.130 (2.853, 2.986) | +0.392 (NS) | |
| DHC | Forearm | 2.444 (2.194, 2.551) | 2.940 (2.449, 2.846) | +0.496 (NS) | |
| DBW | Forearm | 2.735 (2.544, 2.859) | 2.894 (2.495, 2.784) | +0.159 (NS) | |
| Product | Body site | Taxonomic changes (D29 vs D0) | Category | p value |
| DFCD | Face | NSD | - | - |
| DFCN | Face | NSD | - | - |
| DFC-SPF | Face | NSD | - | - |
| WPF | Face |
Corynebacterium tuberculostearicum ↓ Frankia nodulisporulans ↑ |
Species Species |
0.003 0.002 |
| DBL | Forearm | ASV_0009 – Xanthomonas campestris ↑ Xanthomonas campestris ↑ Veillonella ↑ Xanthomonas ↑ Veillonellaceae ↑ Xanthomonadaceae ↑ Firmicutes_C ↑ |
ASV Species Genus Genus Family Family Phylum |
<0.001 <0.001 <0.001 <0.001 <0.001 <0.001 <0.001 |
| DBH | Forearm | Haemophilus_D parainfluenzae ↑ | Species | 0.001 |
| DHC | Forearm | Epilithonimonas ↑ Rothia ↑ Xanthomonas ↑ |
Genus Genus Genus |
0.010 0.007 0.008 |
| WPB | Forearm | NSD | - | - |
| Species | Body site | Sex | Average Count (SD) | Lower CI | Upper CI | N |
| S. epidermidis | Face | Female | 4250.56 (5083.19) | 3352.22 | 5148.90 | 62 |
| Male | 7150.76 (9486.93) | 4643.50 | 9658.03 | 28 | ||
| Forearm | Female | 1607.17 (2122.55) | 1232.06 | 1982.28 | 63 | |
| Male | 2441.20 (2658.55) | 1744.85 | 3137.54 | 28 | ||
| S. aureus | Face | Female | 87.57 (317.14) | 14.31 | 160.83 | 36 |
| Male | 9.38 (38.85) | -6.81 | 25.57 | 11 | ||
| Forearm | Female | 91.43 (370.03) | 4.12 | 178.75 | 36 | |
| Male | 0.00 (0.00) | 0.00 | 0.00 | 11 | ||
| C. acnes | Face | Female | 21448.19 (18560.41) | 18168.06 | 24728.32 | 62 |
| Male | 31771.31 (19974.18) | 26492.41 | 37050.21 | 28 | ||
| Forearm | Female | 6461.51 (9054.49) | 4861.34 | 8061.69 | 63 | |
| Male | 8494.68 (9603.19) | 5979.45 | 11009.91 | 28 | ||
| Other | Face | Female | 32.28 (391.06) | 29.51 | 35.06 | 62 |
| Male | 36.86 (535.64) | 31.08 | 42.63 | 28 | ||
| Forearm | Female | 39.48 (440.08) | 36.34 | 42.62 | 63 | |
| Male | 46.26 (480.83) | 41.13 | 51.39 | 28 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





