1. Introduction
Hepatitis B virus (HBV) infection remains a huge health problem despite the introduction of the HBV vaccine in the late 1990s. From the discovery of the disease until the beginning of the 21st century, approximately two billion cases of the disease have been reported worldwide, with more than 400 million of these cohorts developing into chronic carriers of HBV [1]. Chronic HBV infection is considered an important cause of many liver-related medical complications, such as hepatocellular carcinoma, liver cirrhosis, and liver failure [2,3,4]. Due to the virus complexity, the genome of HBV contains an overlapping region that encodes four different genes (S, C, P and X). The S gene encodes exclusively the HBV surface envelope protein in its long open reading frame [6]. The presence of start codons allows the gene to be assembled into small, medium and large polypeptides, which are typically used either as a single target gene or in combination with other HBV genes to detect HBV DNA [5]. HBV core antigen is traditionally encoded by the C gene, whereas the X gene encodes protein X [7]. The P gene encodes a polymerase protein, an integral component of the reverse transcription process during HBV replication. The HBV replication cycle lacks proofreading properties and therefore typically results in progeny with high genomic variability [5].
Due to significant genetic diversity, HBV is classified into ten genotypes (A-J) with intergroup variants accounting for 7.5% [8]. In addition to E and G, all genotypes are divided into 25 subgenotypes with 4% amino acid variability [9,10]. HBV genotypes are distributed differently depending on geographic location: HBV-B, HBV-C and HBV-E are most common in Oceania and East Asia, while HBV-E is most common in Central and West Africa. HBV-F and HBV H are found only in Alaska and Latin America. In contrast, HBV-D is a global pandemic. In Australia, Europe, Indonesia, North Africa and Western Asia, HBV-D1 is the most common virus, while HBV-D2 is found in Albania, Japan, Malaysia, North-Eastern Europe, Russia and the UK [11–15].
The progression and natural history of the disease varies among different HBV genotypes. These factors can make the treatment of HBV infection very difficult, as the effectiveness of known therapeutic drugs becomes ineffective against certain genotypes and new genotypic variants [8,9]. Thus, there is a great need worldwide for constant update in genotypic information and studies of people infected with HBV [15].
2. Materials and Methods
Samples, to test for HBV markers using СLIA/ELISA and PCR methods, we used 884 blood serum/plasma samples obtained from patients and blood donors with acute and chronic hepatitis B from different regions of the country. The age of the patients ranged from 4 years to 78 years. 404 samples were obtained from males, including 250 females. For 230 cases, gender and age were unknown as haven’t been reported. Serum/plasma samples were collected and analyzed during 2014-2023.
HBsAg was detected using a commercial test system CLIA Architect HBsAg Qualitative II Reagent Kit, Abbott, USA. Confirmation of a positive result was carried out using the Architect HBsAg Qualitative II Confirmatory Reagent Kit, Abbott, USA. ELISA “Vectogep B-HBs antigen”, produced by “Vector-Best”, Novosibirsk, Russia.
The polymerase chain reaction to determine the DNA of the hepatitis B virus was carried out using test systems, produced by Vector-Best CJSC: “RealBest HBV DNA (quantitative”, “RealBest HIV RNA (quantitative”) in accordance with the manufacturer’s instructions.
The synthesis of specific primer pairs for the P and S gene regions of the HBV genome was carried out at ODO "Primetech", Minsk. For sequence of the HBV genome in conservative regions, subsequently for P and S regions, the specific pairs of primers were synthesized, which made it possible to carry out HBV genotyping in full [16].
Isolation of viral RNA/DNA from blood serum/plasma samples was performed using the “Kit of reagents for the isolation of NK” (manufactured by “Vector-Best”, Russia) and the “Kit of reagents for the isolation of RNA/DNA from clinical material” “RIBO- sorb", "RNA-prep", Russia. All manipulations were carried out according to the manufacturers’ instructions.
PCR was performed in a nested version using previously described primer pairs p1/pR5 (position 1197-1178) and p4/pR2 (1017-997) [16] according to the following protocol in a volume of 25µl: 2,5 µl 10х buffer+ MgCl2; 0,25 µl 25mM dNTPs; 0,5 µl 10 µM p1; 0,5 µl 10 µM pR5; 0,8 10 µM Taq polymerase; 18,45 µl bdH20. The first round of PCR was carried out according to the following protocol: one denaturation cycle at 95oC 2 minutes and 30 cycles of amplification were performed with denaturation at 95°C for 30 sec, annealing at 53°C for 30 sec, and extension at 68°C for 1 minutes, then one cycle of 68oC for 5 minutes and storage at 10oC. In the second round of PCR, the annealing temperature was 50°C, the rest of the parameters were not changed.
Amplified DNA fragments were analyzed on a 2% agarose gel. Electrophoresis was carried out at 10 w/cm of gel in TRIS-acetate buffer, pH 8.0. DNA was visualized using the Vitran Photo gel documentation system (Biocom Company LLC, Russia). The fragment size was determined according to the molecular weight of marker of 100-1000 bp (Fermentas, Lithuania).
The resulting DNA fragments were purified using the NimaGen ExS-Pure kit, Holland.
Sequencing PCR was carried out using the second round primer pair p4/pR2 in a volume of 10 µl according to the following protocol: 5x seqbuf - 2 µl; BigDye Terminator v. 3.1 (applied biosystems) - 1 µl; p4/pR4 - 2 µl; bdH20 - 3 bdH20; DNA - 2 µl.
Electrophoresis of HBV DNA fragments obtained and purified after sequencing PCR was performed on AB 3500 genetic analyzer, USA.
Phylogenetic analysis of the obtained HBV DNA fragments was carried out using the computer programs: Sequencing Analysis v.6, BioEdit, SeqScape v3, MEGAX and Genious 8.1. Phylogenetic trees were constructed using the ML (maximum likelihood) algorithm in the PHYML program. The SH-aLRT test was used to calculate the statistical significance of the clusters. Clusters with a support node ≥0.9 were considered to be reliable.
3. Results
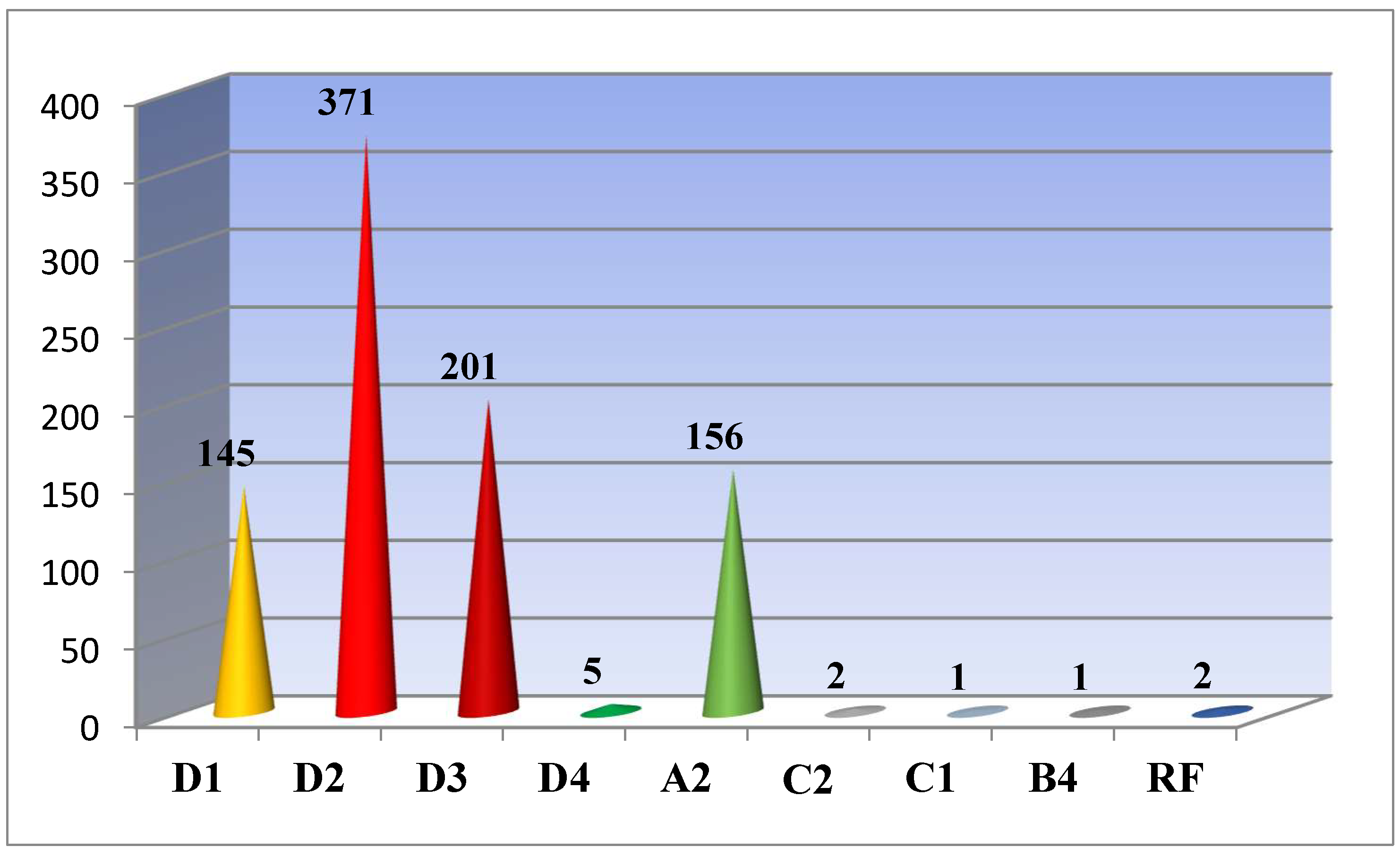
As studies have shown, out of all 884 sequenced and analyzed samples, 722 (81.7%) of them have genotype D; 156 (17.7%) - genotype A; 3 (0.3%) - genotype C and the only 1 (0.1%) – genotype B. For two cases (0.2%) a recombinant form of the hepatitis B virus was detected. Genotype D was found representing by subgenotypes D1 (145/20.1%), D2 (371/51.4%), D3 (201/27, 8%) and D4 (5/0.7%); A – A2; C – C1 and C2; B – B4; recombinant forms C2/ D and A2/ С2 were detected (
Figure 1).
In 12 cases, a virus with mutations of resistance to reverse transcriptase inhibitors Lamivudine, Zeffix® and Telbivudine, Tyzeka®, Sebivo® and partial resistance to Entecavir, Baraclude® was identified. Moreover, in 4 cases, resistance of mutations was identified in subgenotypes A2 and D2, in 3 cases- in the genome of subgenotype D1, and in one case - in subgenotype D3. Mutations were most often recorded at positions 180M and 204V - in 8 cases, 204I - in three cases, 80I and 173L - 2 times each, and 80V and 181T in one case (
Table 1).
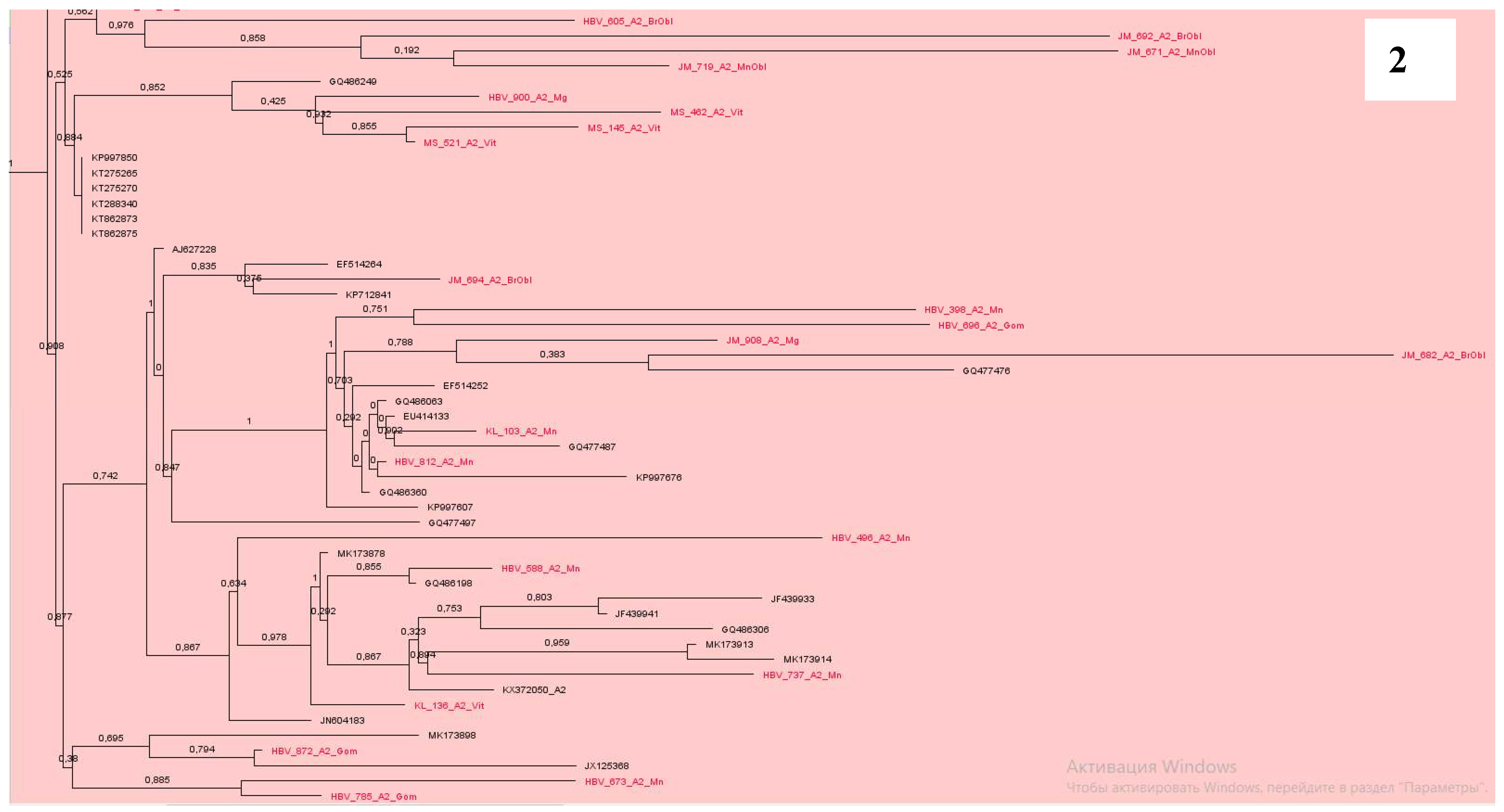
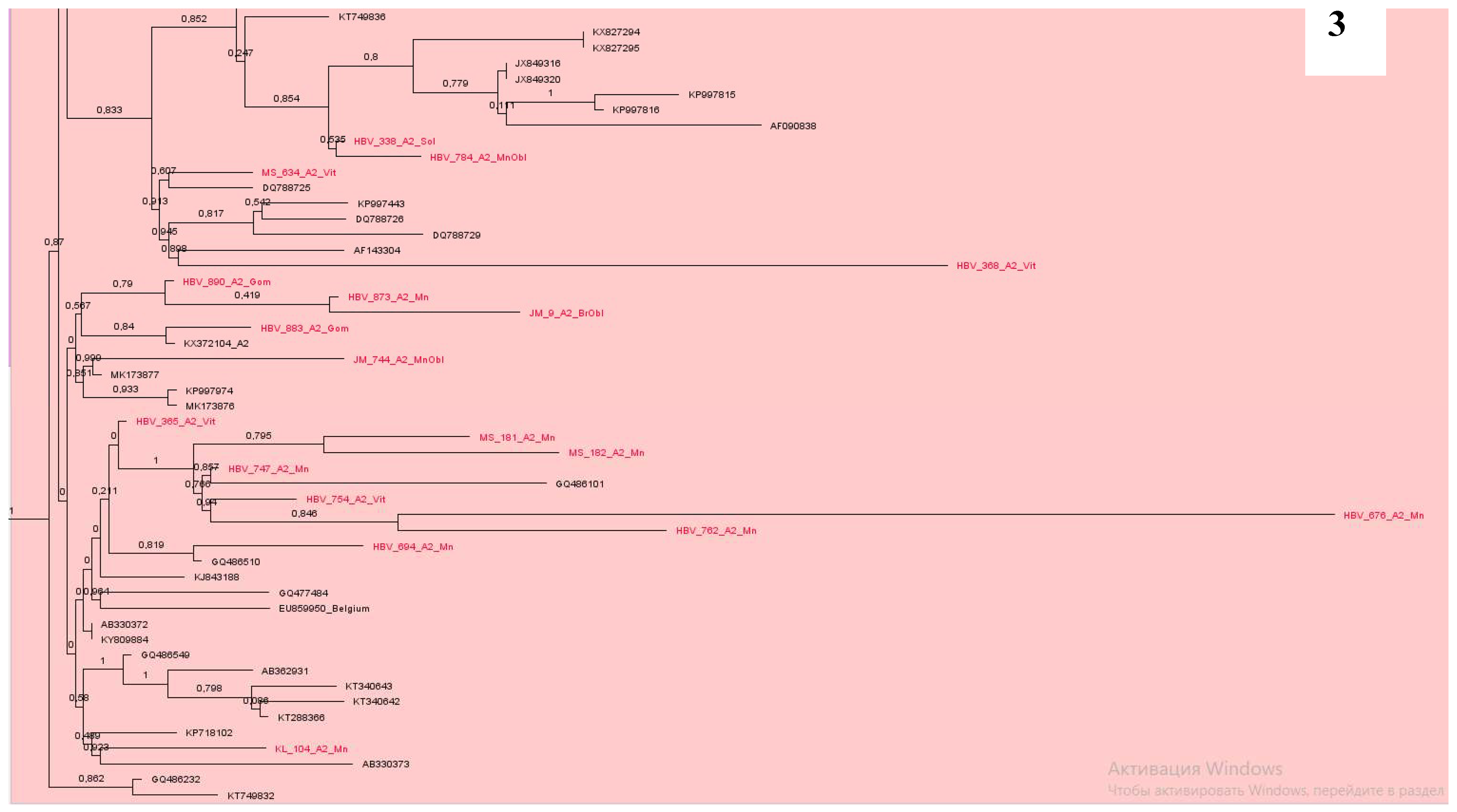
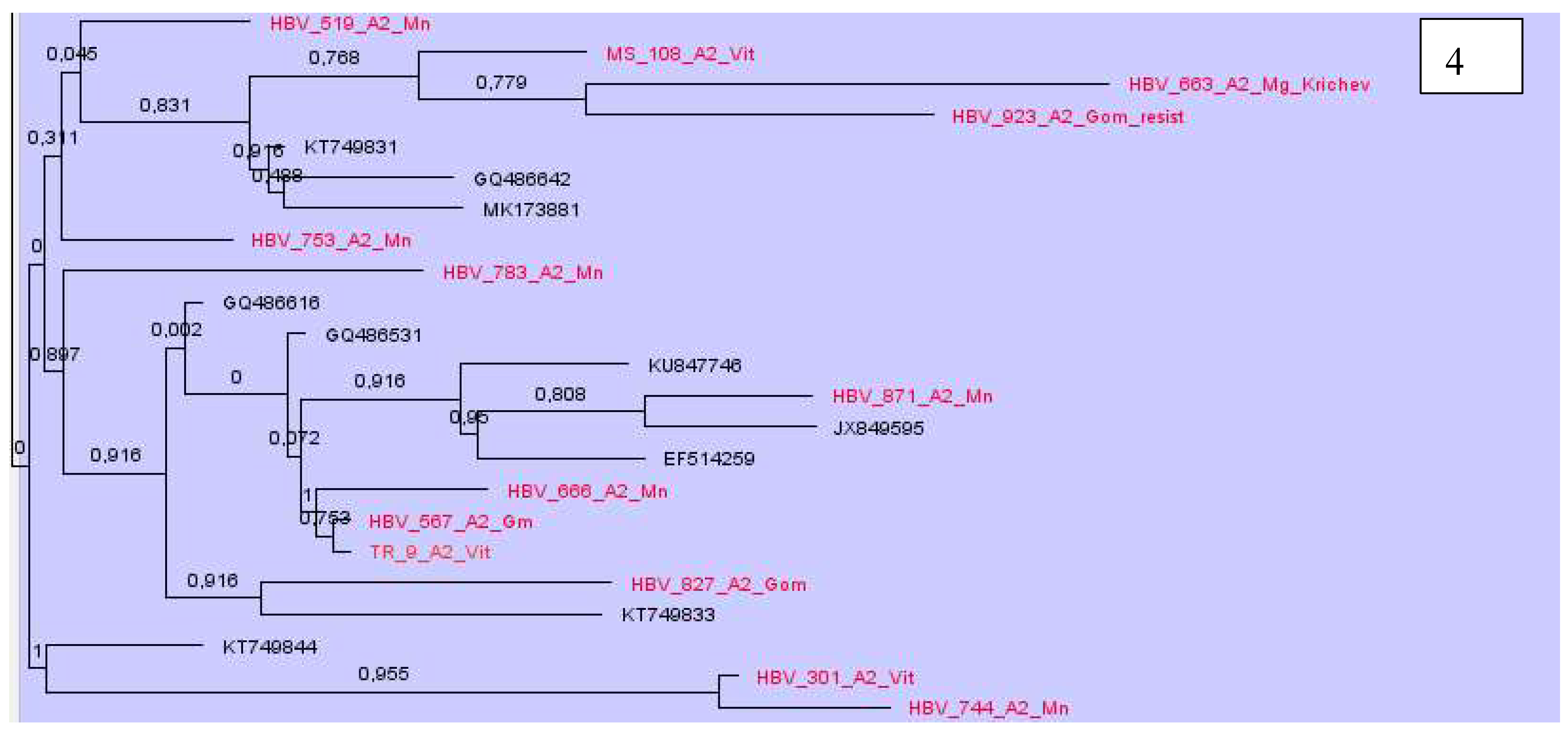
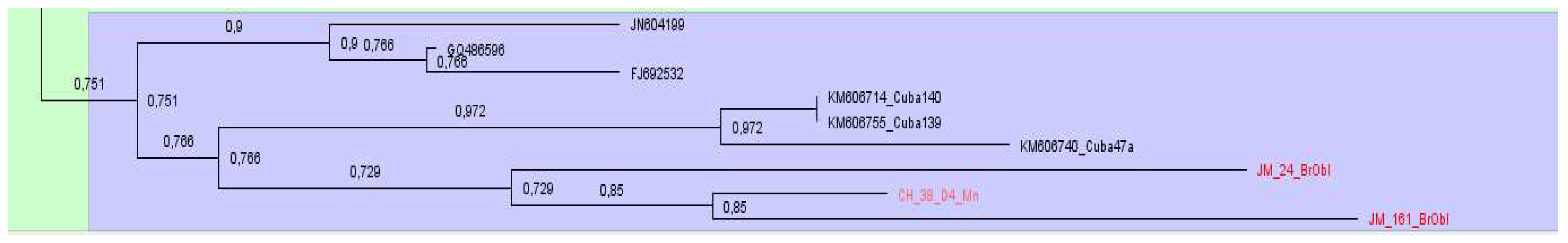
Phylogenetic analysis of DNA sequences of subtype A2 showed that most samples from Belarus were located nearby and formed at least 7 clusters (
Figure 2,
Figure 3 and
Figure 4). At the same time, some samples were in different groups and formed clusters with samples from the USA, Cuba, Brazil, similarly to Western European countries, mainly from Poland, Italy, France, Belgium, Germany and some others (
Figure 2,
Figure 3 and
Figure 4).
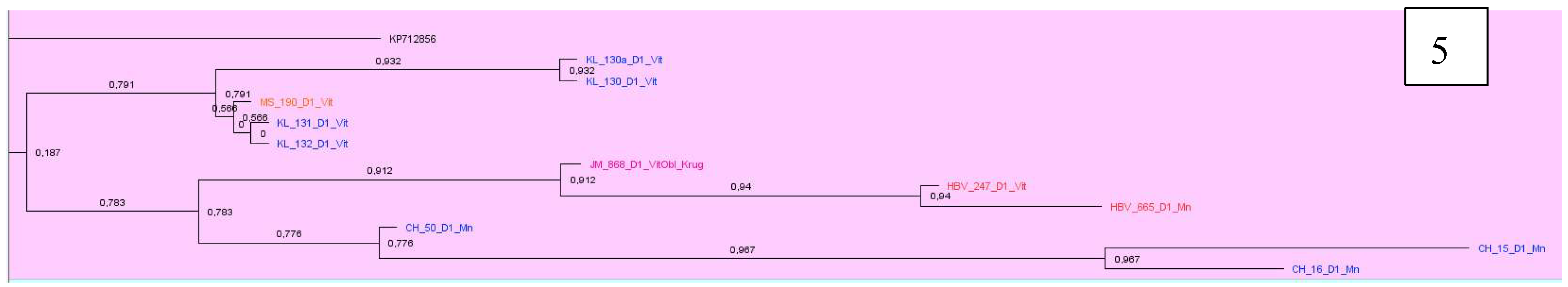
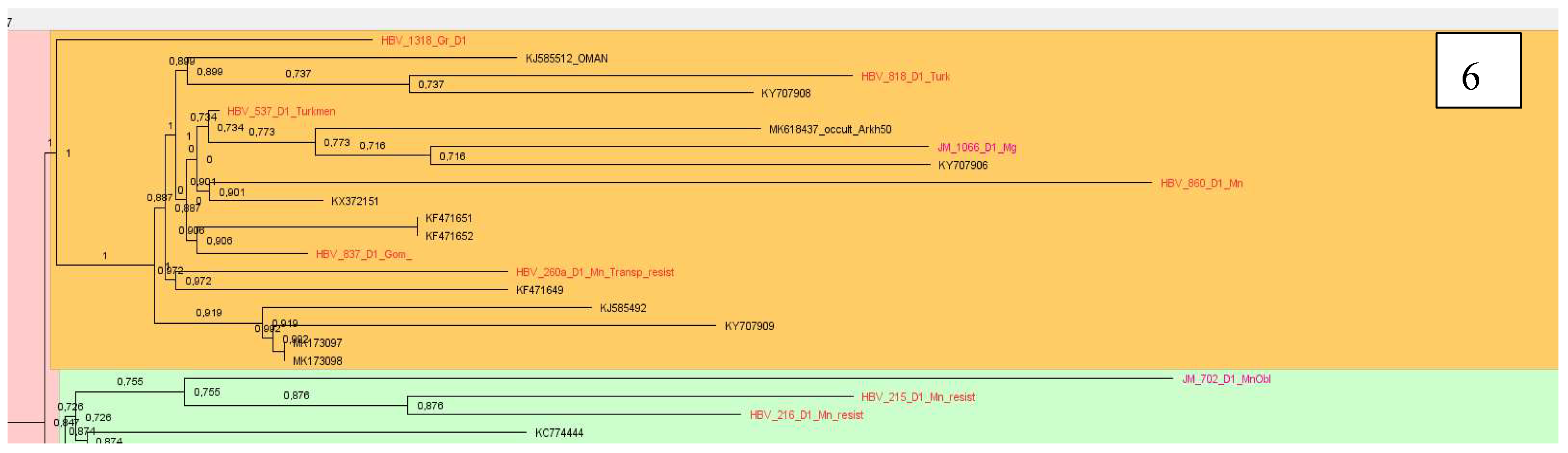
Analyzing samples of subtype D1, 12 clusters were identified. Most of the sequenced and analyzed DNA samples of the hepatitis B virus were identified with sequences from Pakistan, India, Iran, Bangladesh, some European countries as France, Holland, Italy. Two large clusters, consisting mainly of samples from Belarus, were found with sequences from Iran and Italy, as well as Pakistan and France, Oman, Holland and Russia (
Figure 5 and
Figure 6).
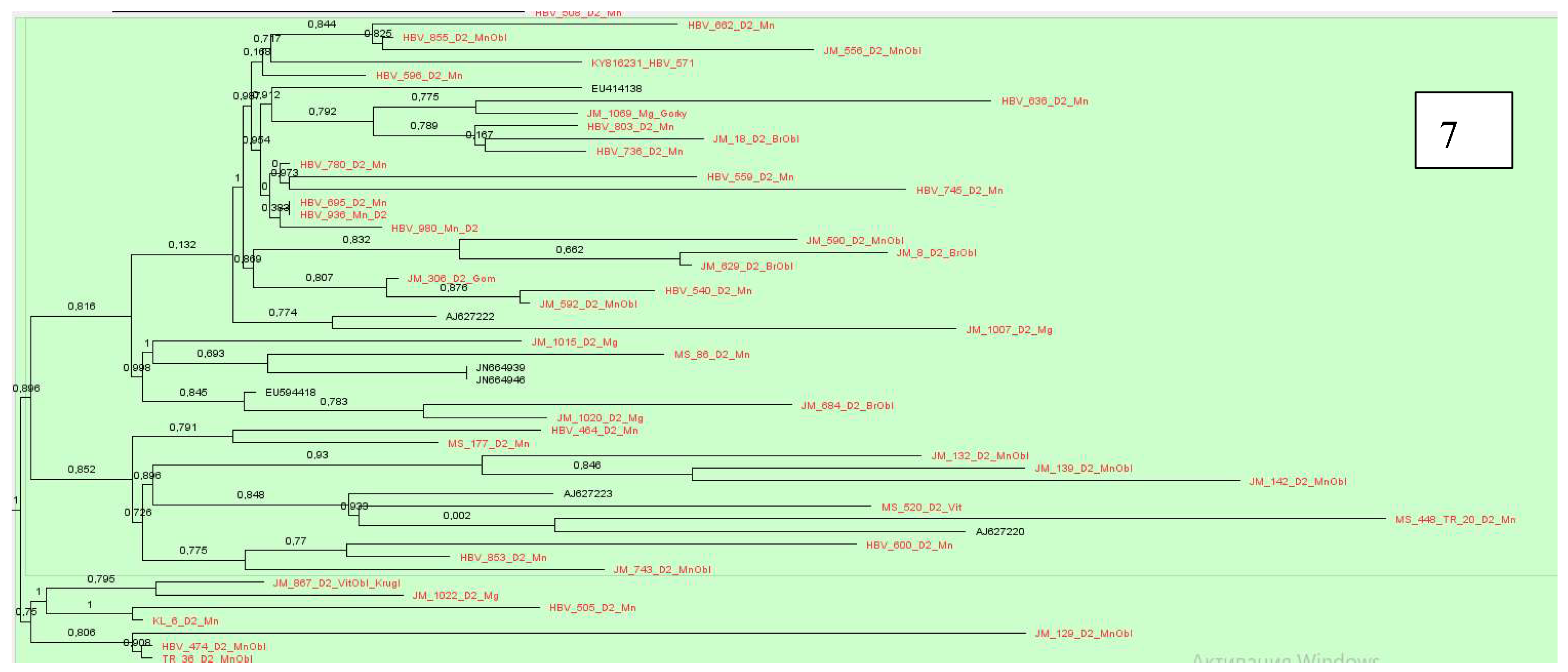
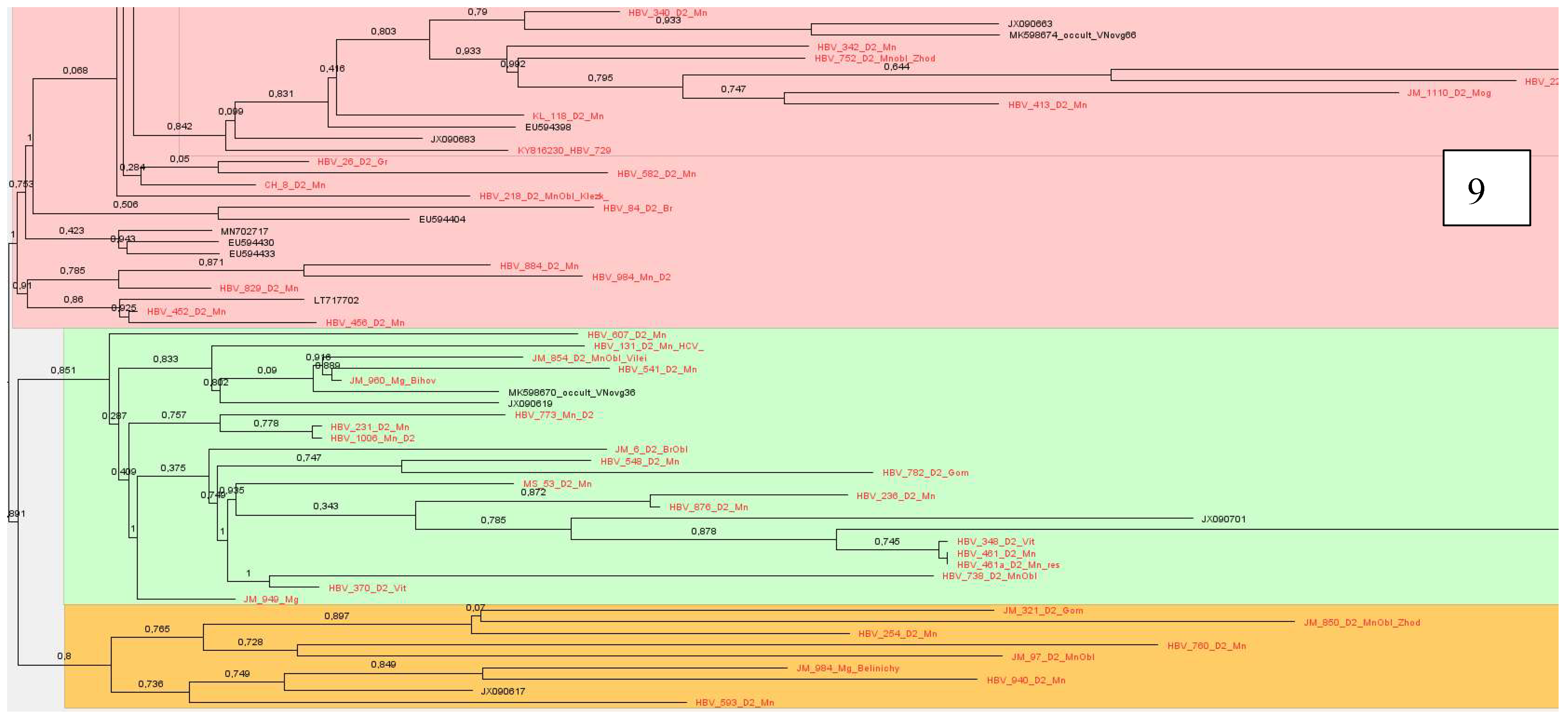
Sequences of subtype D2 formed 14 clusters, which were located with samples from Belarus, we have described earlier, showing similarities with samples from Estonia, USA, India, Spain, Russia, Cameroon, Latvia, Iran and Belgium,
Figure 7,
Figure 8 and
Figure 9.
Phylogenetic analysis of the D3 subtype showed that the samples formed 14 clusters mainly originated from India, Croatia, Russia, Italy, Germany and even Brazil. The largest cluster included samples from Belarus, with the sequences we have previously described, were originated from Poland, Russia, Rwanda, Estonia and Albania,
Figure 10.
The phylogenetic analysis of the samples of subtype D4 showed that they all have been found in the same group and clustered with HBV sequences from Cuba and Haiti,
Figure 11.
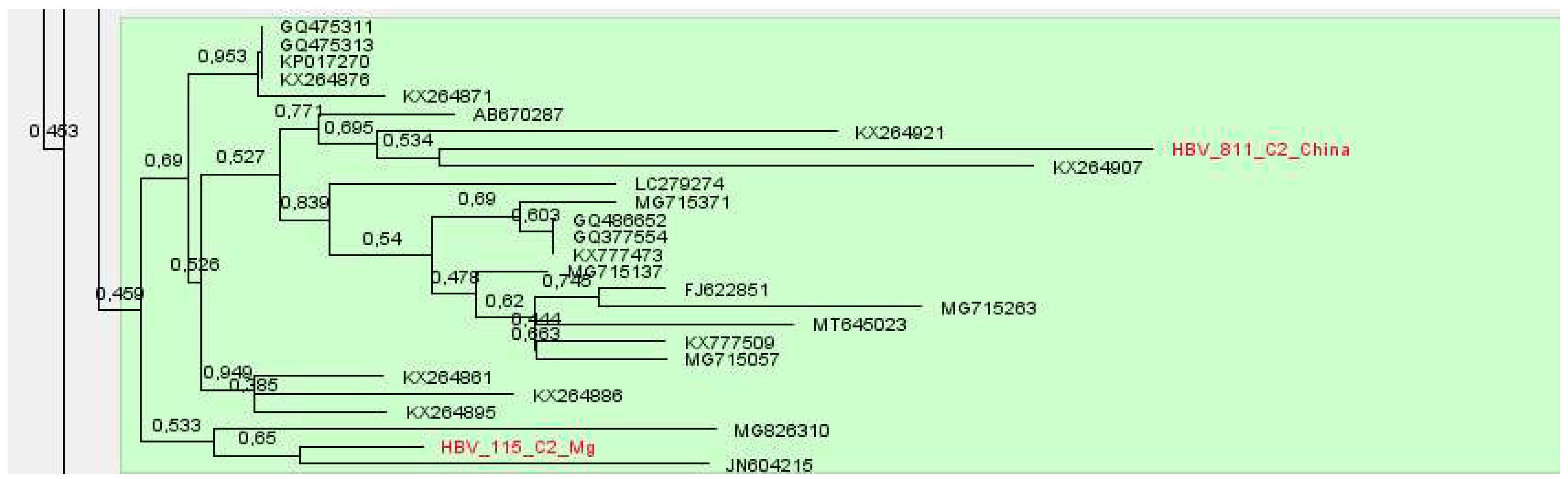
Samples of subgenotypes C1 and C2 were identified in adult patients and a child from China. Sequences of subtype C1 formed a cluster, with samples from the USA and Vietnam, and subtype C2 with reference samples from South Korea, USA and China,
Figure 12 and
Figure 13.
A sample of subtype B4 was discovered in a student from Vietnam who came to study in our country.
Finally, both samples with recombinant forms of hepatitis B virus were identified in residents of Belarus
4. Discussion
Every year in the Republic of Belarus, 800-900 new cases of chronic forms of hepatitis B are registered. Compulsory vaccination, introduced in the mid-90s, certainly had a positive effect on reducing the detection of new cases of hepatitis B virus. According to estimates, there are approximately 75,000 patients with chronic hepatitis B living in the country.
In a previous study, we showed that subgenotype D2 (55.8%) was dominant in Belarus, followed by D3 (18,3%), D1 (11,6%) and A2 (11,6%) [17]. The conducted studies showed that the structure of genotypes/subgenotypes in the country has remained virtually unchanged. Genotype D still dominates in the country and accounts for 81.7% of all analyzed cases. Genotype D was found representing by subgenotypes D1 (145/20.1%), D2 (371/51.4%), D3 (201/27, 8%) and D4 (5/0.7%); A – A2; C – C2; B – B4; recombinant forms C2/ D and A2/ С2 were detected. We determined the possible sources of infection of patients. Namely, occurring due to the circulation of “domestic” variants of the virus and/or the introduction of the virus from outside.
Our data show, the epidemic transmission of viral hepatitis B in the country is maintained mainly due to the circulation of local variants of the virus. This can be seen in the examples of subgenotypes A2, D1, D2 and D3, where the largest clusters were formed by samples from Belarus. At the same time, it should be noted that new variants of HBV are constantly being introduced into the country. The main directions of introduction of the A2 subgenotype are Western Europe countries and even the USA, Cuba, Brazil; for D1 - Pakistan, India, Iran, Bangladesh, Italy, Holland and Russia; D2 - Estonia, USA, India, Spain, Russia, Cameroon, Latvia, Iran and Belgium; D3 - Poland, Russia, Rwanda, Estonia and Albania. All samples of D4 subgenotype were clustered with sequences from Cuba and Haiti, C1 and C2 – from China, and B4 – from Vietnam. Two recombinant forms C2/D and A2/D were identified within the residents of our country.
Comparing the distribution of different genotypes of the hepatitis B virus in neighboring countries and even in different regions of the same country, it can be noted that it changes to one degree or another. So, for example, in the Russian Federation, genotype D significantly dominates in the European part of the country, while, for example, in Yakutia, genotype A occupies one of the leading positions (36.4% - A and 58,6% - D) [18]. In Asia, particularly in its eastern part, genotypes B and C dominate, and the C2 subgenotype is epidemic in China [19,20]. In Western Europe, in particular in Portugal HBV genotype A (HBV/A) was the most prevalent genotype (41.5%), followed by D [HBV/D; (33.8%)], and E [HBV/E; (24.6%)]. Subgenotypes A1 (HBV/A1) and D4 (HBV/D4) were almost equally prevalent with 23.1% and 22.3%, respectively, followed by A2 (HBV/A2) with 16.2% and D3 (HBV/D3) with 11.5% [21].
The Republic of Belarus is located in the geographic center of Europe, residents of the country actively travelling to different countries of the world, many people from different countries of the world come to the country for work and study, and this can explain the emergence of new variants of HBV in the Republic.
Thus, given a molecular genetic characteristic of HBV, detected in the territory of the Republic of Belarus, identified possible directions for the introduction of new variants of the virus, and detected the main mutations in the P region of the virus genome leading to resistance to antiviral drugs.
Funding
We are grateful to the Abbott Transfusion Medicine Medical affairs team for financial support of the publication.
References
- Thun M.J., DeLancey J.O., Center M.M., Jemal A., Ward E.M. The global burden of cancer: Priorities for prevention. Carcinogenesis. 2010;31:100–110. [CrossRef]
- Philips C.A., Ahamed R., Abduljaleel J.K., Rajesh S., Augustine P. Critical Updates on Chronic Hepatitis B Virus Infection in 2021. Cureus. 2021;13:e19152. [CrossRef]
- Kao J.-H., Chen P.-J., Lai M.-Y., Chen D.-S. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology. 2000;118:554–559.
- Aghakhani A., Hamkar R., Zamani N., Eslamifar A., Banifazl M., Saadat A., Sofian M., Adibi L., Irani N., Mehryar M. Hepatitis B virus genotype in Iranian patients with hepatocellular carcinoma. Int. J. Infect. Dis. 2009;13:685–689. [CrossRef]
- Beck J., Nassal M. Hepatitis B virus replication. World J. Gastroenterol. 2007;13:48–64.
- Pollicino T., Cacciola I., Saffioti F., Raimondo G. Hepatitis B virus PreS/S gene variants: Pathobiology and clinical implications. J. Hepatol. 2014;61:408–417. [CrossRef]
- Ie S.I., Thedja M.D., Roni M., Muljono D.H. Prediction of conformational changes by single mutation in the hepatitis B virus surface antigen (HBsAg) identified in HBsAg-negative blood donors. Virol. J. 2010;7:326.
- Lin C.-L., Kao J.-H. The clinical implications of hepatitis B virus genotype: Recent advances. J. Gastroenterol. Hepatol. 2011;26((Suppl. 1)):123–130.
- Zhu S.S., Zhang H.F., Wang H.B., Dong Y., Chen D., Jia W., Gan Y., Chen J. Relation of viral genotypes to clinical features in children with chronic hepatitis B. Chin. J. Exp. Clin. Virol. 2008;22:192–194.
- Shamseer L., Moher D., Clarke M., Ghersi D., Liberati A., Petticrew M., Shekelle P., Stewart L.A. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: Elaboration and explanation. BMJ. 2015;349:g7647. [CrossRef]
- George B.J., Aban I.B. An application of meta-analysis based on DerSimonian and Laird method. J. Nucl. Cardiol. 2016;23:690–692.
- Fletcher J. What is heterogeneity and is it important? BMJ. 2007;334:94–96.
- Higgins J.P.T., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. [CrossRef]
- Wallace B.C., Dahabreh I.J., Trikalinos T.A., Lau J., Trow P., Schmid C.H. Closing the Gap between Methodologists and End-Users: R as a Computational Back-End. J. Stat. Softw. 2012;49:2–15.
- Munn Z., MClinSc S.M., Lisy K., Riitano D., Tufanaru C. Methodological guidance for systematic reviews of observational epidemiological studies reporting prevalence and cumulative incidence data. Int. J. Evid. Based Healthc. 2015;13:147–153.
- Serfaty L., Thabut D., Zoulim F., Andreani T., Eres O. C., Carbonell N., Loria A., and Poupon R. / Hepatology – 2001 – V.34 – N.- 3 – P.573-577.
- Olinger C.M., Lazouskaya N.V., Eremin V.F., Muller C.P. Multiple genotypes of hepatitis B and C viruses in Belarus: similarities with Russia and European influences. Clin Microbiol Infect – 2008; 14:575-581. [CrossRef]
- Anastasia A Karlsen, Karen K Kyuregyan, Olga V Isaeva, Vera S Kichatova2, Fedor A Asadi Mobarkhan, Lyudmila V Bezuglova, Irina G Netesova, Victor A Manuylov , Andrey A Pochtovyi, Vladimir A Gushchin, Snezhana S Sleptsova, Margarita E Ignateva, Mikhail I Mikhailov Different evolutionary dynamics of hepatitis B virus genotypes A and D, and hepatitis D virus genotypes 1 and 2 in an endemic area of Yakutia, Russia // /BMC Infect Dis – 2022; 12;22:452.
- Kizito Eneye Bello, Tuan Nur Akmalina Mat Jusoh, Ahmad Adebayo Irekeola, Norhidayah Abu, Nur Amalin Zahirah Mohd Amin, Nazri Mustaffa, Rafidah Hanim Shueb / A Recent Prevalence of Hepatitis B Virus (HBV) Genotypes and Subtypes in Asia: A Systematic Review and Meta-Analysis // Healthcare (Basel) – 2023; 1;11(7):1011.
- Bing Sun, Aida Andrades Valtueña, Arthur Kocher, Shizhu Gao, Chunxiang Li, Shuang Fu, Fan Zhang, Pengcheng Ma, Xuan Yang, Yulan Qiu, Quanchao Zhang, Jian Ma, Shan Chen, Xiaoming Xiao, Sodnomjamts Damchaabadgar, Fajun Li, Alexey Kovalev, Chunbai Hu, Xianglong Chen, Lixin Wang, Wenying Li, Yawei Zhou, Hong Zhu Johannes Krause, Alexander Herbig, Yinqiu Cui/ Origin and dispersal history of Hepatitis B virus in Eastern Eurasia // Nat Commun, 2024; 5;15:2951.
- Rute Marcelino, Ifeanyi Jude Ezeonwumelu, André Janeiro, Paula Mimoso, Sónia Matos, Veronica Briz, Victor Pimentel, Marta Pingarilho, Rui Tato Marinho, José Maria Marcelino, Nuno Taveira, Ana Abecasis / . Phylogeography of hepatitis B virus: The role of Portugal in the early dissemination of HBV worldwide // PLoS One. 2022; 17(12):.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).