Graphical Abstract
1. Introduction
Biological microchips (microarrays) with immobilized protein molecules are a rapidly developing field of biotechnology for the creation medical test systems and for use in basic research. The development of rapid diagnostics for socially significant diseases, including the detection of allergic reactions, is currently an important task. An example of the successful use of biological microchips with immobilized allergen molecules in allergy diagnostics is the technology ImmunoCAP
® ISAC [
1,
2,
3].
When immobilizing protein molecules (allergens, immunoglobulins and other proteins) as probes, it is very important to create conditions in which these molecules would retain their native properties and do not denature during the production and storage of microchips. A suitable medium for proteins is a low-density acrylamide gel [
4,
5].
Protein microchips, containing protein molecules immobilized in three-dimensional (3D) polyacrylamide gel elements on glass surface are currently produced only for experimental studies [
6,
7,
8,
9]. Unlike protein microchips, biological microchips with DNA molecules immobilized in 3D gel elements on glass or plastic have found wide application for the diagnosis of socially significant diseases, especially for the diagnosis of resistance of Mycobacterium tuberculosis to antibiotics used in the treatment of this disease. In Russian Federation, a TB biochip containing 77 gel cells has passed the appropriate certification and has been included in the laboratory diagnostic complex [
10]). TB biochip is successfully used in more than 20 tuberculosis facilities. A further development of the TB biochip was the biochip-based TB Test system, which is capable of detecting the resistance of Mycobacterium tuberculosis to the main drugs used to treat tuberculosis – rifampicin, isoniazid, ethambutol, fluoroquinolones, aminoglycosides and capreomycin [
11].
In an alternative method of immobilization of biomolecules on microchips, which has been demonstrated for hybridization analysis of DNA molecules, probe molecules are attached to biopolymers grown on the basis of acrylic acid and acrylamide in the form of brushes on a plastic surface [
12,
13].
In order to use protein microchips as a diagnostic tool for various socially significant diseases, for example, for allergy diagnostics based on the analysis of immunoglobulins E in patient serum for the specificity of binding to immobilized biochip allergens, it is necessary to improve the available technology in several parameters and to overcome a number of drawbacks.
One such significant drawback is the slow kinetics of the interaction of allergen and other antigen molecules immobilized in the biochip gel elements with specific human serum immunoglobulins (sIgE). In previous studies, the incubation time of allergens and other biochip antigens with blood serum was 20 hours, which was necessary for further detection of fluorescent signals from the biochip elements [
9,
14,
15,
16]. However, the duration of the immunoassay in a commercial test system should not exceed one working day, or approximately eight hours.
The second significant drawback is associated with the need to switch to the use of a plastic substrate consisting of a polybutylene terephthalate polymer as a biochip substrate, which has been successfully proven to be commercially suitable for the production of DNA microchips [
17]. The fact is that such a substrate is produced by high-pressure casting at high temperatures, as a result of which the surface of such a substrate becomes largely inert. The content of chemical groups necessary for reliable attachment of gel elements with protein to such a surface turns out to be negligible.
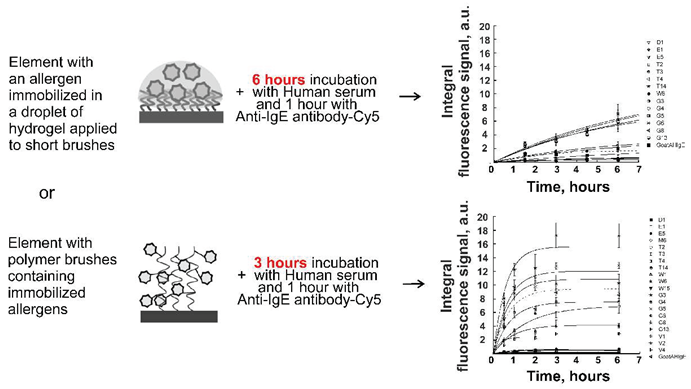
In our work, we describe a 3D hydrogel microchip containing hydrogel droplets with protein molecules (allergens, immunoglobulins and others) disposed on elements consisting of short polymer brushes grafted from a surface made of polybutylene terephthalate polymer. Protein molecules were immobilized during copolymerization induced by short-wave UV radiation. On such a biochip, the kinetics of allergen-sIgE complex formation reached 70% of saturation for 6 hours, and the immunoassay took 7.5 hours. We also offer a 3D brush microchip containing on the surface of a polyethylene terephthalate polymer in brush elements with protein molecules (allergens, immunoglobulins and others) covalently immobilized by opening oxirane cycles with nucleophilic groups contained in the protein (amino and thiol groups). In the case of 3D brush microchip the kinetics of allergen-sIgE complex formation reached 100% of saturation for 3 hours, and the immunoassay was performed for 5 hours.
2. Results
2.1. Immunofluorescence Analysis on a 3D Hydrogel Biochip
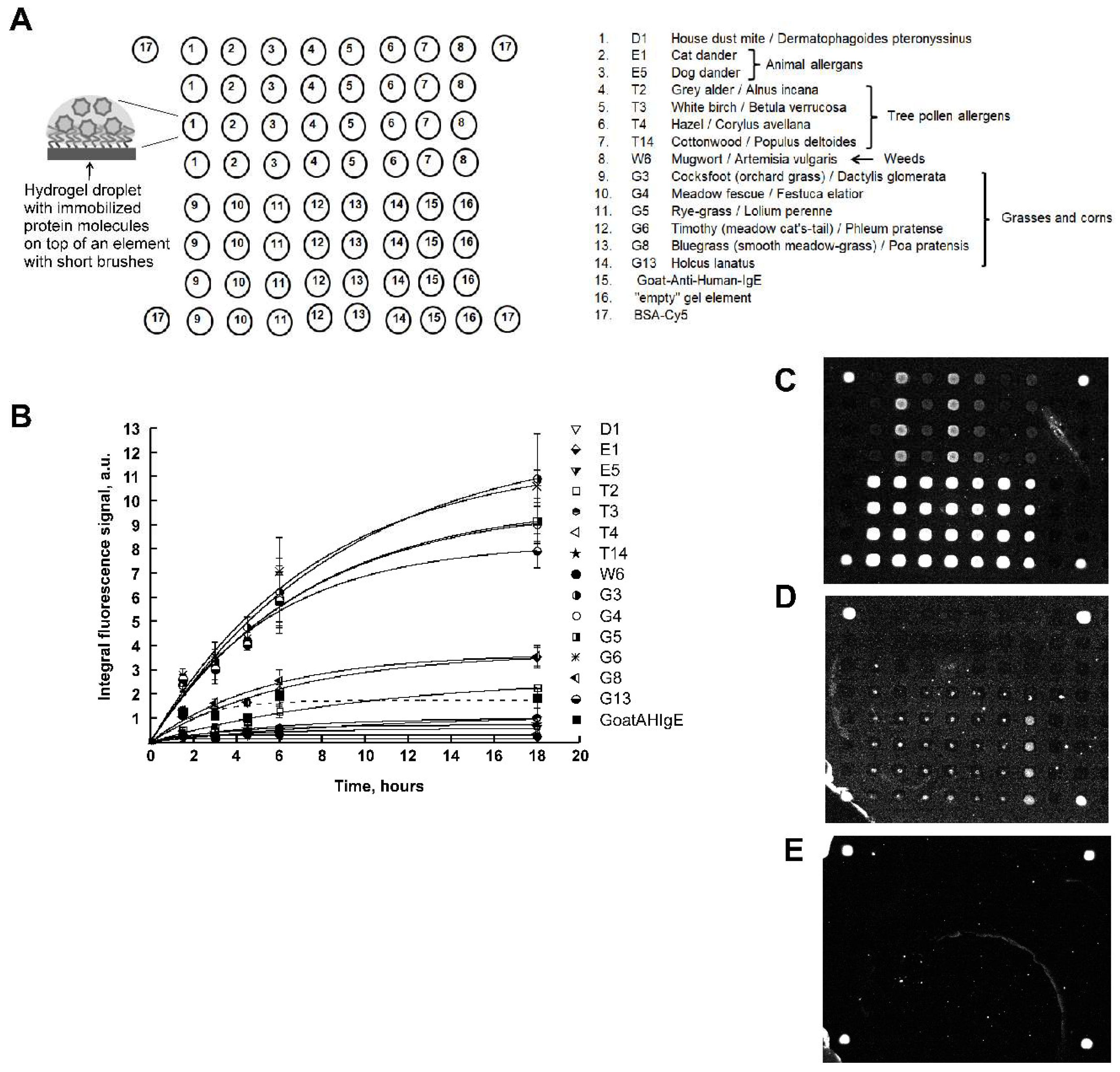
Figure 1A shows a scheme of the developed 3D hydrogel biochip. To manufacture such a biochip, at first, by photolithography, a matrix of polymers in the form of short brushes was made from brush polymers based on polyethelene glycol methacrylate. Polymer brushes were covalently fixed at one end on the surface of polybutylene terephthalate plastic [
12,
13]. Then, using a robot with a pin, an aqueous methacrylamide-glycerin-salt solution of the protein was applied, with the addition of TEMED as a polymerization catalyst for the gel elements of the biochip. The immobilization of biomolecules occurred in the process of co-polymerization induced by short-wave UV radiation with a maximum at a wavelength of 312 nm.
As
Figure 1A shows, the biochip contained 14 immobilized allergens:
house dust mite D1,
animal allergens E1 (cat dander), E5 (dog dander),
tree pollen allergens T2 (grey alder), T3 (white birch), T4 (hazel), T14 (cottonwood),
weeds W6 (mugwort),
grasses and corns G3 (cocksfoot), G4 (meadow fescue), G5 (rye-grass), G6 (timothy), G8 (bluegrass) and G13 (holcus lanatus),
each of which was immobilized in four repeats. Also, the biochip contained goat polyclonal antibodies to human IgE (Goat Anti-Human-IgE), “empty” gel elements and the protein BSA-Cy5 as a marker.
Figure 1B shows the kinetics curves of the interaction of allergens with specific IgE of human blood serum. As one can see, according to the kinetics curves, the time of saturation of the fluorescent signals was 10 hours and the time for the fluorescent signal to reached 70% from saturation was approximately 6 hours for each allergen.
Also in
Figure 1B, a broken line shows the kinetics curve of the interaction of immobilized specific IgG (Goat Anti-Human-IgE) with serum IgE. As can be seen from the figure, the saturation time for the formation of complexes from antibodies was 4 hours.
Figure 1C-E shows images of hydrogel biochips after 6-hour incubation with a control positive (inhalation) serum (CS+) (
Figure 1C), a control negative serum (CS-) (
Figure 1D) or with a zero calibration sample (СS0) (
Figure 1E), each followed by a 1-hour incubation in the presence of Goat Anti-Human-IgE-Cy5.
In the case of incubation with CS+, it is seen that elements containing allergens and Goat Anti-Human-IgE fluoresced with different intensities. Thus, CS+ contains sIgE to biochip allergens.
In the case of incubation with CS-, the gel elements of the biochip containing immobilized Goat Anti-Human-IgE antibodies fluoresced (
Figure 1D), which is consistent with the composition of CS-.
In the case of CS0, only marker elements containing immobilized BSA–Cy5 fluoresced (
Figure 1E), which is consistent with the composition of CS0.
It should also be noted that in all images of biochips in
Figure 1 C-E there are no signals from “empty” gel elements.
2.2. Immunofluorescence Analysis on a 3D Brush Biochip
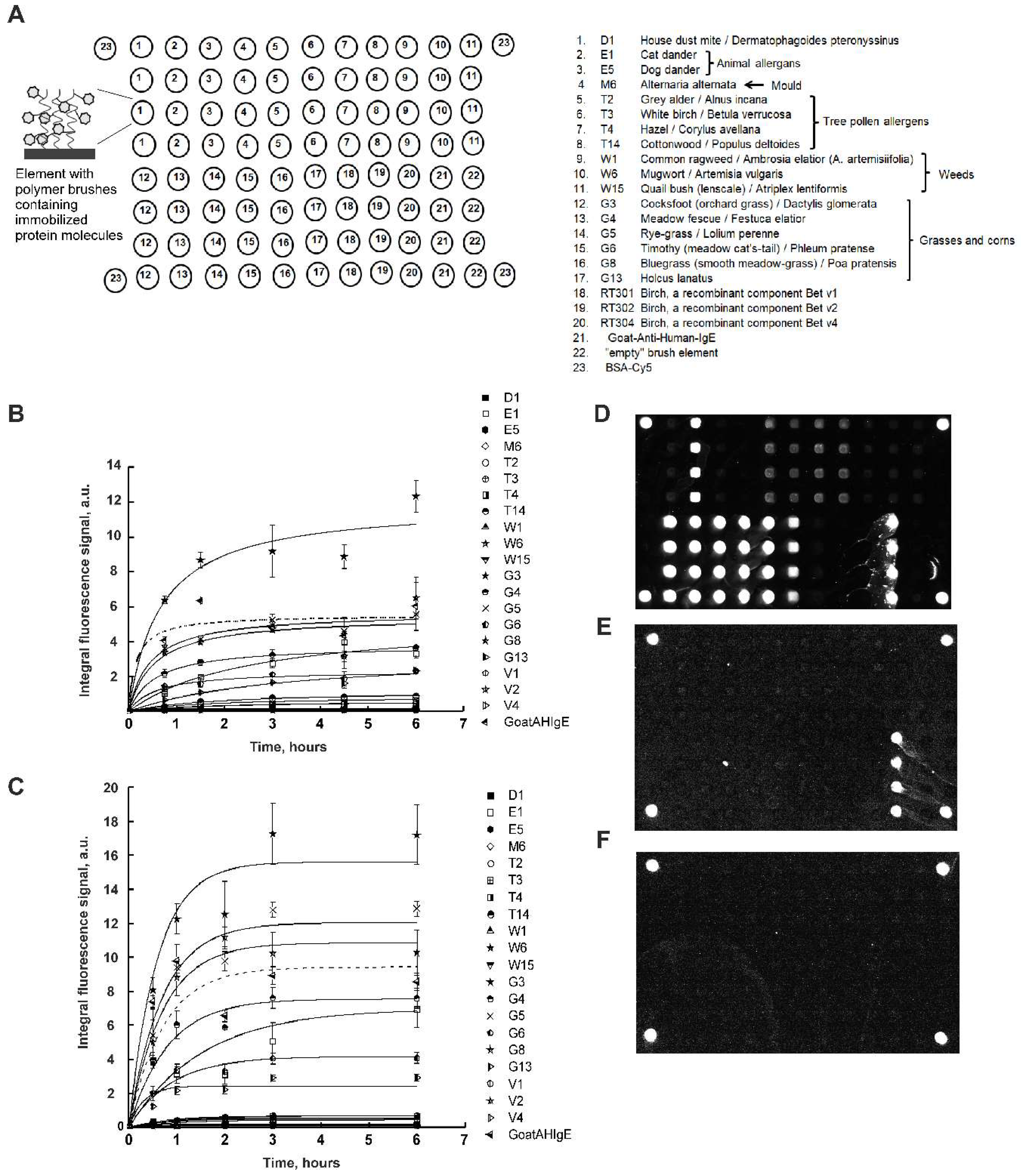
Figure 2A shows a scheme of the developed 3D brush biochip. To manufacture such a biochip, at first, by photolithography, a matrix of polymers in the form of brushes was made from monomers of glycidyl methacrylate. Epoxy groups are able to react with amino and thiol groups in the protein without additional activation. Then, using a robot with a pin an aqueous glycerin protein solution was applied, also containing triethylamine, as a catalyst for the opening of the oxirane cycle.
As
Figure 2A shows, the biochip contained 20 immobilized allergens:
house dust mite D1,
animal allergens E1 (cat dander), E5 (dog dander),
mould M6 (Alternaria alternate)
tree pollen allergens T2 (grey alder), T3 (white birch), T4 (hazel), T14 (cottonwood),
weeds W1 (common ragweed), W6 (mugwort), W15 (quail bush (lenscale)),
grasses and corns G3 (cocksfoot), G4 (meadow fescue), G5 (rye-grass), G6 (timothy), G8 (bluegrass) and G13 (holcus lanatus),
recombinant components of birch Bet v1, Bet v2 and Bet v4,
each of which was immobilized in four repeats. Also, the biochip contained goat polyclonal antibodies to human IgE (Goat Anti-Human-IgE), “empty” gel elements and the protein BSA-Cy5 as a marker.
Figure 2B shows the kinetics curves of the interaction of allergens with specific IgE of human blood serum. As one can see, according to the kinetics curves, the time of saturation of the fluorescent signals was from 3 to 6 hours.
Figure 2C shows the kinetics curves of the interaction of allergens with specific IgE of human blood serum obtained with mixing. As one can see, according to the kinetics curves, the time of saturation of the fluorescent signals was 3 hours. At the same time, the fluorescent saturation signals increased 1.5 times, which made it possible to increase the sensitivity of the method.
Figure 2D-F shows images of hydrogel biochips after 3-hour incubation with a control positive (inhalation) serum (CS+) with mixing (
Figure 2D), a control negative serum (CS-) with mixing (
Figure 2E) or with a zero calibration sample (СS0) with mixing (
Figure 2F), as well as a 1-hour incubation in the presence of Goat Anti-Human-IgE-Cy5.
In the case of incubation with CS+, it is seen that elements containing allergens and Goat Anti-Human-Age fluoresced with different intensities. Thus, the biochip revealed sIgE in CS+.
In the case of incubation with CS-, the gel elements of the biochip containing immobilized Goat Anti-Human-IgE antibodies fluoresced (
Figure 2E), which is consistent with the composition of CS-.
In the case of CS0, only marker elements containing immobilized BSA–Cy5 fluoresced (
Figure 2F), which is consistent with the composition of CS0.
It should also be noted that in all images of biochips in
Figure 2 D-F there are no signals from “empty” gel elements.
2.3. Analysis of Fluorescent Signals on 3D Hydrogel and 3D Brush Biochips, Comparison with ELISA Results
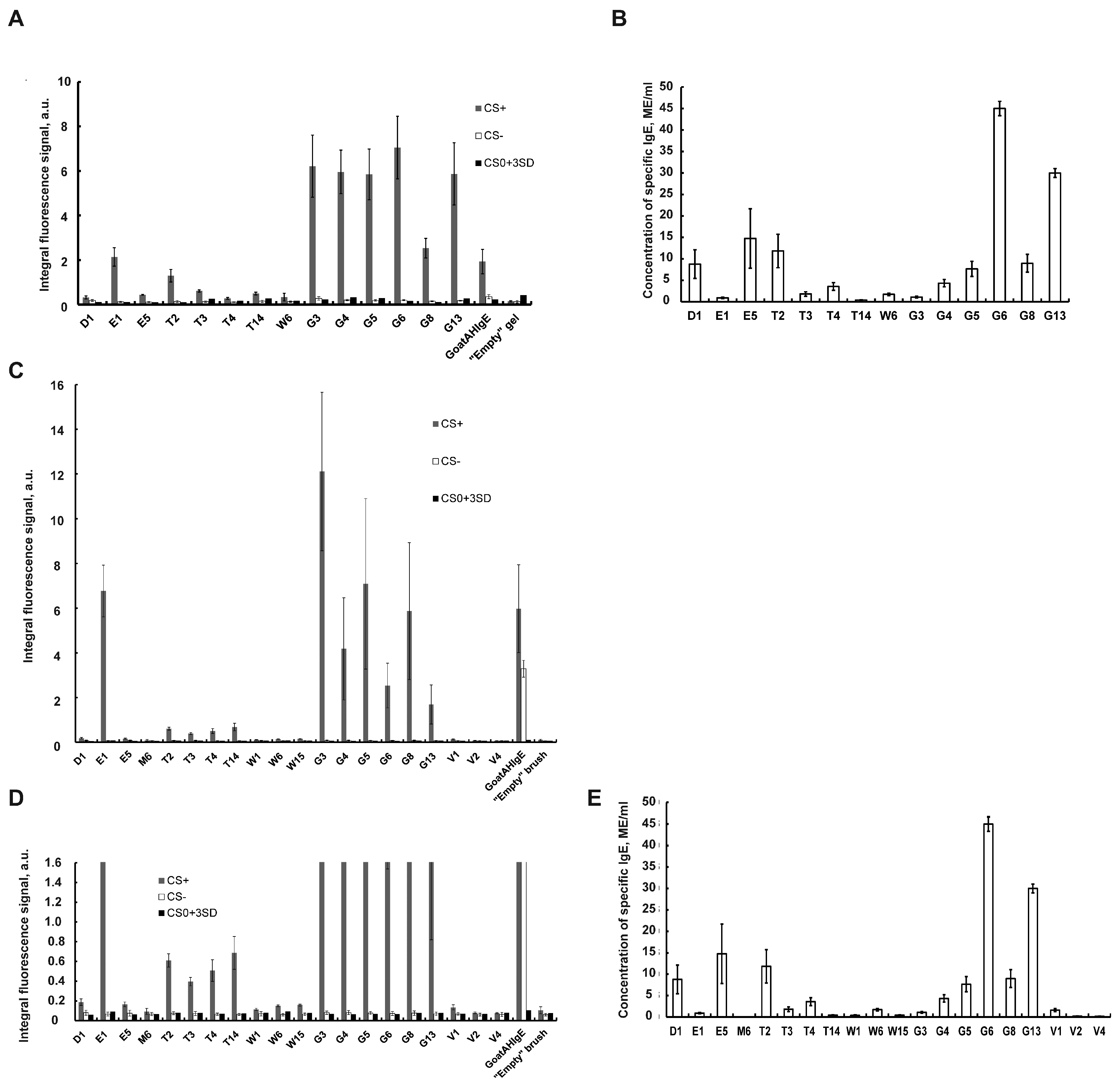
Figure 3A shows a comparative histogram of the averaged fluorescent signals of gel elements with immobilized allergens, goat polyclonal antibodies and an “empty” gel in the case of incubation for 6 hours with a control positive inhalation, CS+ (gray columns), with a control negative serum (CS-) (white columns) and with a zero calibration sample CS0 (black columns) for each allergen of the 3D hydrogel biochip. As can be seen from the histogram, the values of fluorescent signals for gel elements with 14 biochip allergens after incubation with CS+ exceed the values of fluorescent signals in the same gel elements after incubation with CS- or with CS0, as well as the values of signals from “empty” gel elements. Consequently, CS+ contains sIgE to all biochip allergens.
Figure 3B shows a histogram of the concentrations of sIgE in CS+ determined by enzyme immunoassay (ELISA) for the same 14 allergens. As can be seen from the histogram, the sIgE concentration values for all 14 allergens exceeded the value of 0.35 IU/ml. Thus the ELISA method confirmed the result obtained using a 3D hydrogel biochip, namely the presence of sIgE in CS+ to all 14 allergens considered.
Thus, the analysis of human blood serum using a hydrogel biochip (in which the fluorescent signal of the biochip gel elements reaches 70 percent saturation in 6 hours of serum incubation) and the ELISA analysis gave the same qualitative result on the presence of antibodies in the control positive serum to 14 allergens. Therefore, achieving 70% saturation of the fluorescent signals in the cells of the biochip is sufficient to determine the presence of sIgE in the serum.
Taking into account the 1-hour development of allergen-sIgE complexes using Goat Anti-Human-IgE-Cy5 antibody, the total time for the analysis of blood serum for sIgE to allergens using a hydrogel biochip is 7.5 hours. At the same time, the amount of serum required for the analysis of 14 allergens on a biochip (100 µl) is seven times less than for the analysis of 14 allergens by the ELISA method (50µl x 14 = 700 µl).
Figure 3C shows a histogram of the averaged fluorescent signals of brush elements with immobilized allergens, goat polyclonal antibodies and an “empty” gel in the case of incubation for 3 hours with a control positive inhalation, CS+ (gray columns), with a control negative serum (CS-) (white columns) and with a zero calibration sample CS0 (black columns) with mixing for each allergen of the 3D brush biochip. Additionally
Figure 3D shows the increased y-axis scale at low values of fluorescence signals. As can be seen from the histogram, the values of fluorescent signals for gel elements with 17 biochip allergens after incubation with CS+ exceed the values of fluorescent signals in the same gel elements after incubation with CS- or with CS0, as well as the values of signals from “empty” gel elements. Consequently, CS+ contains sIgE to 17 biochip allergens.
Meanwhile, fluorescence signals from brush elements with three allergens, M6, Bet v2 and Bet v4, do not exceed the values of fluorescent signals in the same gel elements after incubation with CS- or with CS0, as well as the values of signals from “empty” gel elements (
Figure 3D). Thus, CS+ doesn’t contain sIgE to allergens M6, Bet v2 and Bet v4.
Figure 3E shows a histogram of the concentrations of sIgE in CS+ determined by enzyme immunoassay (ELISA) for the same 20 allergens. As can be seen from the histogram, the sIgE concentration values for 17 allergens exceeded the value of 0.35 IU/ml. Meanwhile, concentration of three allergens, M6, Bet v2 and Bet v4, is lower than 0.35 IU/ml. Thus the ELISA method confirmed the result obtained using 3D brush biochip.
3. Discussion
In this work, two possibilities to produce biochips with immobilized molecules of allergens, antibodies and other proteins were demonstrated. In the case of 3D hydrogel biochip, proteins are immobilized in the environment of acrylamide gel elements under short-wave UV irradiation. This variant of biochip manufacturing is similar to the previously used technique (4-9, 14-16), but has a number of changes. If previously hydrogel droplets were attached to the surface of glass, then in our technique hydrogel droplets with protein are strongly attached to short brushes “grown” on the surface of plastic made of polybutylene terephthalate polymer.
The second significant difference is the shorter wavelength of irradiation during polymerization of gel cells with protein. If earlier (4-9, 14-16) a wavelength of 350 nm was used, then in our work gel elements were irradiated at a wavelength of 312 nm, which contributed to a more optimal polymerization of hydrogel elements, since the saturation time of fluorescent signals in gel elements was reduced from 20 hours (9,14-16) to 10 hours (
Figure 1B).
In the second variant of manufacturing a protein biochip, allergens, antibodies and other proteins are immobilized in the volume of acrylamide brushes due to the formation of covalent bonds in one-step immobilization of protein molecules, greatly simplifying the manufacturing stages. It should be noted that under UV irradiation used in 3D hydrogel biochips manufacturing, there is a risk of partial denaturation of proteins, which can affect their structural integrity and activity. Compared with 3D hydrogel biochips, protein macromolecules of 3D brush biochips are immobilized in more gentle conditions, without exposure to short-wave radiation.
Also, brush polymers made it possible to achieve 2-3 times higher fluorescent signals in the elements of 3D brush biochip than in the elements of 3D hydrogel biochip.
Another significant advantage of 3D brush biochip is the faster kinetics of the interaction of biomolecules in the brush elements of the biochip. In the brush elements, the fluorescent saturation signal is achieved in 3 hours (
Figure 2B), while in hydrogel elements, the saturation time of the fluorescent signal is 10 hours (
Figure 1B). With mixing, the saturation time of the fluorescent signal in the brush elements increase in two times or remains approximately the same for some allergens, while the fluorescence signals increase by 1.5 times. The growth of signals in the brush elements during mixing apparently occurs due to a more optimal penetration of biomolecules of the solution to the immobilized probes.
Thus, our work demonstrates an updated version for the manufacture of a 3D hydrogel protein biochip, and a new variant for the manufacture of a 3D brush protein biochip. The possibility of conducting immunofluorescence analysis on these biochips has also been demonstrated. The results obtained on 3D hydrogel and brush biochips concerning the detection of sIgE to allergens qualitatively coincide with the results obtained by the ELISA method. It is shown that the 3D brush biochip is the most optimal, since the saturation time of the fluorescent signal in the brush elements is reduced to 3 hours while the fluorescent signals are more than two times higher.
It can be concluded that 3D brush biochips are more suitable as the instrument for further analysis of protein-protein and other biomolecular interactions.
4. Materials and Methods
4.1. Manufacture of 3D Hydrogel Protein Biochips
Polymer brushes of polyethylene glycol methacrylate were grafted from a plastic substrate made of polybutylene terephthalate polymer by UV-induced (λ=254 nm) photolithography using quartz photomask.
To the resulting matrix of brush elements (200 x 200 microns) using a robot QArray 2 (Genetix, UK) the solutions of allergens D1, E1, E5, T2, T3, T4, T14, W6, G3, G4, G5, G6, G8 or G13, (0.4-3 mg/ml, Greer Laboratories, USA) or polyclonal antibodies to human IgE (0.3 мг/мл, Bethyl Laboratories, USA) in the volume of an acrylamide gel with the addition of glycerin and TEMED (3.7% methacrylamide, 0.11% N,N′-methylenebisacrylamide, 50% glycerin, 3.6 % TEMED) were applied. Similar solutions containing BSA-Cy5 (0.3 mg/ml, Sigma, USA), were applied in the corners of the biochip, see the scheme of the biochip in
Figure 1A.
Next, the biochips were irradiated at a wavelength of 312 nm using Vilber UV-B T-15M lamp (Vilber, USA) for 20 minutes in a nitrogen atmosphere at 22°С. Then, the biochips were washed in the PBST buffer (0,1% Tween, 150 mM NaCl, 10mM Na-Phosphate buffer, pH 7,2) for 20 minutes with stirring at 150 r.p.m. at room temperature. The biochips were washed with distilled water and dried in an air stream. After these procedures, the biochips were ready for use.
4.2. Conducting Immunoassay on 3D Hydrogel Biochips
On 3D hydrogel biochips in individual chambers, 100 µl of a solution of control positive inhalation serum (CS+, Alkorbio, Russia) was applied. On other 3D hydrogel biochips in individual chambers, 100 µl of a solution of control negative serum (CS-, Alkorbio, Russia) or 100 µl of a zero calibration sample (CS0, Hema, Russia) were applied.
The biochips were incubated with CS+ for 1.5, 3, 4.5, 6 and 18 hours at 37 °C. The biochips were incubated with CS- and CS0 for 6 hours at 37 °C. Then the biochips were washed with distilled water and dried in an air stream.
Next, the biochips were incubated in 100 µl of polyclonal goat antibodies on human IgE (Bethyl Laboratories, USA), dissolved in PBS buffer (150 mM NaCl, 10mM Na-Phosphate buffer, pH 7,2) at 0.01 mg/ml. Preliminary, goat antibodies were labeled with Cy5 fluorescent dye according to the procedure described earlier [
19]. The biochips were incubated in individual chambers at 37°C for 1 hour.
At the next stage, individual chambers were removed from the biochips, it were rinsed in distilled water and placed in the tubes containing 25 ml of a PBST solution diluted 8 times with water, and incubated in this solution for 20 minutes with stirring 150 r.p.m., thus destroying non-specific interactions of goat antibody.
Then, the biochips were washed with distilled water and dried in an air stream.
4.3. Manufacturing of 3D Brush Protein Biochips
Elements of 3D brush biochips consisted of brush polymers with active epoxy groups on the plastic surface and were obtained on a plastic substrate made of polybutylene terephthalate polymer by UV-induced (λ=254 nm) photolithography using a quartz photomask with 200 x 200 microns holes and monomers based on glycidylmethacrylate.
Aqueous solutions of allergens D1, E1, E5, M6, T2, T3, T4, T14, W1, W6, W15, G3, G4, G5, G6, G8, G13, Bet v1, Bet v2 and Bet v4 (3 mg/ml, Alkorbio, Russia) or polyclonal goat antibodies to human IgE (0.3 мг/мл, Bethyl Laboratories, USA) with the addition of glycerin and triethylamine (TEA) (30% of glycerin, 2% TEA) were applied to the resulting matrix of brush elements using a robot QArray 2 (Genetix, UK). Similar solutions containing BSA-Cy5 (0.3 mg/ml, Sigma, USA) were applied in the corners of the biochip, see the biochip scheme shown in
Figure 2A.
After immobilization, the biochips were washed with a stream of deionized water and washed in a PBST buffer for 20 minutes with stirring at a speed of 150 r.p.m. at room temperature. The biochips were washed with distilled water and dried in an air stream. After these procedures, the biochips were ready for use.
4.4. Conducting Immunoassay on 3D Brush Biochips
For immunoassay, 100 µl of control positive inhalation serum (CS+, Alkorbio, Russia) was applied to the biochip chambers. In order to evaluate the binding kinetics, 3D brush biochips were incubated with a control positive serum at a temperature of 37 °C for 45 minutes, 1.5, 3, 4.5, and 6 hours both without stirring and with stirring at a speed of 300 rpm.
As a control, brush biochips were incubated with 100 µl of a control negative serum solution (CS-, Alkorbio, Russia) or 100 µl of a zero calibration sample (CS0, Hema, Russia) for 3 hours.
After incubation, the biochips were washed with distilled water and dried in an air stream. Next, the biochips were incubated with polyclonal goat antibodies and further treated as described for 3D hydrogel biochips.
4.5. Recording of Fluorescent Images of Biochips
Biochips were recorded in the fluorescence range of Cy5 dye (λabsmax = 648 nm, λemmax = 670 nm) using a fluorescent LED microscope equipped with a CCD camera and software (IMB RAS, Moscow, Russia). Further, the fluorescent images of the biochips were analyzed using the Image Express program (IMB RAS, Moscow, Russia), which made it possible to calculate the total fluorescent signal (integral fluorescent signal) for each element of the biochip.
4.6. CS+ Analysis by Enzyme Immunoassay (ELISA).
Concentrations of IgE specific to allergens D1, E1, E5, M6, T2, T3, T4, T14, W1, W6, W15, G3, G4, G5, G6, G8, G13, Bet v1, Bet v2 and Bet v4 in the control positive inhalation serum, CS+, were determined by the ELISA method using the Alkor bio ELISA kit (Alkor Bio, Russia).
5. Conclusions
Our work demonstrates an updated version of the manufacture of a 3D hydrogel protein biochip, and a new variant for the manufacture of a 3D brush protein biochip. The results obtained using 3D hydrogel and brush biochips concerning the detection of sIgE to allergens qualitatively coincide with the results obtained by the ELISA method. It is shown that the 3D brush biochip is the most optimal, since the saturation time of the fluorescent signal in the brush elements is reduced to 3 hours while the fluorescent signals are more than two times higher.
It can be concluded that 3D brush biochips are more suitable as the instrument for further analysis of protein-protein and other biomolecular interactions.
Author Contributions
Conceptualization, A.V.C. and O.A.Z.; methodology A.V.C., O.A.Z., V.I.B., V.E.B.; investigation, R.A.M., G.F.S., D.A.K., I.Yu.S, V.A.V., S.A.P, V.E.K. and V.E.S.; writing—original draft preparation, O.A.Z, R.A.M., G.F.S.; writing—review and editing, O.A.Z. and A.V.C; supervision, A.V.C.; funding acquisition, A.V.C.. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Russian Science Foundation, grant number 20-14-00287.
Acknowledgments
The authors thank Dr. Marina A. Filippova and Dr. Olga. A. Ivashkina for help in 3D hydrogel microchips manufacturing.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lejeune, S., Bouazza, N., Nicaise, P.R.; Jolaine, V.; Roditis, L.; Marguet, C.; Amat, F.; Berger, P.; Fayon, M.; Dubus, J.C.; Valois, S.; Reix, P.; Pellan, M.; Brouard, J.; Chiron, R.; Giovannini-Chami, L.; de Blic, J.; Deschildre, A. and Lezmi, G.; COBRAPed Study Group. COBRAPed cohort: Do sensitization patterns differentiate children with severe asthma from those with a milder disease? Pediatr Allergy Immunol. 2024, 35, e14112. [CrossRef] [PubMed]
- Nösslinger, H.; Mair, E.; Oostingh, G.J.; Ahlgrimm-Siess, V.; Ringauf, A. and Lang, R. Multiplex Assays in Allergy Diagnosis: Allergy Explorer 2 versus ImmunoCAP ISAC E112i. Diagnostics (Basel). 2024, 14, 976. [CrossRef]
- Rambo, I.M; Kronfel, C.M.; Rivers, A.R.; Swientoniewski, L.T.; McBride, J.K.; Cheng, H.; Simon, R.J.; Ryan, R.; Tilles, S.A.; Nesbit, J.B.; Kulis, M.D.; Hurlburt, B.K. and Maleki, S.J. IgE and IgG4 epitopes of the peanut allergens shift following oral immunotherapy. Front Allergy. 2023, 4, 1279290. [CrossRef]
- Rubina, A.Y.; Dementieva, E.I.; Stomakhin, A.A.; Darii, E.L.; Pan’kov, S.V.; Barsky, V.E.; Ivanov, S.M.; Konovalova, E.V. and Mirzabekov, A.D. Hydrogel-based protein microchips: manufacturing, properties, and applications. Biotechniques. 2003, 34, 1008-1014, 1016-20, 1022. [CrossRef]
- Zubtsov, D.A.; Savvateeva, E.N.; Rubina, A.Y.; Pan’kov, S.V.; Konovalova, E.V.; Moiseeva, O.V.; Chechetkin, V.R. and Zasedatelev A.S. Comparison of surface and hydrogel-based protein microchips. Anal Biochem. 2007, 368, 205-213. [CrossRef]
- Smoldovskaya, O.; Feyzkhanova, G.; Voloshin, S.; Arefieva, A.; Chubarova, A.; Pavlushkina, L.; Filatova, T.; Antonova, E.; Timofeeva, E.; Butvilovskaya, V.; Lysov, Y.; Zasedatelev, A. and Rubina, A. Allergen-specific IgE and IgG4 patterns among patients with different allergic diseases. World Allergy Organization Journal. 2018, 11, 35. [CrossRef]
- Feyzkhanova, G.U.; Filippova, M.A.; Talibov, V.O.; Dementieva, E.I.; Maslennikov, V.V.; Reznikov, Y.P.; Offermann, N.; Zasedatelev, A.S.; Rubina, A.Y. and Fooke-Achterrath, M. Development of hydrogel biochip for in vitro allergy diagnostics. J Immunol Methods. 2014, 406, 51-57. [CrossRef]
- Butvilovskaya, V.I.; Smoldovskaya, O.V.; Feyzkhanova, G.U.; Filippova, M.A.; Pavlushkina, L.V.; Voloshin, S.A. and Rubina A.Yu. Modification of Anti-Glycan IgG and IgM Profiles in Allergic Inflammation. Molecular biology (Moscow). 2018, 52, 634-643. [CrossRef]
- Buianova, A. A.; Yukina, M. Y.; Cheranev, V. V.; Suchalko, O. N.; Shmitko, A. O.; Samitova, A. F.; Nuralieva, N. F.; Kulagina, E. V.; Savvateeva, E. N.; Troshina, E. A.; Rebrikov, D. V.; Gryadunov, D. A. and Korostin, D. O. Trio-Based Exome Sequencing and High-Resolution HLA Typing in Families of Patients with Autoimmune Adrenal Insufficiency and Autoimmune Polyglandular Syndrome. PloS one. 2024, 19, e0312335. [CrossRef]
- Gryadunov, D.; Dementieva, E.; Mikhailovich, V.; Nasedkina, T.; Rubina, A.; Savvateeva, E.; Fesenko, E., Chudinov, A.; Zimenkov, D.; Kolchinsky, A. and Zasedatelev A. Gel-based microarrays in clinical diagnostics in Russia. Expert review of molecular diagnostics. 2011, 11, 839-853. [CrossRef]
- Bespyatykh, Yu.A.; Shitikov, E.A.; Zimenkov, D.V.; Kulagina, E.V.; Gryadunov, D.A.; Nosova, E.Yu.; Bukatina, А.A.; Shulgina, M.V.; Zhuravlev, V.Yu. and Ilyina E.N. Identification of drug resistance and genotyping of clinical isolates of Mycobacterium tuberculosis using the TB-TEST. Pulmonology, Moscow. 2013, 4, 77-81.
- Miftakhov, R.A.; Ikonnikova, A.Y.; Vasiliskov, V.A.; Lapa, S.A.; Levashova, A.I.; Kuznetsova, V.E.; Shershov, V.E.; Zasedatelev, A.S.; Nasedkina, T.V. and Chudinov, A.V. A DNA Biochip with Cells of Carboxylated Brush Polymers. Russ J Bioorg Chem. 2023, 49, 1143–1150. [CrossRef]
- Miftakhov, R.A.; Nasedkina, T.V.; Zasedatelev, A.S.; Shershov, V.E.; Kuznetsova, V.E.; Levashova, A.I.; Lapa, S.A.; Vasiliskov, V.A; Ikonnikova, A.Yu. and Chudinov, A.V. Biochip with Cells of Brush Polymers Carrying Carboxyl Groups for DNA Analysis. Russ J Bioorg Chem. 2023, 49, 641-648. [CrossRef]
- Smoldovskaya, O.; Feyzkhanova, G.; Arefieva, A.; Voloshin, S.; Ivashkina, O. Allergen extracts and recombinant proteins: comparison of efficiency of in vitro allergy diagnostics using multiplex assay on a biological microchip. Allergy, Asthma & Clinical Immunology. 2016, 12, 1-5. [CrossRef]
- Feyzkhanova, G.; Voloshin, S.; Smoldovskaya, O.; Arefieva, A.; Filippova, M.; Barsky, V.; Pavlushkina, L.; Butvilovskaya, V.; Tikhonov, A.; Reznikov, Y. and Rubina, A. Development of a microarray-based method for allergen-specific IgE and IgG4 detection. Clin Proteomics. 2017, 14, 1. [CrossRef]
- Smoldovskaya, O.; Feyzkhanova, G.; Voloshin, S.; Arefieva, A.; Chubarova, A.; Pavlushkina, L.; Filatova, T.; Antonova, E.; Timofeeva, E.; Butvilovskaya, V.; Lysov, Y.; Zasedatelev, A. and Rubina, A. Allergen-specific IgE and IgG4 patterns among patients with different allergic diseases. World Allergy Organ J. 2018, 11, 35. [CrossRef]
- Shaskolskiy, B.; Kandinov, I.; Kravtsov, D.; Vinokurova, A.; Gorshkova, S.; Filippova, M.; Kubanov, A.; Solomka, V.; Deryabin, D.; Dementieva, E. and Gryadunov, D. Hydrogel Droplet Microarray for Genotyping Antimicrobial Resistance Determinants in Neisseria gonorrhoeae Isolates. Polymers. 2021, 13, 3889. [CrossRef]
- Zasedateleva, O.A.; Vasiliskov, V.A.; Surzhikov, S.A.; Sazykin, A.Y.; Putlyaeva, L.V.; Schwarz, A.M.; Kuprash, D.V.; Rubina, A.Y.; Barsky, V.E. and Zasedatelev, A.S. UV fluorescence of tryptophan residues effectively measures protein binding to nucleic acid fragments immobilized in gel elements of microarrays. Biotechnol J. 2014, 9, 1074-1080.
- Shtylev, G.F.; Shishkin, I.Y.; Shershov, V.E.; Kuznetsova, V. E.; Kachulyak, D. A.; Butvilovskaya, V. I.; Levashova, A. I.; Vasiliskov, V. A.; Zasedateleva, O. A. and Chudinov, A. V. Immobilization of Protein Probes on Biochips with Brush Polymer Cells. Russ J Bioorg Chem. 2024, 50, 2036–2049. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).