1. Introduction
Banana (
Musa sp.), an herbaceous genus of plants native to tropical Indomalaya and Australia, is an important food crop world-wide and is valued for its flavours and nutritional values. Banana is widely grown in the tropics and is consumed as staple food in many countries, having contributed to a total global production of 135 million tons in 2022, with world exports totaling 24 million tons worth approximately 13 billion USD [
1].
Cavendish (genome AAA), including the cultivar known as 'Williams', is the most dominant dessert banana in the market accounting for 47% of the global banana production [
1]. The over-reliance on so few cultivars and lack of genetic diversity makes banana crops vulnerable to pests and diseases, leading to major outbreaks that put constrains on production at both commercial and small-scale levels [
2]. One such disease is the Fusarium wilt of banana (FWB), caused by the soil-borne fungus
Fusarium oxysporum f. sp.
cubense (
Foc). The
Fusarium oxysporum species complex (FOSC) contains many plant pathogens that infect economically important crop species, identified as crop specialist forms (
formae speciales) or defined by the hosts on which they cause disease. Within
formae speciales, isolates are classified using a race structure based on the cultivars to which they are pathogenic [
3,
4,
5]. However, the relationships of different
formae speciales of
F. oxysporum is complex with evidence of a polyphyletic history and horizontal gene transfer [
5,
6].
Within the banana-infecting Fusaria,
Foc race 1 and
Foc race 4 have had the greatest impact on global banana production.
Foc race 1caused the demise of 'Gros Michel' (AAA) banana in the 1950s leading to its replacement by the
Foc race 1 resistant Cavendish cultivars [
7].
Foc race 1 causes disease on the cultivars ‘Gros Michel’ (AAA), ‘Maqueño’ (AAB), ‘Silk’ (AAB), ‘Pome’ (AAB) and ‘Pisang Awak’ (ABB) [
8]. The emergence of
Foc race 4 from Southeast Asia in 1960s saw Cavendish cultivars and certain other cultivated forms succumbing once again to Fusarium wilt [
9].
Foc race 4 is divided into; strains that infect Cavendish after a period of relatively cooler temperatures in the subtropics known as Subtropical race 4 (STR4) and strains known as Tropical race 4 (TR4) that cause infection under both tropical and subtropical conditions [
10,
11].
Foc TR4 has now spread across all major continents, decimating banana plantations in its wake and finally reaching the major banana production regions of Latin America in 2019 [
12].
Foc TR4 was first recorded in the early 1990s in Southeast Asia and soon after in 1997 in Northern Territory, Australia [
13]. However, it was almost another two decades before it was detected in Australia's main banana production area, North Queensland, when in 2015, TR4 was reported in Tully, resulting in an unprecedented increase in regional biosecurity monitoring and strict quarantine control [
14].
Fusarium wilt is not a disease that can be easily eradicated, so it is important to minimise the dissemination of infection through quarantine control or the use of clean planting materials [
15]. For locations where the disease has spread to, there are no effective chemical control methods against FWB, hence the focus has been on developing cultivars carrying genetic resistance to
Foc [
16,
17,
18]. Somaclonal variants with enhanced resistance to
Foc TR4 and
Foc STR4 have been identified from Cavendish derived plants [
9,
19]. The resistance of banana genotypes to both
Foc STR4 and TR4 have been investigated in field and pot trials [
20,
21,
22,
23]. Rhizomes appear to play an important role in determining a plant's response to
Foc [
20]. Some of these somaclonal variants, particularly the Giant Cavendish tissue culture variants (GCTCV), have shown promising results in TR4-infested field trials in the Philippines [
24], Australia [
21], Africa [
25] and China [
22] as well as in pot trials [
20,
22].
Conventional crossing has also been used to develop
Foc resistant cultivars. The Honduran Foundation for Agricultural Research (FHIA) established a banana breeding program to develop resistant banana types, to combat a range of different diseases as alternatives to the predominantly susceptible Cavendish cultivars [
26]. These varieties include dwarf desserts banana, plantain and cooking bananas hybrids, some of which were resistant to
Foc Race 4 types. Field evaluations suggest that some of these FHIA varieties can be as productive and robust as natural hybrids [
27]. Despite their high yields and resistances to Fusarium wilt, the FHIA hybrids have not met the needs and preferences of the market [
28]. However, these hybrids were derived in turn from a set of diverse progenitor hybrids (SH hybrids) that have been used extensively in the breeding programs at the International Institute for Tropical Agriculture (IITA) in East and West Africa, the National Agricultural Research Organisation (NARO) in Uganda, as well as the Brazilian Agricultural Research Corporation (Embrapa) in Brazil and The Centre for International Cooperation in Agricultural Research (CIRAD) in France. So far, FHIA lines have been used for dissecting disease responses against
Foc TR4 in field and pot trials [
20,
21].
Foc produces three different types of asexual spores; the short-lived microconidia and macroconidia, and the long-lived chlamydospores. All three types of spores have the ability to cause infection on susceptible banana plants [
29].
Foc is known to survive in the soil for decades, persisting in the soil on dead host plant tissues in the absence of a suitable host either as chlamydospores or as saprophytes [
30].
Foc spores germinate and directly adhere to the roots and gain entry through the root epidermis or open wounds [
31,
32], followed by the movement into the xylem vessels of the roots and rhizomes. As the fungus colonises the plant, extensive mycelial networks occlude the xylem vessels, obstructing nutrient and water uptake, which results in plant wilting and eventual plant death [
33]. External symptoms of FWB include pseudostem splitting, leaf yellowing, and necrosis and stunted growth. The fungus is not transferred to the fruit parts, but the infected plants will produce less fruit [
7]. Recently it has been demonstrated that
Foc TR4 can colonise the fruit peduncle in
Foc TR4-infected banana plants as improvements in agronomic practices to manage the disease have resulted in affected plants producing bunches [
34].
Previous studies in many different host species, have demonstrated a series of plant defense reactions against
Fusarium pathogens, including papilla formation, production of antimicrobial substances, cell wall lignification, occlusion by gums, gels or tyloses within xylem vessels and vessel crushing [
35,
36]. The roles of each of these plant reactions in resistance are unclear, but they all contribute to the overall resistance capacity in plants. For instance, tyloses formation is considered a protective response of trees and herbaceous plants to vascular damage whether from mechanical injury or fungal or bacterial infections [
37]. Tyloses formation occurs in the vessel lumens of root xylem in infected plants. They are formed from the extension of parenchyma cells through the pit membrane of the inner xylem wall and can fill up the entire xylem lumen. The amount of tyloses accumulation can vary depending on the resistance level of a cultivar. VanderMolen et al. (1987) showed that tyloses formation occurs in both susceptible and resistant cultivars, where rapid occlusion with tyloses occurred in infected root xylem vessels of resistant cultivars, whereas susceptible cultivars showed a similar tyloses initiation with plant growth reduced at later stages [
38]. Tyloses formation has been shown to have a significant role in plant defense mechanisms in relation to the susceptibility of banana plants.
Green Fluorescent Protein (GFP)-tagged
Foc strains have been used to observe the movement of
Foc in banana hosts resistant and susceptible to
Foc TR4 [
39,
40] and
Foc STR4 [
33]. In the study presented here, histological process of
Foc infection was investigated using five banana cultivars: 'Williams' Cavendish (AAA), its somaclone 'GCTCV119' (AAA), as well as Pome type cultivar known in Australia as 'Lady Finger' (AAB), 'FHIA02' (AAAA), and 'FHIA25' (AAB) hybrids. We observed that host susceptibility is associated with the active proliferation of the fungus in the xylem vessels of the rhizomes. In comparison, the fungus was restricted in the rhizomes of the
Foc resistant cultivars 'FHIA25' and 'GCTCV119', although, the ability of
Foc to still enter the roots appeared unhindered in these cultivars. Given that
Foc TR4 resistant cultivars have already been deployed in some of the
Foc TR4 affected regions around the world, it will be important to understand the level of resistance such cultivars possess. It is equally important to address what constitutes resistance specifically relating to plant host's response to
Foc and whether the fungal presence can still be detected in resistant cultivars. Thus, addressing some of these questions allows the suitability of deploying apparent resistant genotypes in
Foc TR4 affected regions to be assessed.
2. Materials and Methods
2.1. The GFP-tagged Fusarium Oxysporum f. sp. Cubense Isolate
The GFP-tagged
Foc STR4 (GFP-
Foc-STR4) strain UQ6817 was derived from the strain BRIP40389 (Queensland Plant Pathology Herbarium) and has been described in previous studies with respect to its pathogenicity on
Musa [
20,
41]. The strain was single-spored and stored in the form of water-agar plugs in sterile water at 4°C.
2.2. The banana Cultivars and Plant Growth Conditions
Tissue-culture banana plantlets of the known Foc STR4 susceptible lines 'FHIA02', 'Lady Finger', 'Williams', and the Foc STR4 tolerant/resistant lines 'GCTCV119' (somaclonal variant of 'Williams') and 'FHIA25' were de-flasked into 30 cell potting trays of 35 cm (length) x 29 cm (width) x 5.5 cm (depth). The trays were placed on a lab bench and were positioned under fluorescent light set at a day/night cycle of 16 h / 8 h and temperature of 20-22°C. The soil mix, UQ23, contained 70% composted pine bark 0-5 mm in size and 30% coco peat and has a pH range of 5.5-6.5. After 4 weeks of hardening-off post tissue culture, plants were repotted into 140 mm diameter pots and to which a teaspoon of a balanced fertiliser (Osmocote) was added. Plants were transferred to a temperature-controlled glasshouse with day and night temperatures set at 22°C/26°C and were grown for 6 weeks under a regime of watering to field capacity once every 2-3 days.
2.3. Preparation of Spore Suspension For Plant Inoculation
The GFP-Foc-STR4 strain was grown on full strength potato dextrose agar (PDA), supplemented with 100 mg/L hygromycin B, and was incubated at 25°C for 4 days. Four 5 mm2 mycelial blocks of GFP-Foc-STR4 were cut from a fully colonised PDA plate and were used to inoculate 500 mL of sterile potato dextrose broth (PDB), supplemented with 50mg/L hygromycin B. The cultures were placed on a shaker and agitated at 28°C, 180 rpm for 4 days. The culture was then filtered through four layers of sterile Miracloth and the spores were collected and then washed with sterile distilled water (SDW). For root dipping, the concentration of the spore suspension was adjusted to 2×106 spores per mL with SDW. Additional spore suspension was used to drench the soil at a concentration of approximately 50,000 spores per gram of soil.
For plants subjected to scanning electron microscopy, the plants were inoculated with 45 g of GFP-
Foc-STR4 infested Japanese millet (
Echinochloa esculenta) variety 'Shirohie', as previously described [
20]. A spore suspension of the same concentration and application method as above was also directly poured onto the root zone of the plants. Non-inoculated plants served as controls.
2.4. Plant Inoculation
After 6 weeks, 30 plants of each 'FHIA25', 'FHIA02', 'Lady Finger', 'Williams' and 'GCTCV119' were inoculated by root dipping, followed by soil-drenching using the GFP-Foc-STR4 spore suspension. Plants with five to six healthy leaves and a stem height of 30 cm were selected and plant roots were washed with SDW and then dipped in the spore suspension for 2 hours. Instead of a spore suspension, non-inoculated control plants were root-dipped in SDW for 2 hours. All plants were then transplanted to 250 mm diameter pots containing the soil mix UQ23.
2.5. Plant Harvest and Visualisation of GFP Colonised Plants
Plants were harvested at weekly time points up to 70 days throughout this experiment. External symptoms were visually assessed on plant leaves and pseudostems at harvest. Internal symptoms including discolouration within the roots, rhizome and stems were assessed on the day when confocal microscopy was performed to detect the localisation of GFP-Foc-STR4 inside the host. For visualisation under a confocal microscope, a double-edged razor blade was used for sectioning plant tissues (transverse and longitudinal), including roots, stem, rhizomes and leaf parts. Sections were mounted in sterile deionised water.
GFP was detected using Zeiss 700 Laser Scanning Microscope (Zeiss, Oberkochen, Germany) and a laser at an excitation wavelength of 488 nm. The Z-stack function was used to capture 3D images consisting of 10-30 optical slices taken at intervals of 1-5 μm. The T-PMT (transmission detector setting) was also used to see the sectioned plant tissues in an overlay of brightfield.
2.6. Reisolation of GFP-Foc-STR4 from Symptomatic Plants
Diseased plant materials were surface sterilised using 0.5% bleach for 30 seconds and then washed twice in SDW before blotting dry on sterile paper towel under laminar flow hood. Tissues including leaf, petiole, stem just above the rhizome, central cylinder of the rhizome and rhizome node connecting roots, were cut into 2-5 mm sections using a sterile scalpel and then embedded into water agar. The plates were incubated at 25°C for 10 days. The presence of Fo-like spores and mycelia were identified using a dissecting microscope. The Fo-like colonies (white mycelia with pink to mauve staining of agar) were then sub-cultured onto half strength PDA, supplemented with 100 mg/L hygromycin B. Plates were scored for the presence or absence of Fo-like growth after 10 days of incubation at 25°C. These colonies were further examined under confocal microscope to confirm GFP fluorescence.
2.7. Sample Preparation and Scanning Electron Microscopy
Primary roots and rhizomes at the base of the banana plants were sectioned for scanning electron microscopy (SEM) at 2- and 6-weeks post inoculation. The sample preparation protocol was adapted from a previous study [
42]. Sections of 0.5 to 1 cm in size were fixed in a buffer containing 2.5% glutaraldehyde, 0.1 M monobasic sodium phosphate and 0.1 M dibasic sodium phosphate. The samples were embedded in a specimen tube containing agarose (50%, w/v) and were sliced at a depth of 100 μm using a vibratome (VT1000 S, Leica Biosystems, Wetzlar, Germany). Sectioned samples were stored in a 6% sodium azide buffer.
The primary root and rhizome were washed twice by the same 0.1 M sodium phosphate buffer to replace the glutaraldehyde. Following this, all processing was performed in a Biowave microwave (Ted Pella, Redding, USA). Each step consisted of a one minute on, one minute off, one minute on process using the Biowave operated at 150W. Dehydration in ethanol used a graded series of 60 %, 70 %, 80 %, 90 % and two 100 % steps. This was followed by chemical drying using hexamethyldisilazane (HMDS) added in steps as 1:1 diluted with absolute ethanol, then twice in 100 ethanol. The 100% HMDS was left overnight to evaporate.
Once dried, each section was mounted onto an aluminium stub using double-sided sticky carbon tabs, and was sputter coated with platinum to create electrical conductivity on the sample surface. Approximately 400 sections were imaged by the SEM: HitachiTM4000Plus Bench top (Hitachi High-Tech, Tokyo, Japan) in a high-vacuum mode operating at 15 kV and the working distance of 15 mm to 18 mm. The presence or absence of the tyloses within the tissue due to
Foc inoculation were analysed by SEM comparing with previous studies [
38,
43].
4. Discussion
Green fluorescent proteins have been a critical tool in understanding pathogen and host plant interactions
in planta [
20,
45,
46]. Being able to visualise the fungus
in plantae means that the progress of infection can be analysed in great detail. In
F. oxysporum, the production of microconidia and macroconidia are critical for the fungus to proliferate inside the hosts. Understanding the accumulation of spores in specific compartments of plants has relevance for the management and containment of
Foc in the field. The plant responses associated with the pathogen attack are not well known in resistant/tolerant cultivars such as 'FHIA25' and 'GCTCV119'.
The movement of
Foc through the roots was observed in all five cultivars, including 'Williams', 'Lady Finger', 'FHIA02', 'FHIA25', and 'GCTCV119'. Despite having good levels of resistance against
Foc STR4, chlamydospores and microconidia of the inoculated GFP-STR4 isolate were observed to attach and germinate on the surface of the root tip and fine root hairs in 'FHIA25'. This was followed by the penetration of the root surface and the movement of hyphae in the root vascular systems. These observations suggest that
Foc can actively enter banana hosts via the root systems regardless of the level of resistance the hosts carry. In both 'FHIA02' and 'FHIA25', the intercellular movement of the fungus was observed on the epidermis and then in the elongation zones of the roots. This is consistent with the characteristic movement of a biotrophic pathogen and has been observed in the infection process of other
Fusarium species [
47,
48]. The presence of the fungus was consistently detected in the xylem vessels of both the roots and root nodes connecting to the rhizome. This suggests that the vascular streams facilitate the movement of the fungus inside the plant host in previously described susceptible and resistant lines.
Our previous study showed that the plant host's response, in the rhizome, plays an important role in inhibiting the fungus from spreading to the rest of the plant [
20]. In this study, a differential response was observed in the rhizome of 'FHIA02' and 'FHIA25' plants. In the symptomatic rhizomes of 'FHIA02', proliferation of spores and mycelia was clearly detected in the xylem vessels. Interestingly, only a few hyphae were detected in these regions of 'FHIA25', and they appeared to be confined in the xylem. The formation of tyloses wase detected in these regions of both cultivars. Furthermore, in the susceptible cultivars 'Lady Finger' and 'Williams', the observations were that they were extensively colonised by the fungus in the rhizome, a characteristic of
Foc susceptibility, whereas the fungus displayed limited movement and appeared confined in the rhizome of 'GCTCV119'. Taken together, these observations suggest that the rhizome is essential in the interplay between the pathogen and plant host.
A similar pattern of restricted colonisation in
Dianthus caryophyllus by
F. oxysporum f. sp.
dianthi has been reported and further characterisation revealed that the infected regions of the xylem became compartmentalised by cell wall thickening, hyperplasia of parenchyma cells and the built-up of vascular occluded materials [
36]. In the current study, vascular occluded xylem vessels were detected in the rhizome of both resistant and susceptible cultivars inoculated with the fungus. Vascular occlusion in the rhizome was detected as early as 36 dpi in 'FHIA25' and was detected for the first time in the 'FHIA2' at 68 dpi. Although SEM showed that vascular occlusion occurred as early as 14 dpi in the main roots of both 'FHIA2' and 'FHIA25' inoculated with the fungus. Occluded vessels were similarly visualised in 'GCTCV119' and 'Williams' at 56-59 dpi. These results collectively suggest that vascular occlusion is an inducible plant mechanism to prevent the spread of an invading pathogen. While it may contribute to deterring the movement of the pathogen inside the host, tyloses formed as extensions of contact parenchyma cells [
38] do not fully explain the host resistances observed in 'FHIA25' and 'GCTCV119'. Formation of tyloses is typically triggered by infections [
49], wounding [
50], heartwood formation [
51] and abscission [
52]. The ability to form tyloses was an important factor in resistance to
F. oxysporum in cotton [
53] and has been shown to be upregulated by an exogenous chemical application in banana [
54]. Therefore, the presence of tyloses observed in this study is consistent with their roles in growth and development, as well as in the regulation of stresses including pathogen attacks and activation of oxidative stress.
Another aspect of pathogen deterrence is the formation of vascular coatings in or around infected regions observed in this study. These regions have been identified as plant physico-chemical barriers induced against xylem vascular wilt pathogens [
44]. Particularly, gel and lignin depositions have been found to be associated with vertical and horizontal restrictions, respectively, of
F. oxysporum f. sp.
cubense in banana hosts [
38,
55]. These mechanisms involving the formation of gels, gums, or mucilage in and around the vascular systems have been shown to limit fungal growth in banana [
43], tomato [
56], carnation [
57], pea [
35,
58], cotton [
45,
53], and bean plants [
59]. These barriers are mostly composed of carbohydrates like pectin, polyphenols, and sometimes phytoalexins, lignin-like compounds or lipoidal substances [
35]. It was observed in peas that the production of carbohydrates and polyphenolics by vascular parenchyma cells progressively accumulated in the lumen of xylem cells, highlighting important functions of these compounds in defense against
F. oxysporum f. sp.
pisi [
35].
The presence of chlamydospores and hyphae on the petiole and outer leaf sheath was detected in both 'FHIA02' and 'FHIA25'. Despite 'FHIA25' being resistant to
Foc STR4, the fungus was able to move through the outer leaf sheaths and reach the aerial parts of the plant. Furthermore, sporodochia were detected in the pseudostem of 'GCTCV119' as well as around the stomata in the leaves of 'William' and 'Lady Finger'. These results are consistent with a previous study which showed the movement of
Foc through the leaf sheaths [
40] and capability of reaching the outside of the leaf sheath via stomata [
33]. However, the transmission of
Foc through the aerial part of a
Foc resistant plant has not been previously reported.
To date, conventional breeding efforts have not yielded varieties resistant to
Foc TR4 that are also agronomically viable [
12]. 'GCTCV119', is one of the Cavendish tissue culture derived somaclones that have been shown to carry improved tolerance to
Foc TR4 [
9]. Although it should be noted that 'GCTCV119' and the improved variant 'GCTCV218', also known as 'Formosana', are not truly resistant to
Foc TR4 [
12]. 'GCTCV218' did seem to be moderately resistant to
Foc TR4 in one study [
25] and intermediately resistant only in the first cropping cycle in another study [
21]. 'GCTCV218' has replaced Cavendish cultivars in TR4-affected regions in the Philippines and Mozambique [
25]. Similarly, 'FHIA25' has previously shown to be highly resistant to both
Foc STR4 and TR4 [
20,
21]. Given the evidence in this study to suggest that
Foc can colonise resistant/tolerant cultivars such as 'GCTCV119' and 'FHIA25', it is important then to understand the inoculum threshold levels these and other resistant cultivars can withstand before they succumb to the disease. Further stress testing of these cultivars under different conditions will need to be performed to guide the deployment of these cultivars in
Foc TR4-affected regions and to maintain and improve appropriate management strategies to control
Foc TR4.
Author Contributions
Conceptualization, A.C. and E.A.B.A.; methodology, A.C., T.Y.C., Y.C. and S.M.A.F.; software, A.C., T.Y.C., Y.C. and S.M.A.F.; validation, A.C., J.A., J.S., H.C., S.G., B.F., A.S., D.M.G. and E.A.B.A.; formal analysis, A.C., T.Y.C., Y.C. and S.M.A.F.; investigation, T.Y.C., Y.C. and S.M.A.F.; resources, A.C. and E.A.B.A.; data curation, A.C.; writing—original draft preparation, A.C.; writing—review and editing, A.C., T.Y.C., Y.C., S.M.A.F., J.A., J.S., H.C., S.G., B.F., A.S., D.M.G. and E.A.B.A.; visualization, A.C.; supervision, A.C. and E.A.B.A.; project administration, A.C. and E.A.B.A.; funding acquisition, A.C. and E.A.B.A. All authors have read and agreed to the published version of the manuscript.
Figure 1.
Localisation of GFP-Foc-STR4 in the root systems of 'FHIA02', 'FHIA25', 'Williams', 'GCTCV-119' and 'Lady Finger' at the onset of infection, 5 to 9 dpi. (A) Macroconidia and microconidia of GFP-Foc-STR4 observed under a confocal microscope. (B) 'FHIA02' at 5 dpi, showing mycelia moving in the xylem of a lateral root, in the presence of microconidia. (C) 'FHIA02' at 5dpi, and with GFP tagged spores and mycelia clearly visualised between cortical cells of the elongation zone and the xylem vessels of the primary root. (D) 'FHIA25' at 5 dpi, strong fluorescence was detected in the xylem parenchyma cells and tracheid of the elongation zone of the fine root. A single mycelium confined to the xylem and multiple microconidia observed in the adjoining xylem vessel. Strong discoloration of parenchyma cells and vascular cylinder was also observed. (E) 'Williams' at 5 dpi, Mycelia networks visualised along the surface of the epidermis of a lateral root and in the cortex region adjacent to a xylem vessel. (F) 'Williams' at 9 dpi, an abundance of microconidia was visualised proliferating in the xylem vessels of a lateral root. (G) 'GCTCV119' at 5 dpi, mycelial networks were detected in the xylem vessels of a lateral root. (H) 'GCTCV119' at 7 dpi, a germ tube clearly visible from spores attempting to penetrate the epidermis of a lateral root. (I) 'GCTCV119' at 7 dpi, movement of GFP tagged mycelia detected within the vasculature of a lateral root. (J) 'Lady Finger' at 8 dpi, GFP mycelia networks were detected in the xylem vessels and the cortex region of a lateral root. (K) A longitudinal section of a lateral root of a non-inoculated 'FHIA02' plant at 5 dpi. (L) A longitudinal section of a lateral root of a non-inoculated 'FHIA25' plant at 6 dpi. Abbreviations are annotated as F2 = 'FHIA02'; F25 = 'FHIA25'; G = 'GCTCV119'; LF = 'Lady Finger'; C = non-inoculated control; ma = macroconidia; c = conidia; h = hyphae; m = mycelium; gt = germ tube. Horizontal bars indicate the scale used to capture the confocal images.
Figure 1.
Localisation of GFP-Foc-STR4 in the root systems of 'FHIA02', 'FHIA25', 'Williams', 'GCTCV-119' and 'Lady Finger' at the onset of infection, 5 to 9 dpi. (A) Macroconidia and microconidia of GFP-Foc-STR4 observed under a confocal microscope. (B) 'FHIA02' at 5 dpi, showing mycelia moving in the xylem of a lateral root, in the presence of microconidia. (C) 'FHIA02' at 5dpi, and with GFP tagged spores and mycelia clearly visualised between cortical cells of the elongation zone and the xylem vessels of the primary root. (D) 'FHIA25' at 5 dpi, strong fluorescence was detected in the xylem parenchyma cells and tracheid of the elongation zone of the fine root. A single mycelium confined to the xylem and multiple microconidia observed in the adjoining xylem vessel. Strong discoloration of parenchyma cells and vascular cylinder was also observed. (E) 'Williams' at 5 dpi, Mycelia networks visualised along the surface of the epidermis of a lateral root and in the cortex region adjacent to a xylem vessel. (F) 'Williams' at 9 dpi, an abundance of microconidia was visualised proliferating in the xylem vessels of a lateral root. (G) 'GCTCV119' at 5 dpi, mycelial networks were detected in the xylem vessels of a lateral root. (H) 'GCTCV119' at 7 dpi, a germ tube clearly visible from spores attempting to penetrate the epidermis of a lateral root. (I) 'GCTCV119' at 7 dpi, movement of GFP tagged mycelia detected within the vasculature of a lateral root. (J) 'Lady Finger' at 8 dpi, GFP mycelia networks were detected in the xylem vessels and the cortex region of a lateral root. (K) A longitudinal section of a lateral root of a non-inoculated 'FHIA02' plant at 5 dpi. (L) A longitudinal section of a lateral root of a non-inoculated 'FHIA25' plant at 6 dpi. Abbreviations are annotated as F2 = 'FHIA02'; F25 = 'FHIA25'; G = 'GCTCV119'; LF = 'Lady Finger'; C = non-inoculated control; ma = macroconidia; c = conidia; h = hyphae; m = mycelium; gt = germ tube. Horizontal bars indicate the scale used to capture the confocal images.
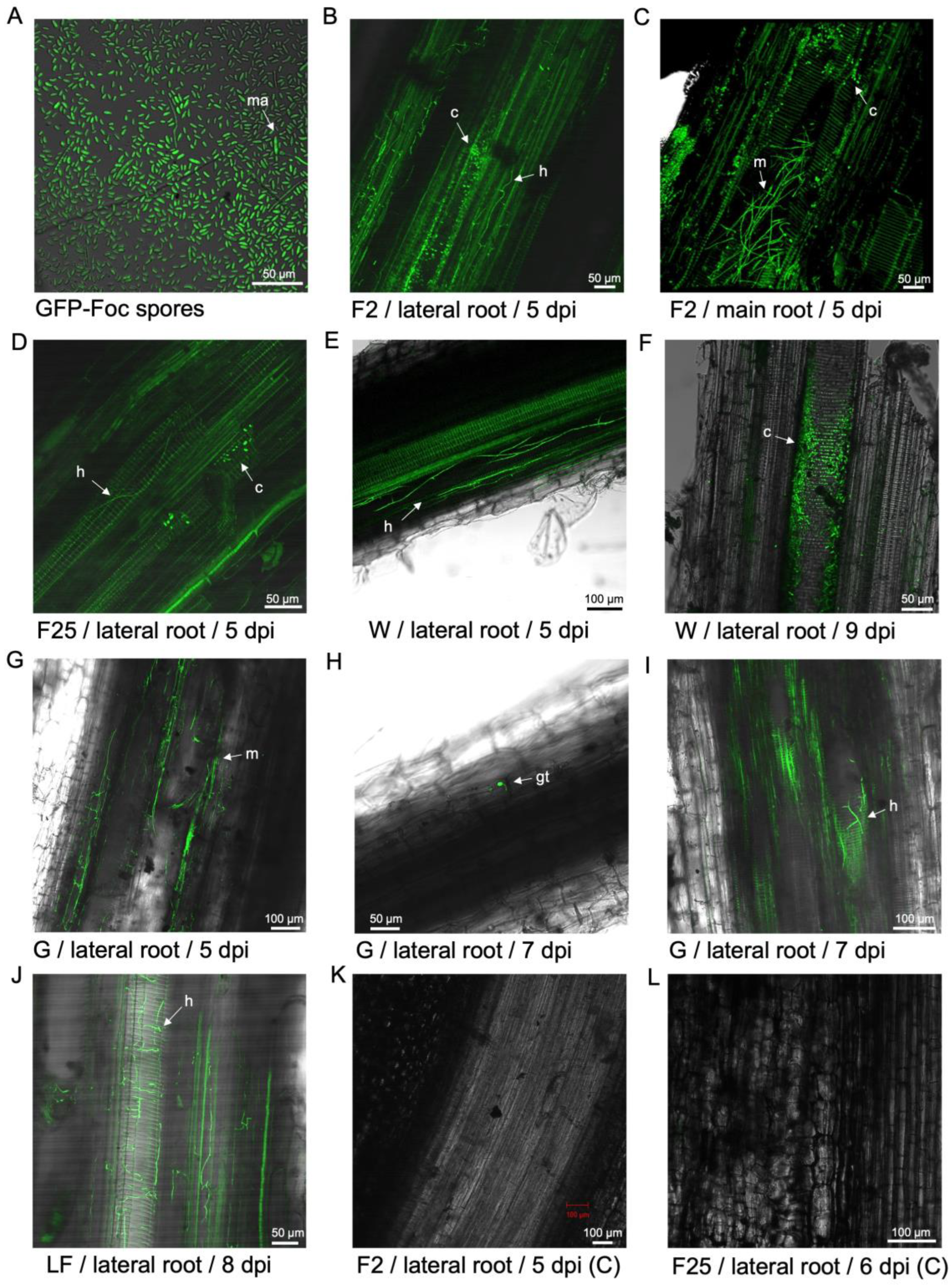
Figure 2.
Localisation of GFP-Foc-STR4 in the susceptible cultivar 'FHIA02'. (A) Microconidia and mycelia moving inter- and intracellularly in the cortex near the xylem vessels of a fine root at 14 dpi. (B) Germinating microconidia, chlamydospores and mycelia present in the epidermis of a lateral root at 14 dpi. (C) Mycelia moving intercellularly in the cortex region of a lateral root. Microconidia produced in false heads on a long monophialide were observed. (D) A wound site was penetrated by the fungus on a lateral root and its movement into the adjacent xylem vessels at 19 dpi. (E) Spore gemination and mycelia colonising the xylem vessels of a fine root at 26 dpi. (F) Spore germination and mycelia movement in the xylem vessels of a primary root at 29 dpi. (G) Spore germination in the xylem of a primary root near a rhizome node, in a region with red discolouration. (H) The movement of mycelia was detected in the cortex region of a main root at 41 dpi. (I) Spore germination and localised mycelia movement in the xylem vessels of the corm joining the main root. (J) Xylem vessels completely colonised by the fungus at 42 dpi. False heads on monophialides were observed. (K) Non-inoculated control of a fine root at 18 dpi. (L) Non-inoculated control of a fine root at 62 dpi. Abbreviations are annotated as F2 = 'FHIA02'; C = non-inoculated control; ma = macroconidia; ch = chlamydospores; c = conidia; h = hyphae; m = mycelium; gt = germ tube. Horizontal bars indicate the scale used to capture the confocal images.
Figure 2.
Localisation of GFP-Foc-STR4 in the susceptible cultivar 'FHIA02'. (A) Microconidia and mycelia moving inter- and intracellularly in the cortex near the xylem vessels of a fine root at 14 dpi. (B) Germinating microconidia, chlamydospores and mycelia present in the epidermis of a lateral root at 14 dpi. (C) Mycelia moving intercellularly in the cortex region of a lateral root. Microconidia produced in false heads on a long monophialide were observed. (D) A wound site was penetrated by the fungus on a lateral root and its movement into the adjacent xylem vessels at 19 dpi. (E) Spore gemination and mycelia colonising the xylem vessels of a fine root at 26 dpi. (F) Spore germination and mycelia movement in the xylem vessels of a primary root at 29 dpi. (G) Spore germination in the xylem of a primary root near a rhizome node, in a region with red discolouration. (H) The movement of mycelia was detected in the cortex region of a main root at 41 dpi. (I) Spore germination and localised mycelia movement in the xylem vessels of the corm joining the main root. (J) Xylem vessels completely colonised by the fungus at 42 dpi. False heads on monophialides were observed. (K) Non-inoculated control of a fine root at 18 dpi. (L) Non-inoculated control of a fine root at 62 dpi. Abbreviations are annotated as F2 = 'FHIA02'; C = non-inoculated control; ma = macroconidia; ch = chlamydospores; c = conidia; h = hyphae; m = mycelium; gt = germ tube. Horizontal bars indicate the scale used to capture the confocal images.

Figure 3.
Localisation of GFP-Foc-STR4 observed in the rhizome of 'FHIA02' during the infection process. (A) Germ tubes of chlamydospores and hyphae moving through cortical cells of a discoloured rhizome at 14 dpi. (B) A discoloured region of a rhizome was associated with a light fluorescence, with chlamydospores and hyphae moving through cortical cells at 14 dpi. (C) Strong fluorescence signals, along with mycelia and microconidia detected in the cortex and the vascular bundles at 14 dpi. (D) Substantial presence of microconidia, germ tubes, hyphae and chlamydospores detected in a vascular bundle at 36 dpi. (E) Extensive mycelia networks established in the xylem vessels at 36 dpi. (F) Tyloses were detected occluding the xylem vessel lumen of a rhizome node, with microconidia and hyphae clearly present in this region at 68 dpi. (G) Two xylem vessels fully occluded by mature tyloses, while microconidia, chlamydospores and mycelia colonised the vessel lumen and along the boundaries of occluding tyloses. (H) Non-inoculated rhizome at 14 dpi. (I) Non-inoculated rhizome showing weak autofluorescence in the vascular regions at 68 dpi. Abbreviations are annotated as F2 = 'FHIA02'; C = non-inoculated control; ch = chlamydospores; c = conidia; h = hyphae; m = mycelium; gt = germ tube; ty = tyloses. Horizontal bars indicate the scale used to capture the confocal images.
Figure 3.
Localisation of GFP-Foc-STR4 observed in the rhizome of 'FHIA02' during the infection process. (A) Germ tubes of chlamydospores and hyphae moving through cortical cells of a discoloured rhizome at 14 dpi. (B) A discoloured region of a rhizome was associated with a light fluorescence, with chlamydospores and hyphae moving through cortical cells at 14 dpi. (C) Strong fluorescence signals, along with mycelia and microconidia detected in the cortex and the vascular bundles at 14 dpi. (D) Substantial presence of microconidia, germ tubes, hyphae and chlamydospores detected in a vascular bundle at 36 dpi. (E) Extensive mycelia networks established in the xylem vessels at 36 dpi. (F) Tyloses were detected occluding the xylem vessel lumen of a rhizome node, with microconidia and hyphae clearly present in this region at 68 dpi. (G) Two xylem vessels fully occluded by mature tyloses, while microconidia, chlamydospores and mycelia colonised the vessel lumen and along the boundaries of occluding tyloses. (H) Non-inoculated rhizome at 14 dpi. (I) Non-inoculated rhizome showing weak autofluorescence in the vascular regions at 68 dpi. Abbreviations are annotated as F2 = 'FHIA02'; C = non-inoculated control; ch = chlamydospores; c = conidia; h = hyphae; m = mycelium; gt = germ tube; ty = tyloses. Horizontal bars indicate the scale used to capture the confocal images.
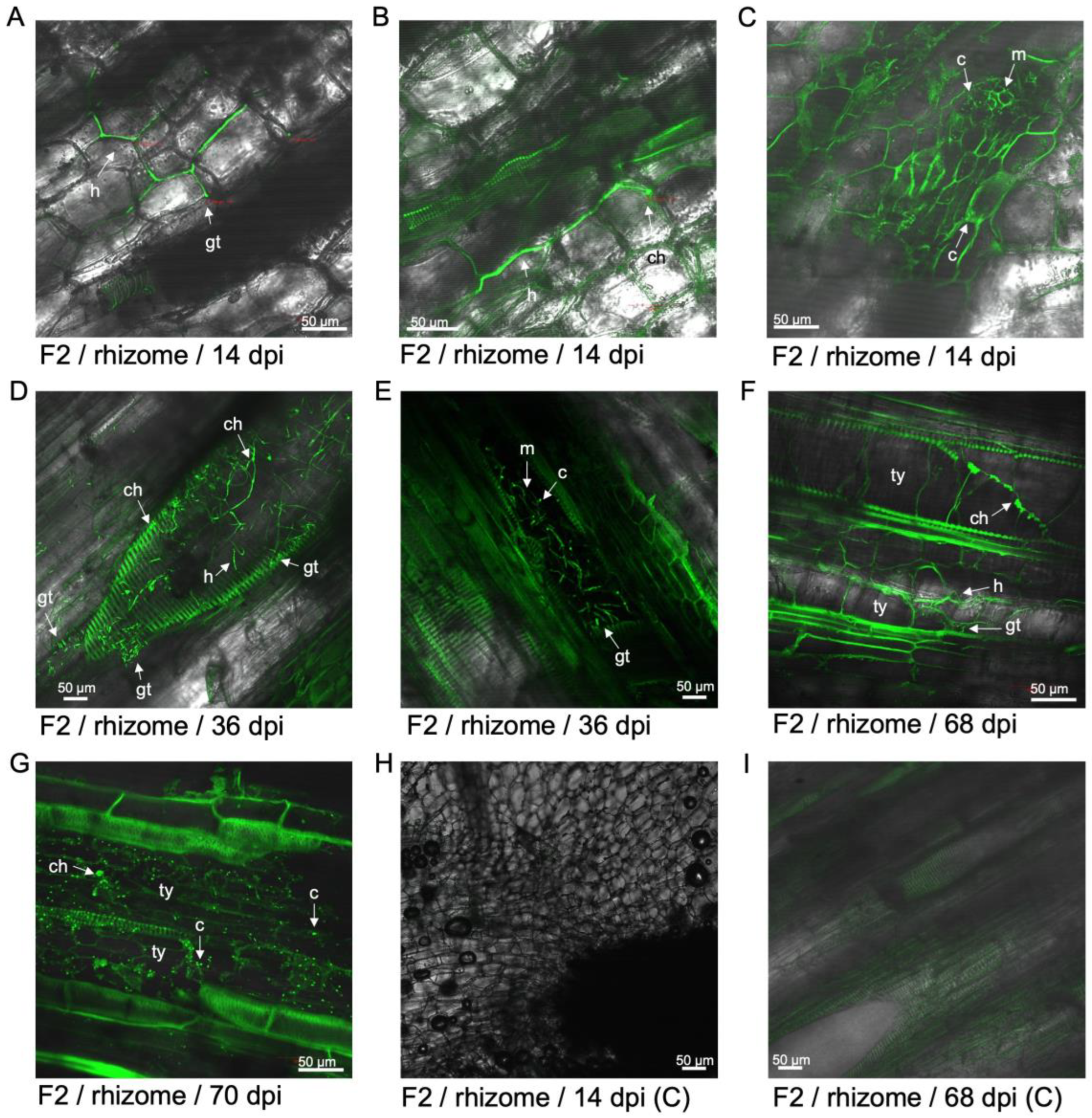
Figure 4.
Localisation of GFP-Foc-STR4 observed in the roots of 'FHIA25' during the infection process. (A) Microconidia and hyphae moving through the epidermis of fine roots in an intercellular manner at 12 dpi. (B) Spore germination and hyphae movement in the tracheid and xylem vessel of fine roots at 12 dpi. (C) Germinating spores and hyphae moving through the xylem vessels of fine roots at 21 dpi. (D) The presence of spores and hyphae detected in the xylem vessels in a lateral root to corm joint at 26 dpi. (E) GFP-Foc-STR4 completely colonised the epidermis of a primary root tip at 29 dpi. (F) Hyphae and germinating spores moving through the xylem vessels of fine roots at 29 dpi. (G) Hyphae and a single terminal chlamydospore detected in the vasculature of a fine root at 36 dpi. (H) GFP fluorescence localised in the vasculature with individual hyphae moving through the cortex. (I) GFP fluorescence associated with hyphae in both the cortex and the vasculature of a root to corm joint at 49 dpi. (J) Non-inoculated lateral roots at 21 dpi. (K) Non-inoculated lateral roots at 36 dpi. (L) Non-inoculated lateral root to corm joint at 51 dpi. Abbreviations are annotated as F25 = 'FHIA25'; C = non-inoculated control; ch = chlamydospores; c = conidia; h = hyphae; m = mycelium; gt = germ tube. Horizontal bars indicate the scale used to capture the confocal images.
Figure 4.
Localisation of GFP-Foc-STR4 observed in the roots of 'FHIA25' during the infection process. (A) Microconidia and hyphae moving through the epidermis of fine roots in an intercellular manner at 12 dpi. (B) Spore germination and hyphae movement in the tracheid and xylem vessel of fine roots at 12 dpi. (C) Germinating spores and hyphae moving through the xylem vessels of fine roots at 21 dpi. (D) The presence of spores and hyphae detected in the xylem vessels in a lateral root to corm joint at 26 dpi. (E) GFP-Foc-STR4 completely colonised the epidermis of a primary root tip at 29 dpi. (F) Hyphae and germinating spores moving through the xylem vessels of fine roots at 29 dpi. (G) Hyphae and a single terminal chlamydospore detected in the vasculature of a fine root at 36 dpi. (H) GFP fluorescence localised in the vasculature with individual hyphae moving through the cortex. (I) GFP fluorescence associated with hyphae in both the cortex and the vasculature of a root to corm joint at 49 dpi. (J) Non-inoculated lateral roots at 21 dpi. (K) Non-inoculated lateral roots at 36 dpi. (L) Non-inoculated lateral root to corm joint at 51 dpi. Abbreviations are annotated as F25 = 'FHIA25'; C = non-inoculated control; ch = chlamydospores; c = conidia; h = hyphae; m = mycelium; gt = germ tube. Horizontal bars indicate the scale used to capture the confocal images.
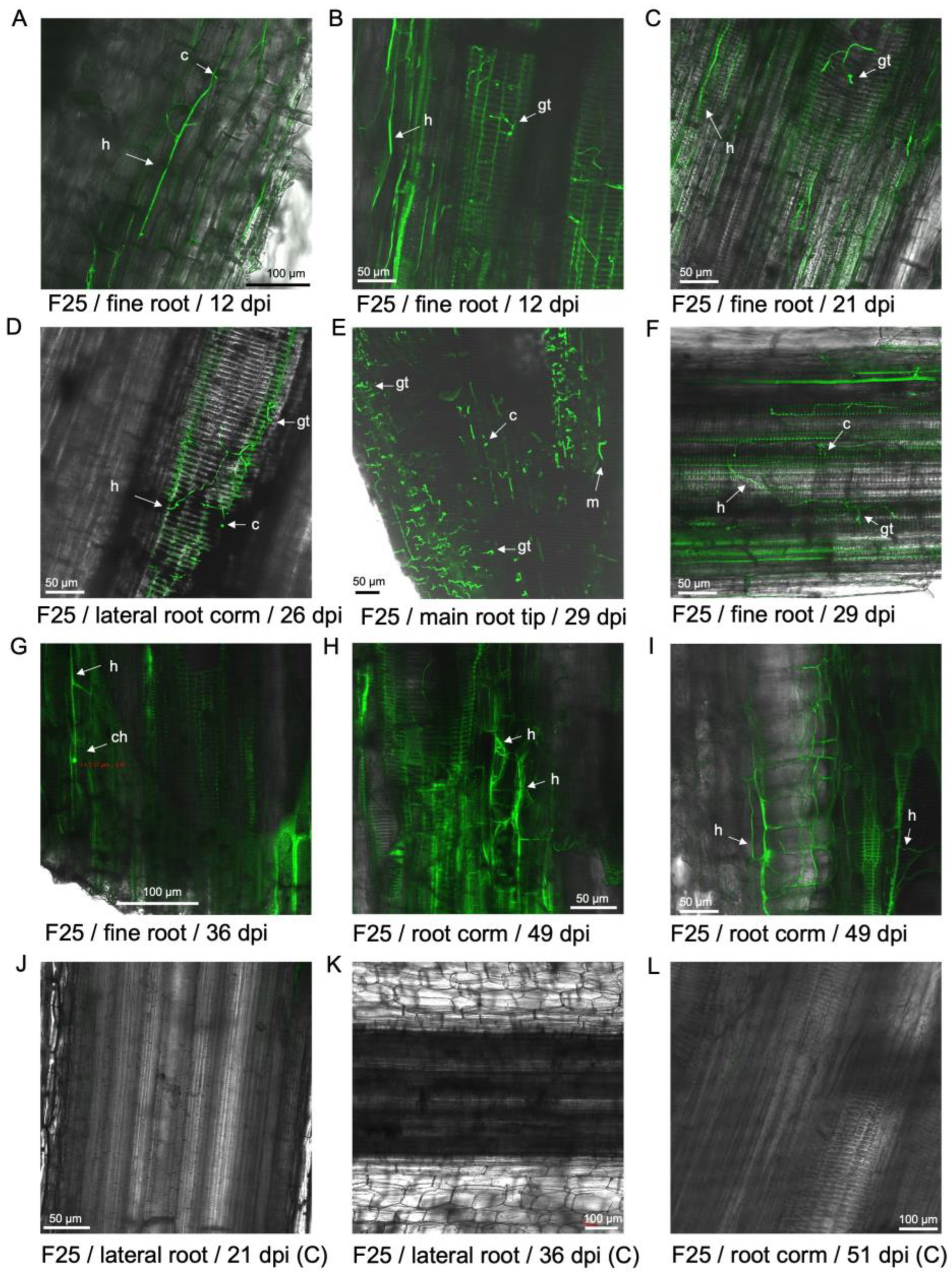
Figure 5.
Examination of GFP-Foc-STR4 in the rhizome of 'FHIA25' during the infection process. (A) Strong fluorescence associated with the rhizome cortex and vascular bundles at 14 dpi. (B) Hyphae was detected in the rhizome cortex at 14 dpi. (C) Individual hyphae were visualised in the vasculature of a rhizome node, with a strong GFP fluorescence signal detected in this region at 36 dpi. (D) Tyloses were visualised in the vascular bundles at 36 dpi. Microconidia were also present. (E) Tyloses were visualised in the xylem vessels of a corm node at 41 dpi. Strong fluorescence in the vasculature and the presence of hyphae were detected. (F) Mycelia networks were visualised in the rhizome node to a primary root at 41 dpi. (G) Tyloses were visualised to occlude a xylem vessel in the rhizome node joining a primary root at 42 dpi. (H) Tyloses were visualised to occlude a xylem vessel in the rhizome node joining a primary root at 62 dpi. (I) Tyloses were visualised to occlude multiple xylem vessels in the rhizome node joining a primary root at 70 dpi. (J) Non-inoculated rhizome at 16 dpi. Weak autofluorescence in the vasculature was detected. (K) Non-inoculated rhizome at 36 dpi. (L) Non-inoculated rhizome at 70 dpi. Abbreviations are annotated as F25 = 'FHIA25'; C = non-inoculated control; c = conidia; h = hyphae; m = mycelium; gt = germ tube; ty = tyloses. Horizontal bars indicate the scale used to capture the confocal images.
Figure 5.
Examination of GFP-Foc-STR4 in the rhizome of 'FHIA25' during the infection process. (A) Strong fluorescence associated with the rhizome cortex and vascular bundles at 14 dpi. (B) Hyphae was detected in the rhizome cortex at 14 dpi. (C) Individual hyphae were visualised in the vasculature of a rhizome node, with a strong GFP fluorescence signal detected in this region at 36 dpi. (D) Tyloses were visualised in the vascular bundles at 36 dpi. Microconidia were also present. (E) Tyloses were visualised in the xylem vessels of a corm node at 41 dpi. Strong fluorescence in the vasculature and the presence of hyphae were detected. (F) Mycelia networks were visualised in the rhizome node to a primary root at 41 dpi. (G) Tyloses were visualised to occlude a xylem vessel in the rhizome node joining a primary root at 42 dpi. (H) Tyloses were visualised to occlude a xylem vessel in the rhizome node joining a primary root at 62 dpi. (I) Tyloses were visualised to occlude multiple xylem vessels in the rhizome node joining a primary root at 70 dpi. (J) Non-inoculated rhizome at 16 dpi. Weak autofluorescence in the vasculature was detected. (K) Non-inoculated rhizome at 36 dpi. (L) Non-inoculated rhizome at 70 dpi. Abbreviations are annotated as F25 = 'FHIA25'; C = non-inoculated control; c = conidia; h = hyphae; m = mycelium; gt = germ tube; ty = tyloses. Horizontal bars indicate the scale used to capture the confocal images.

Figure 6.
Localisation of GFP-Foc-STR4 through the outer leaf sheaths of 'FHIA02' and 'FHIA25'. (A) Microconidia colonising the outer leaf sheaths of 'FHIA02' at 14 dpi. (B) Hyphae moving through xylem vessels of the outer leaf sheaths of 'FHIA02' at 26 dpi. (C) Hyphae moving through the epidermis of outer leaf sheaths of 'FHIA02' at 29 dpi. (D) Edge of a petiole of a senescing showing proliferation of microconidia, hyphae and chlamydospores at 54 dpi. (E) Chlamydospores visualised at the edge of a petiole of a senescing leaf in 'FHIA02' at 62 dpi. (F) Non-inoculated edge of a petiole at 63 dpi. (G) Hyphae and chlamydospore confined to the vasculature of the xylem vessels in the outer leaf sheath of 'FHIA25' at 16 dpi. (H) Hyphae confined to the vasculature in the outer leaf sheath of 'FHIA25' at 21 dpi. (I) Mycelial networks visualised near the vasculatures of a leaf of 'FHIA25' at 29 dpi. (J) Chlamydospores visualised in a discoloured region at the edge of a petiole at the 62 dpi. (K) Chlamydospores and hyphae visualised in the vasculature and surrounding mesophyll cells at the edge of a petiole in 'FHIA25' at 70 dpi. (L) Non-inoculated midrib of a 'FHIA25' leaf at 70 dpi. Abbreviations are annotated as F2 = 'FHIA02'; F25 = 'FHIA25'; C = non-inoculated control; ma = macroconidia; ch = chlamydospores; c = conidia; h = hyphae; m = mycelium; gt = germ tube. Horizontal bars indicate the scale used to capture the confocal images.
Figure 6.
Localisation of GFP-Foc-STR4 through the outer leaf sheaths of 'FHIA02' and 'FHIA25'. (A) Microconidia colonising the outer leaf sheaths of 'FHIA02' at 14 dpi. (B) Hyphae moving through xylem vessels of the outer leaf sheaths of 'FHIA02' at 26 dpi. (C) Hyphae moving through the epidermis of outer leaf sheaths of 'FHIA02' at 29 dpi. (D) Edge of a petiole of a senescing showing proliferation of microconidia, hyphae and chlamydospores at 54 dpi. (E) Chlamydospores visualised at the edge of a petiole of a senescing leaf in 'FHIA02' at 62 dpi. (F) Non-inoculated edge of a petiole at 63 dpi. (G) Hyphae and chlamydospore confined to the vasculature of the xylem vessels in the outer leaf sheath of 'FHIA25' at 16 dpi. (H) Hyphae confined to the vasculature in the outer leaf sheath of 'FHIA25' at 21 dpi. (I) Mycelial networks visualised near the vasculatures of a leaf of 'FHIA25' at 29 dpi. (J) Chlamydospores visualised in a discoloured region at the edge of a petiole at the 62 dpi. (K) Chlamydospores and hyphae visualised in the vasculature and surrounding mesophyll cells at the edge of a petiole in 'FHIA25' at 70 dpi. (L) Non-inoculated midrib of a 'FHIA25' leaf at 70 dpi. Abbreviations are annotated as F2 = 'FHIA02'; F25 = 'FHIA25'; C = non-inoculated control; ma = macroconidia; ch = chlamydospores; c = conidia; h = hyphae; m = mycelium; gt = germ tube. Horizontal bars indicate the scale used to capture the confocal images.

Figure 7.
Scanning electron micrographs of tyloses formation within transverse sectioned primary roots of 'FHIA02'. (A) A transverse section of non-inoculated 'FHIA02' roots at 14 dpi. (B) A magnified view of transverse sectioned roots of non-inoculated 'FHIA02' at 14 dpi. (C) Mycelia and vascular occlusion visualised in the vascular cavities of 'FHIA02' at 14 dpi. (D) Developing tyloses visualised in the vascular cavities of 'FHIA02' at 14 dpi. (E) Extensive mycelial networks visualised in the vascular cavities of 'FHIA02' at 14 dpi. (F) Mature and developing tyloses visualised in the vascular cavities of 'FHIA02' at 14 dpi. (G) Individual occluded xylem cavities, developing tyloses and the presence of hyphae were visualised in the main roots of 'FHIA02' at 42 dpi. (H) Tyloses occluding xylem cavities were visualised in the main roots of 'FHIA02' at 42 dpi. (I) Mature tyloses occluding multiple xylem cavities were visualised in the main roots of 'FHIA02' at 42 dpi. Abbreviations are annotated as F2 = 'FHIA02'; C = non-inoculated control; m = mycelium; ty = tyloses; o = vascular occlusion. Horizontal bars indicate the scale used to capture the confocal images.
Figure 7.
Scanning electron micrographs of tyloses formation within transverse sectioned primary roots of 'FHIA02'. (A) A transverse section of non-inoculated 'FHIA02' roots at 14 dpi. (B) A magnified view of transverse sectioned roots of non-inoculated 'FHIA02' at 14 dpi. (C) Mycelia and vascular occlusion visualised in the vascular cavities of 'FHIA02' at 14 dpi. (D) Developing tyloses visualised in the vascular cavities of 'FHIA02' at 14 dpi. (E) Extensive mycelial networks visualised in the vascular cavities of 'FHIA02' at 14 dpi. (F) Mature and developing tyloses visualised in the vascular cavities of 'FHIA02' at 14 dpi. (G) Individual occluded xylem cavities, developing tyloses and the presence of hyphae were visualised in the main roots of 'FHIA02' at 42 dpi. (H) Tyloses occluding xylem cavities were visualised in the main roots of 'FHIA02' at 42 dpi. (I) Mature tyloses occluding multiple xylem cavities were visualised in the main roots of 'FHIA02' at 42 dpi. Abbreviations are annotated as F2 = 'FHIA02'; C = non-inoculated control; m = mycelium; ty = tyloses; o = vascular occlusion. Horizontal bars indicate the scale used to capture the confocal images.
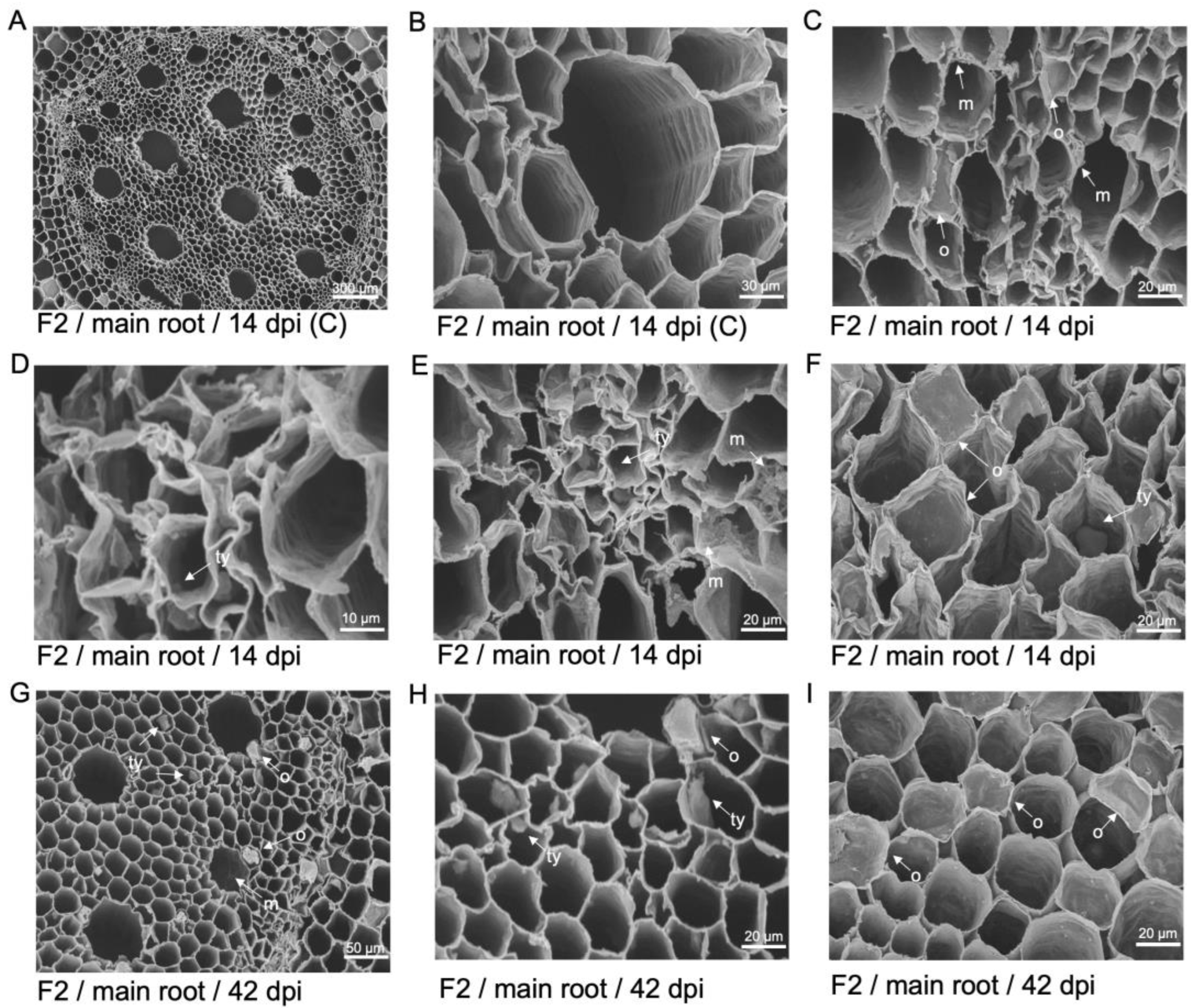
Figure 8.
Scanning electron micrographs of tyloses formation within transverse sectioned primary roots of 'FHIA25'. (A) Vascular bundles of a non-inoculated plant at 14 dpi. (B) A magnified view of the xylem vessels in the primary vascular bundle at 14 dpi. (C) Tyloses formation and mycelia confined to the xylem cavities in the vascular bundles at 14 dpi. (D) Movement of hyphae visualised in the xylem cavities of the vascular bundles at 14 dpi. (E) Tyloses formation and vascular occlusion visualised in the xylem cavities of the vascular bundles at 14 dpi. (F) A developing tylosis and mycelial networks visualised in the xylem cavities at 14 dpi. (G) Extensive mycelia networks visualised in the xylem vessels at 42 dpi. (H) Mature and developing tyloses visualised occluding the xylem cavities at 42 dpi. (I) Mature tyloses occluding multiple xylem cavities at 42 dpi. F25 = 'FHIA25'; C = non-inoculated control; m = mycelium; ty = tyloses; o = vascular occlusion. Horizontal bars indicate the scale used to capture the confocal images.
Figure 8.
Scanning electron micrographs of tyloses formation within transverse sectioned primary roots of 'FHIA25'. (A) Vascular bundles of a non-inoculated plant at 14 dpi. (B) A magnified view of the xylem vessels in the primary vascular bundle at 14 dpi. (C) Tyloses formation and mycelia confined to the xylem cavities in the vascular bundles at 14 dpi. (D) Movement of hyphae visualised in the xylem cavities of the vascular bundles at 14 dpi. (E) Tyloses formation and vascular occlusion visualised in the xylem cavities of the vascular bundles at 14 dpi. (F) A developing tylosis and mycelial networks visualised in the xylem cavities at 14 dpi. (G) Extensive mycelia networks visualised in the xylem vessels at 42 dpi. (H) Mature and developing tyloses visualised occluding the xylem cavities at 42 dpi. (I) Mature tyloses occluding multiple xylem cavities at 42 dpi. F25 = 'FHIA25'; C = non-inoculated control; m = mycelium; ty = tyloses; o = vascular occlusion. Horizontal bars indicate the scale used to capture the confocal images.
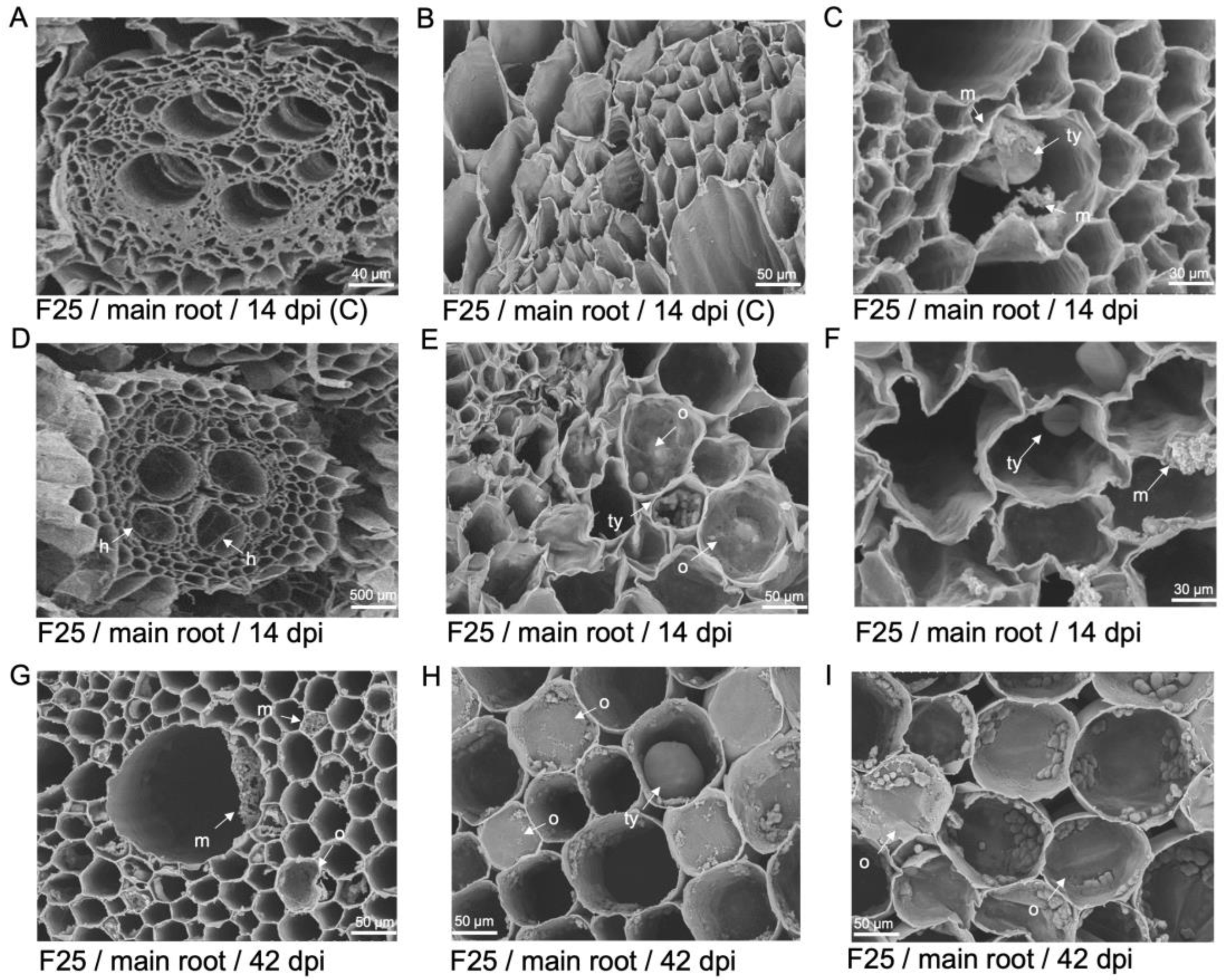
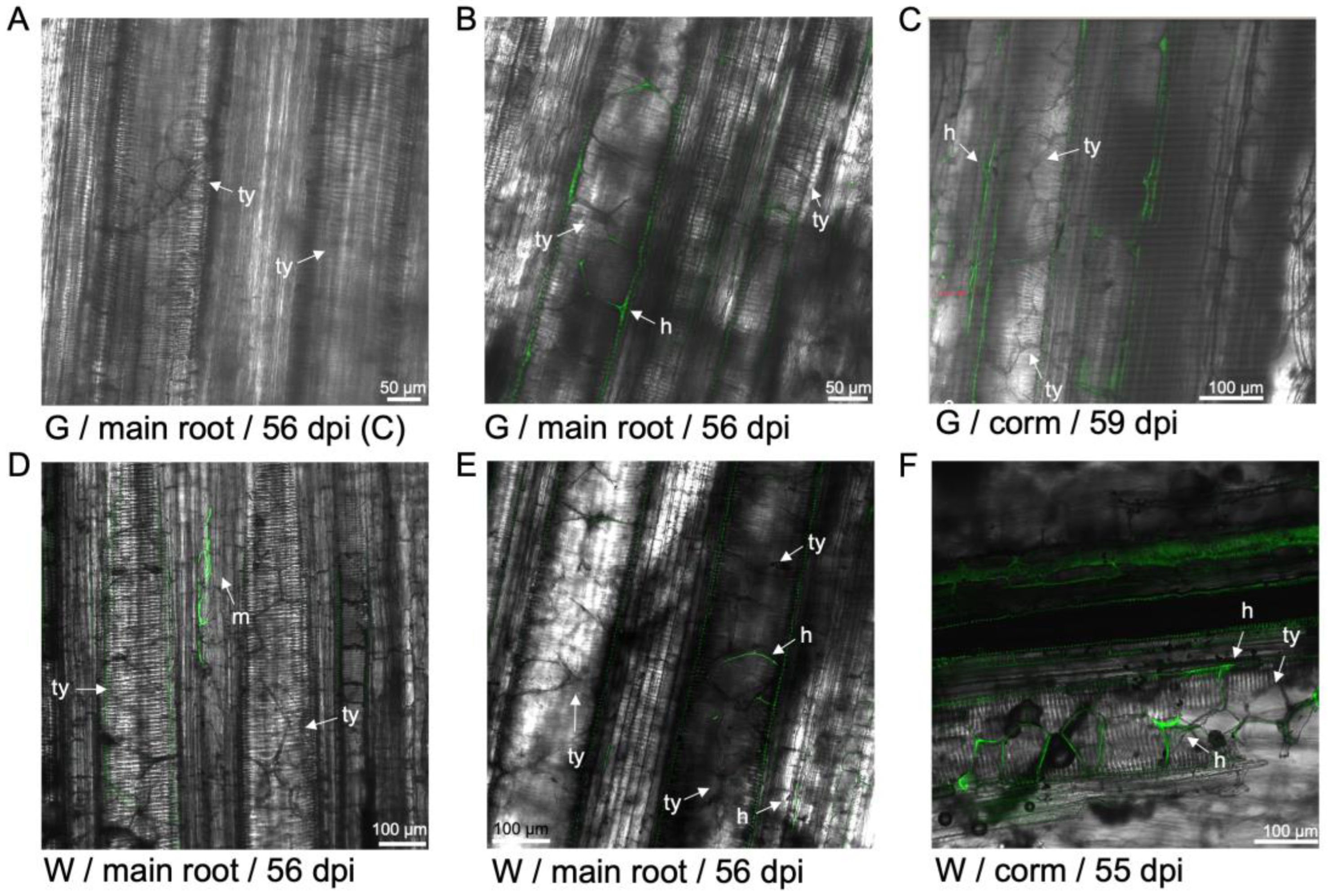
Figure 9.
Tyloses formation and GFP-Foc-STR4 movement in the roots and corm of 'GCTCV119' and 'Williams'. (A) Tyloses visualised in the xylem vessels of a non-inoculated 'GCTCV119' plant at 56 dpi. (B) Movement of hyphae through tyloses was visualised at 56 dpi. (C) Tyloses visualised in the xylem vessels with hyphae present in the same space at 59 dpi. (D) Hyphae confined to the spaces between two xylem vessels in the primary root of 'Williams' at 56 dpi. (E) Hyphae moving in between tyloses in the vasculature of the main root of 'Williams' at 56 dpi. (F) Hyphae moving through the spaces between individual tyloses in the vasculature of 'Williams' at 55 dpi. Abbreviations are annotated as G = 'GCTCV119'; W = 'Williams'; C = non-inoculated control; h = hyphae; m = mycelium; ty = tyloses. Horizontal bars indicate the scale used to capture the confocal images.
Figure 9.
Tyloses formation and GFP-Foc-STR4 movement in the roots and corm of 'GCTCV119' and 'Williams'. (A) Tyloses visualised in the xylem vessels of a non-inoculated 'GCTCV119' plant at 56 dpi. (B) Movement of hyphae through tyloses was visualised at 56 dpi. (C) Tyloses visualised in the xylem vessels with hyphae present in the same space at 59 dpi. (D) Hyphae confined to the spaces between two xylem vessels in the primary root of 'Williams' at 56 dpi. (E) Hyphae moving in between tyloses in the vasculature of the main root of 'Williams' at 56 dpi. (F) Hyphae moving through the spaces between individual tyloses in the vasculature of 'Williams' at 55 dpi. Abbreviations are annotated as G = 'GCTCV119'; W = 'Williams'; C = non-inoculated control; h = hyphae; m = mycelium; ty = tyloses. Horizontal bars indicate the scale used to capture the confocal images.