1. Introduction
Colorectal cancer (CRC) is among the major public health concerns worldwide, with an incidence of approximately 10% of all cancer cases [
1]. Gradually accumulating genetic or epigenetic alterations in healthy colonic epithelial cells underpin the transition from adenomas to invasive adenocarcinomas [
2]. However, while the onset and development of CRC requires genetic modifications that mostly fall into three distinct classes of aberrations, namely chromosomal instability, microsatellite instability and the CpG island methylation phenotype [
3,
4,
5,
6,
7,
8,
9], invasive, or metastatic, these properties follow the acquisition of mesenchymal traits from epithelial cells, a process called the epithelial-mesenchymal transition (EMT) [
10]. By conferring aggressiveness, the EMT triggers apical-basal polarity disruption, tight junction dissolution as well as cytoskeletal rearrangements, thus providing an infiltration and migratory ability to CRC cancer cells [
10,
11,
12,
13,
14,
15,
16,
17,
18,
19,
20,
21,
22]. Extracellular stimuli from the tumor microenvironment (TME) affect the complex molecular network that initiate and support the EMT. Particularly relevant in this context are the Transforming Growth Factor beta 1/Suppressor of Mothers against Decapentaplegic (TFG-β1/SMAD), Wingless/Integrated (Wnt)/β-catenin and Neurogenic locus notch homolog protein 1 (Notch 1) pathways [
23]. Not least, independent groups have shown that a key role in EMT progression and CRC invasion is mediated by the pro-inflammatory cytokines, such as interleukin-1β (IL-1β), released within the TME [
24,
25,
26]. Notoriously, the CRC microenvironment is characterized by elevated levels of several pro-inflammatory cytokines, among which IL-1β, considered the major mediator of inflammation, stands out and fuels tumor invasion through immunosuppressive activities [
27,
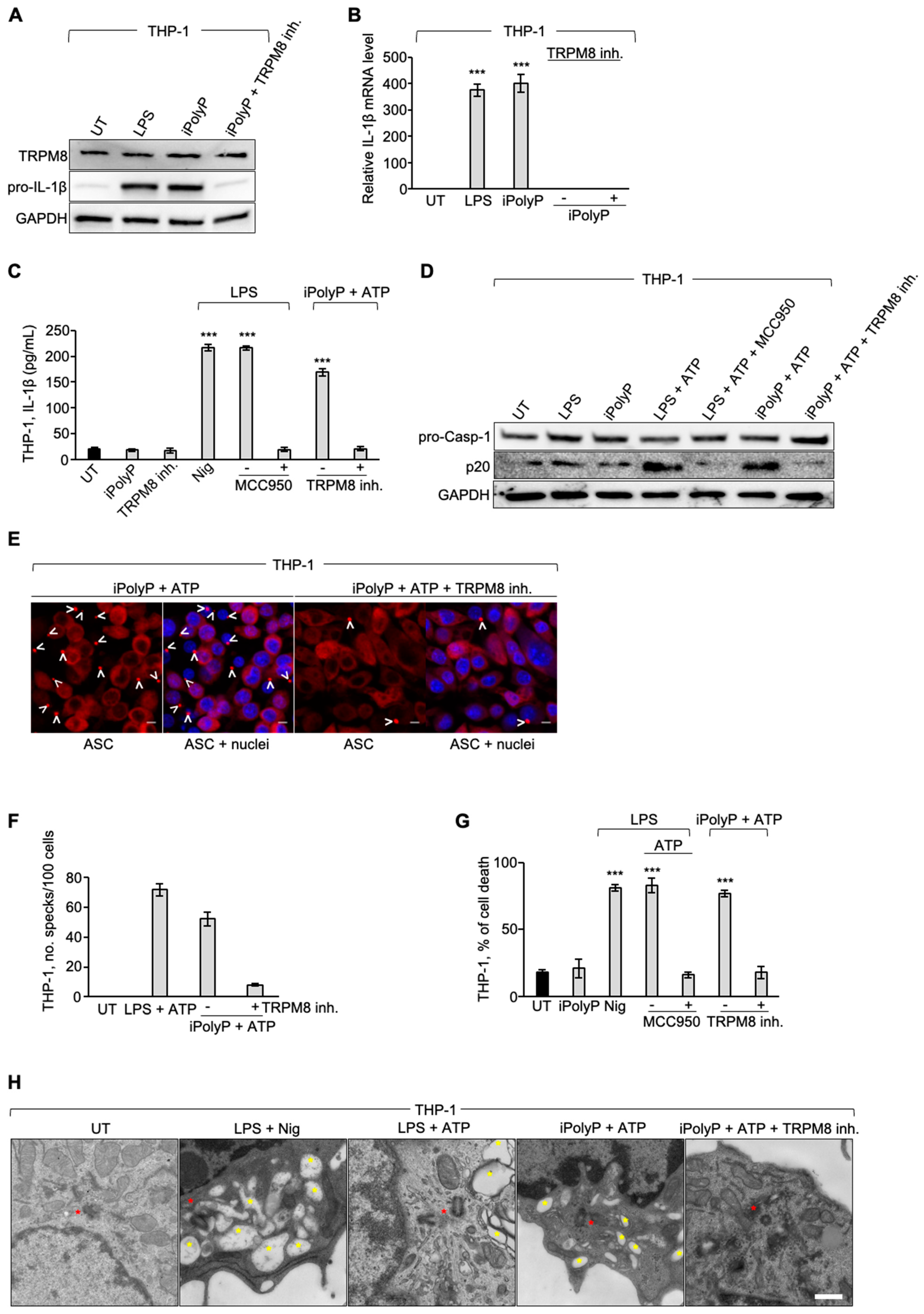
28]. Bioactive IL-1β originates as a direct consequence of inflammasome activation in the innate immune cells [
29]. Inflammasomes are supramolecular cytosolic structures found in all innate immune cells, that auto-assemble into micrometer-size complexes upon endogenous or exogenous insults, and trigger the inflammatory response. Among the several known mammalian inflammasomes, the NLRP3 inflammasome is the best studied. After sensing and binding the threat, monomers of NLRP3 organize into a wheel-shaped multimeric scaffold that signals to an adaptor protein, named apoptosis-associated speck-like protein containing a CARD (ASC) which, in turn, activates pro-Caspase-1, forming the core of the inflammasome. Active Caspase-1 cleaves pro-IL-1β, pro-IL-18 and gasdermin D (GSDMD) into their mature forms (IL-1β, IL-18 and N-terminus GSDMD, respectively). While the N-terminus GSDMD generates plasma membrane pores, allowing the release of IL-1β and IL-18 into the bloodstream, concomitantly an inflammatory form of cell death, called pyroptosis, occurs. Morphologically denominated as a “speck” or punctum, the active NLRP3 inflammasome colocalizes with the centrosome, also known as the microtubule-organizing center (MTOC), located in the perinuclear area [
30]. Aberrant NLRP3 inflammasome activation has been reported to contribute, alongside other multiple risk factors, to the onset of several inflammation-driven diseases, including cancer [
31]. Given the influence on CRC pathophysiology of the inflammasome-dependent and/or -independent EMT, intensive research is now underway. It has been recently reported that the pro-inflammatory molecule, named inorganic polyphosphate (iPolyP), is upregulated in neoplastic CRC tissues compared to the corresponding normal counterpart and contributes to the development and progression of CRC via its binding receptor called transient receptor potential cation channel subfamily M (melastatin) member 8 (TRPM8) [
32]. Being composed by hundreds of orthophosphates linked together by ATP-like bonds, iPolyP is considered an optimal source of energy with roles in disparate pathophysiological processes, including inflammation-driven diseases [
33,
34,
35,
36,
37], tumorigenesis [
38], tumor metastasis [
39,
40] and cellular proliferation [
41,
42]. Both iPolyP and the receptor TRPM8 are gathering attention in CRC pathophysiology nowadays, because while the former might be described as food additive, deriving from the intestinal microbiota or released by cellular organelles within the TME and promote a pro-inflammatory niche favorable for tumor growth [
43], the latter has been associated with poor prognosis in CRC subjects, where it has been found to be overexpressed [
44]. Through
in-vitro and
ex-vivo experimental approaches, the aim of our study was firstly to investigate whether the iPolyP/TRPM8 axis is involved in the EMT program. Additionally, due to its inflammatory properties we wished to rule out the potential role of iPolyP in the activation of the NLRP3 inflammasome in the CRC context. Thus, we tested the expression of several well-characterized EMT markers, such as Vimentin (Vim) [
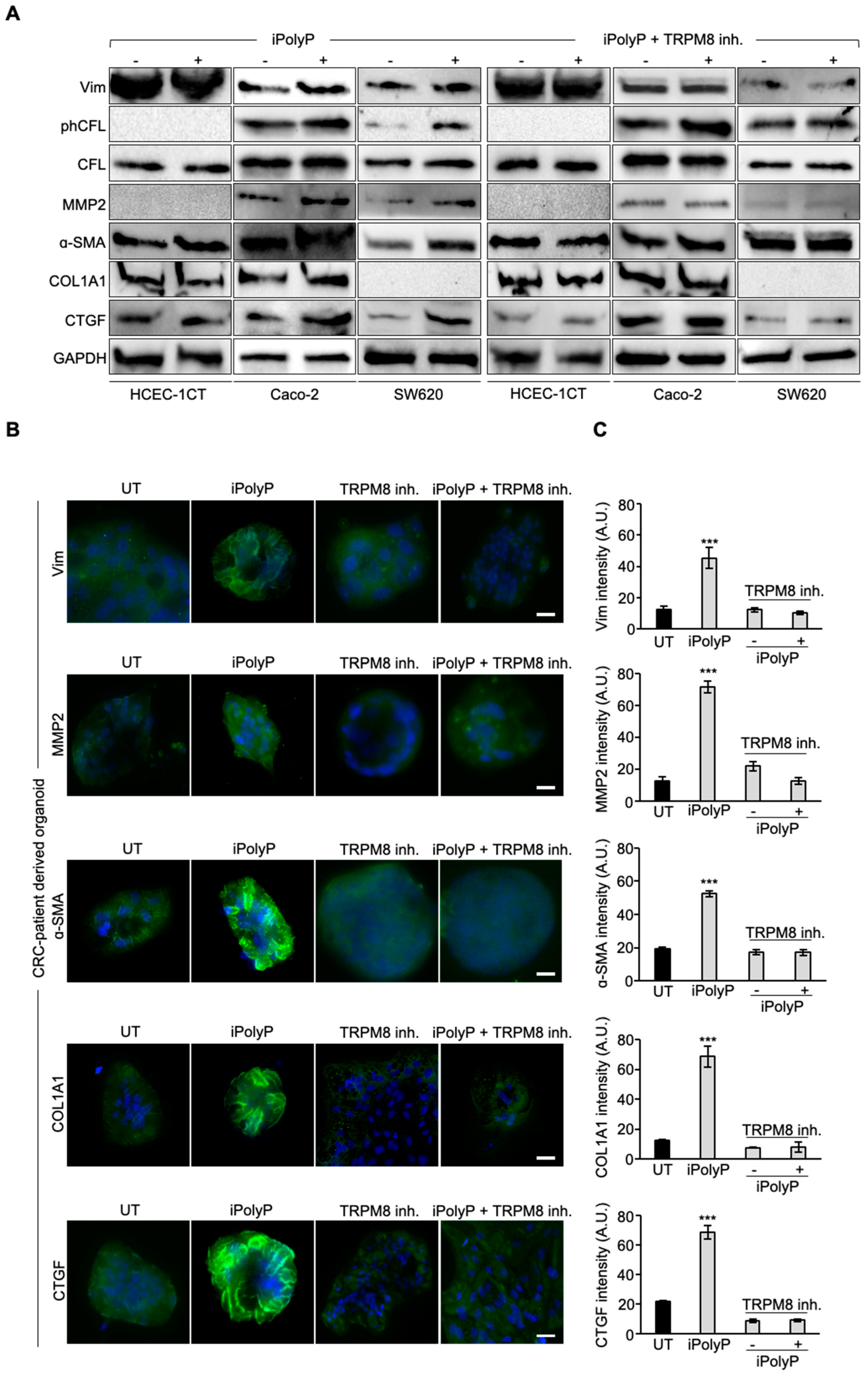
45], phospho-Cofilin (phCFL) [
46], Matrix Metalloprotease-2 (MMP2) [
47], Alpha Smooth Muscle Actin (α-SMA) [
48], Type 1 Collagen (COL1A1) [
49], Connective Tissue Growth Factor (CTGF) [
50], Fibronectin [
51] and Filamentous Actin (F-actin) [
52] in the presence or absence of iPolyP and TRPM8 inhibitor in a CRC cell line and CRC cell line-derived spheroids, as well as in CRC-patient derived organoids. Moreover, using the THP-1 human monocyte cell line, we investigated the possible influence of the iPolyP/TRPM8 axis on NLRP3 inflammasome activation. Together, our findings provide important insights into the involvement of the iPolyP/TRPM8 axis in CRC progression and could potentially pave the way for the development of novel anticancer agents as supplements to conventional chemotherapy.
3. Discussion
Despite significant efforts to develop new therapeutic strategies, colorectal cancer (CRC) remains one of the deadliest cancers in humans. This is largely due to the complex interplay of factors such as age, environmental and genetic predisposition, which underpin each stage of tumorigenesis [
53,
54,
55]. The EMT is a cellular process that plays a crucial role in the development and progression of CRC, thus now considered as one of the major targets for preventing neoplastic cells from acquiring invasive traits. The EMT enables the transformation of polarized epithelial cells, characterized by tight junctions and apical-basal polarity, into motile mesenchymal cells with enhanced invasive properties, thus contributing to tumor heterogeneity as well as to the onset of niches in distant organs [
56]. As a consequence, cancer cells display stem-like characteristics that confer resistance to conventional therapeutic approaches. Multiple extracellular signaling pathways, synergistically or independently, converge toward the EMT. These include agents like: Transforming Growth Factor-β1 (TGF-β1), which promotes the level of several transcription factors like Snail family transcriptional repressor 1 (SNAIL), Twist Family BHLH Transcription Factor 1 (Twist1) and Zinc finger E-box-binding homeobox 1/2 (Zeb1/2); Epidermal Growth Factor (EGF); Hypoxia, which upregulates genes associated with the mesenchymal phenotype; the Wnt Signaling pathway [
57], and inflammatory cytokines, such as IL-1β [
58]. With its ATP-like bonds, accumulating evidence is contextualizing the inorganic polyphosphate in the cancer background as an energy supplier [
59]. iPolyP has been enzymatically linked to two main sources, bacterial and human, that have different lengths. While bacterial iPolyP consists of 100 to 1,000 units, and is synthesized by polyphosphate kinase 1 (ppk1), human-derived iPolyP averages around 60 to 100 phosphate residues of unknown enzymatic origin. Niu and colleague showed in 2023 that the enzyme inorganic polyphosphatase, upregulated in CRC and known to hydrolyze long iPolyP chains into smaller molecules, is capable of activating the phosphatidylinositol 3-kinase/Protein kinase B (PI3K/AKT) pathway [
60] which showed pivotal regulatory tasks in the epithelial to mesenchymal transition development [
61], although the evidence remains indirect. In this study we demonstrate, through
in-vitro and
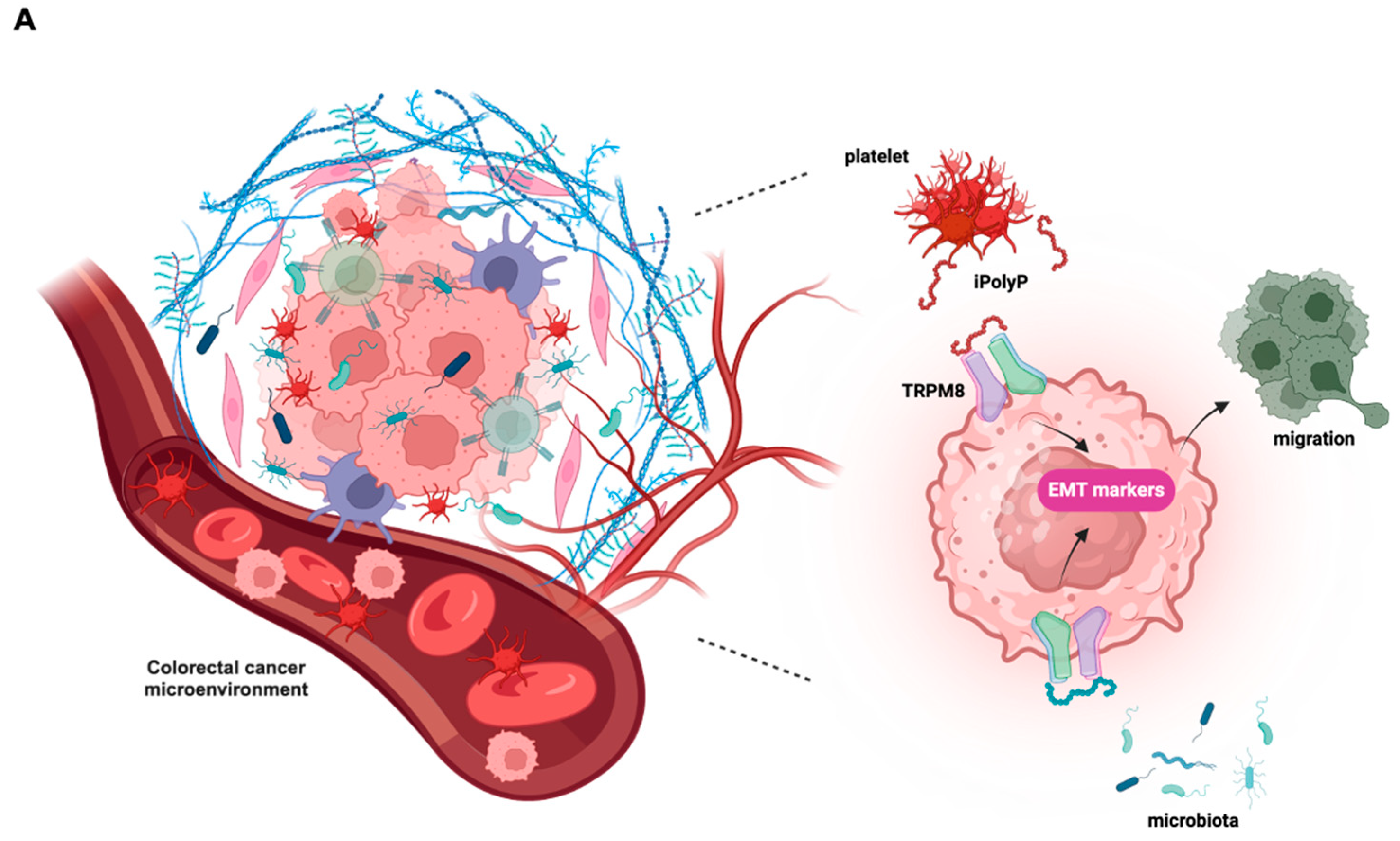
ex-vivo approaches, a direct involvement of the iPolyP/TRPM8 axis in the EMT process, which ultimately confers migratory properties to colorectal cancer cells, as well as a novel and unexpected involvement in the inflammatory pathway through the stimulation of the NLRP3 inflammasome. This conclusion is based on the following evidence, summarized in
Figure 4A: (i) iPolyP upregulates EMT markers, such as Vimentin, phospho-Cofilin, Metallopeptidase-2, Alpha Smooth Muscle Actin, Type 1 Collagen (COL1A1), Connective Tissue Growth Factor (CTGF), Fibronectin and Filamentous Actin (F-actin) by interacting with the TRPM8 receptor in the CRC context; (ii) the iPolyP/TRPM8 axis governs CRC cell migration; (iii) iPolyP is able to shape the tumor microenvironment by priming the NLRP3 inflammasome, which is then fully activated by ATP. Canonical NLRP3 inflammasome activation, a pathway specifically found in innate immune cells, requires two consecutive steps. The priming step (signal 1) is generally mediated by the engagement of the Toll-like receptor (TLR) on the macrophage surface, which recognizes structurally conserved microbial molecules (such as lipopolysaccharide, LPS, from Gram-negative bacteria) and leads to NF-kB-mediated upregulation of intracellular levels of NLRP3, pro-IL-1β and pro-IL-18, whose initial concentrations are inadequate to initiate the assembly of NLRP3 in resting conditions [
62]. Following signal 1, the activation step (signal 2), is provided by a broad range of pathogens-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs), which include particulate matter, extracellular ATP and pore forming toxins. When paired, the two signals allow the oligomerization of NLRP3, Caspase-1 activation as well as the maturation of the executor proteins Gasdermin D, IL-1β and IL-18 into their bioactive, mature forms [
63]. Here we identified a parallel mechanism, existing in innate immune cells, which involves the highly abundant molecule iPolyP and its matched receptor TRPM8, thus mimicking the well-known priming pathway. This novel signal 1 model, in combination with ATP, whose concentration is significantly enhanced within extracellular spaces and interstitial fractions in CRC [
64], is capable, in turn, of allowing the maturation and secretion of IL-1β, inducing a similar efficiency to that seen upon LPS/ATP treatment. Hence, although our findings point out, for the first time, the involvement of the iPolyP/TRPM8 signaling axis in CRC growth and spread, more studies, including those involving
in-vivo approaches, are needed. Therefore, ongoing CRC animal models are currently running in our laboratory aiming to confirm our above-described findings. In addition, we will apply these findings to other cancer types which might help to classify iPolyP/TRPM8-sensitive/insensitive neoplasms. All together, these results uncover a novel, functional axis in the CRC context, shedding light on new directions for study and paving the way for the development of new therapeutic strategies for CRC patients.
4. Materials and Methods
4.1. Patients Samples
Patients provided written informed consent to the collection of biopsy tissue specimens under Prot. No. 397/C.E. of 16/09/2020 of the Local Ethics Committee “Gabriella Serio” IRCCS Istituto Tumori “Giovanni Paolo II”, Bari, Italy. Biopsy tissue specimens were provided by the Histopathology Unit of IRCCS “S. de Bellis”. The inclusion criteria were a confirmed diagnosis by colonoscopy, biopsy, or imaging studies, for which surgery was considered beneficial. Patients with a grade of at least 2, i.e., cancer cells with more abnormal features, were also considered. The patients to undergo surgery also had health conditions sufficiently good to undergo surgery. This included a reasonable performance status and no serious comorbidities that would significantly increase the risk of developing complications post-surgery. However, the patient needed to be willing and able to consent to surgery after being informed of the risks, benefits, and potential outcomes. Samples collected in the operating room were temporarily stored in HypoThermosol FRS (for human cell and tissue preservation—BioLife Solutions Inc, Bothell, Washington, USA.; Cat. No.: 101102), sectioned, passed through liquid nitrogen, and stored dry in −80°C within 3 hours.
4.2. Cell Culture and Reagents
Human colorectal adenocarcinoma Caco-2 and SW620 cells were purchased from the American Tissue Culture Collection (ATCC, Manassas, Virginia, U.S.A.; Cat. No.: HTB-37 and CCL-227, respectively). Human Colonic Epithelial Cells 1 transduced with CDK4 and Telomerase HCEC-1CT cells were purchased from Evercyte GmbH (Vienna, Austria; Cat. No.: CkHT-039-0229). Human embryonic kidney 293T (HEK293T) were purchased from the American Tissue Culture Collection (ATCC, Manassas, Virginia, U.S.A.; Cat. No.: CRL-3216). Human monocytic THP-1 cells, isolated from peripheral blood from an acute monocytic leukemia patient, were purchased from the American Tissue Culture Collection (ATCC, Manassas, Virginia, U.S.A.; Cat. No.: TIB-202). Caco-2, SW620 and HEK293T cells were grown in Dulbecco’s Modified Eagle’s medium (DMEM), (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 11965092), supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: A5256701), 1 mM Sodium Pyruvate (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No: 11360039), 25 mM HEPES (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 15630056) and 100 U/mL Antibiotic-Antimycotic (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 15240062). Human Colonic Epithelial Cells 1 transduced with CDK4 and Telomerase (HCEC-1CT) were grown in ColoUp medium ready to use (Evercyte GmbH, Vienna, Austria; Cat. No.: MHT-039), supplemented with 100 U/mL Antibiotic-Antimycotic (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 15240062). THP-1 cells were maintained in Roswell Park Memorial Institute medium (RPMI), Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 11875093), supplemented with 10% FBS, 100 U/mL Antibiotic-Antimycotic (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 15240062) and 0.05 mM 2-mercaptoethanol (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: M6250-100 mL). All cell lines were maintained in a humidified atmosphere at 37°C with 5% CO2. Cells were passaged and medium was changed every other day.
4.3. Immunoblotting
HCEC-1CT, Caco-2 and SW-620 cell lines were seeded into 6-well plates (Corning, New York, New York, U.S.A.; Cat. No.: 3516) at a density of 0.5 x 106 cells/well in 2 mL of complete cell culture medium. Seeded cells were treated with 0.5 μM of sodium phosphate glass (iPolyP) (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: S4379-500mg), or with 10 μM of TRPM8 receptor inhibitor N-(3-Aminopropyl)-2-[(3-methylphenyl)methoxy]-N-(2-thienylmethyl)benzamide hydrochloride (AMTB), (Santa Cruz Biotechnology, Dallas, Texas, U.S.A.; Cat. No.: 926023-82-7), or in combination for 72 hours. Dimethyl sulfoxide (DMSO), (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: D8418-100mL) was added to the control cells. Pharmacological inhibition of the iPolyP/TRPM8 axis was performed by adding TRPM8 inhibitor to the HCEC-1CT, Caco-2 and SW620 cell lines. THP-1 cells were employed for NLRP3 inflammasome studies. THP-1 cells were seeded into 6-well plates at a density of 2 x 105 cells/mL in 2 mL of complete cell culture medium and treated overnight with 300 ng/mL phorbol myristate acetate (PMA), (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: P8139-5MG). The following day, the priming step was performed by adding 1 μg/mL of lipopolysaccharides (LPS), (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: L4524-5MG), considered as positive control, or 0.5 μM iPolyP, to the cells for 4 hours. Pharmacological inhibition of the priming step of NLRP3 inflammasome was performed by adding 10 μM of TRPM8 inhibitor for 4 hours. NLRP3 inflammasome activation was triggered by adding 5 mM of adenosine 5′-triphosphate (ATP) disodium salt hydrate (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: FLAAS-1VL), or 20 μM of Nigericin sodium salt (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: N7143). Pharmacological inhibition of NLRP3 inflammasome activation step was performed by adding 0.1 μM of the NLRP3 direct inhibitor MCC950 (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: 5.38120) 1 hour before the activation step. Following incubation/activation, adherent cells were detached using 0.05 % Trypsin-EDTA (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 25300-054), centrifuged at 1100 rpm for 5 minutes at 4°C and washed with 1x sterile Dulbecco’s phosphate-buffered saline 1 (1x DPBS) (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 14190-094) twice. Dried pellets were frozen at −80°C or resuspended and lysed in 200 µL of T-PERTM Tissue Protein Extraction Reagent (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 78510) supplemented with Halt™ Protease and Phosphatase Inhibitor Single-Use Cocktail, EDTA-Free (100x) (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 78443), for immunoblotting analysis. Cellular lysates were incubated on ice for 30 minutes and vortexed every 10 minutes. Samples were then centrifuged at 16,000 rpm at 4°C for 20 minutes to clarify and precipitate insoluble debris. Total extracted proteins were assayed to measure concentrations using the Bio-Rad protein assay dye reagent concentrate (Bio-Rad Laboratories, Hercules, California, U.S.A.; Cat. No.: 5000006EDU). Then, proteins were mixed with 4 x Laemmli Sample Buffer (Bio-Rad Laboratories, Hercules, California, U.S.A.; Cat. No.: 1610747) and 10% of β-mercaptoethanol (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: M6250-100mL) and denatured at 95°C for 5 minutes. 25 μg of proteins were loaded onto precast polyacrylamide 4-20% gels (Bio-Rad Laboratories, Hercules, California, U.S.A.; Cat. No.: 4568094), subsequently blotted on a polyvinylidene fluoride (PVDF) membrane (Bio-Rad Laboratories, Hercules, California, U.S.A.; Cat. No.: 1704156) using the trans-blot turbo transfer system (Bio-Rad Laboratories, Hercules, California, U.S.A.; Cat. No.: 1704150). Membranes were blocked using Pierce™ Protein-Free Blocking Buffer (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 37571) for 1 hour and stained overnight with primary antibodies. The next day membranes were washed three times with 1x Tris Buffered saline (1x TBS), (Bio-Rad Laboratories, Hercules, California, U.S.A.; Cat. No.: 1706435 diluted in ddH2O to reach 1x)/TWEEN20 (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: P9416-100mL) incubated for 1 hour with the respective horseradish peroxidase-conjugated secondary antibodies. Proteins were detected using the Clarity Max Western Enhanced Chemiluminescence (ECL) Substrate (Bio-Rad Laboratories, Hercules, California, U.S.A.; Cat. No.: 1705062) and the signals were obtained using the Chemidoc MP Imaging System (Bio-Rad Laboratories, Hercules, California, U.S.A.; Cat. No.: 1708280). The following primary antibodies were used: anti-TRPM8, 1:1000 (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: MA5-35474); anti-Vimentin, 1:1000 (Cell Signaling Technology, Danvers, Massachusetts, U.S.A.; Cat. No.: 57415); anti-phospho-Cofilin (Ser3), 1:500 (Cell Signaling Technology, Danvers, Massachusetts, U.S.A.; Cat. No.: 33115); anti-Cofilin, 1:500 (Cell Signaling Technology, Danvers, Massachusetts, U.S.A.; Cat. No.: 33185); anti-MMP2, 1:500 (Abcam, Cambridge Biomedical Campus, Cambridge, U.K.; Cat. No.: ab86607); anti-α-SMA, 1:500 (Abcam, Cambridge Biomedical Campus, Cambridge, U.K.; Cat. No.: ab5694); anti-COL1A1 1:1000 (Cell Signaling Technology, Danvers, Massachusetts, U.S.A.; Cat. No.: 911445); anti-CTGF, 1:500 (Abcam, Cambridge Biomedical Campus, Cambridge, U.K.; Cat. No.: ab6992); anti-TRPM8, 1:1000 (Thermo Fisher Scientific, Waltham, MA, USA; Cat. No.: MA5-35474); anti-pro-IL-1β, 1:1000 (Abcam, Cambridge Biomedical Campus, Cambridge, U.K.; Cat. No.: ab226918); anti-pro-Caspase-1, 1:1000 (Cell Signaling Technology, Danvers, Massachusetts, U.S.A.; Cat. No.: 3866S); anti-GAPDH, 1:1000 (Santa Cruz Biotechnology, Dallas, Texas, U.S.A.; Cat. No.: SC-47724). GAPDH was used as loading controls. The following secondary antibodies were employed: Anti-rabbit IgG, HRP-linked Antibody, 1:2000 (Cell Signaling Technology, Danvers, Massachusetts, U.S.A.; Cat. No.: 7074S); Goat anti-Mouse IgG (H+L)-HRP Conjugate (Bio-Rad Laboratories, Hercules, California, U.S.A.; Cat. No.: 1706516).
4.4. CRC Tumor Organoids
CRC specimens from surgically resected tumor tissue were washed three times with 1x sterile DPBS (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 14190-094) and digested with collagenase/hyaluronidase mixture (Stemcell Technologies, Vancouver, Canada Cat. No.: 07912), diluted in HBSS solution (with CaCl
2 and MgCl
2, Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 14025-050) for 5 hours under gentle rocking at 37°C. A single cell suspension was obtained and cells were embedded in matrigel matrix basement membrane (Corning, New York, New York, U.S.A.; Cat. No.: 356231), and cultured in IntestiCult-SF (ICT-SF) medium (Stemcell Technologies, Vancouver, British Columbia, Canada; Cat. No.: 100-0340) medium diluted 1:2 in Advanced DMEM/F-12 (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 12634-010), supplemented with N-2 (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 17502-048), B27 supplement (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 12587-010), 0.01% bovine serum albumin (BSA), antibiotic/antimycotic, and HEPES. Medium was renewed every two days until the organoids were fully developed (7-10 days). Mature organoids were split using TrypLE Select dissociation agent (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: A12177-01) according to [
65] and the suspended cells obtained were re-cultured at a lower density in matrigel.
4.5. Immunofluorescence of Tumor Cells Derived from CRC Tumor Organoids
Cells obtained from organoids splitting were seeded at low density on microscopy slides (Nunc Lab-Tek II, Merck Life Sciences, Milan, Italy; Cat. No.: 154534) previously coated with matrigel diluted 1:10 in 1x sterile DPBS (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 14190-094), and left to attach and grow for at least 7 days in ICT-SF medium until the formation of large islets in a humidified incubator set to 37°C and 5% CO2. Cells were treated with Dimethyl sulfoxide (DMSO, indicated as untreated, UT) (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: D8418-100 mL), or 0.5 μM iPolyP, or TRPM8 inhibitor 10 μM or iPolyP + TRPM8 inhibitor for 48 hours, and fixed with 4% paraformaldehyde solution (PFA) (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: P6148-500G) in 1x sterile DPBS (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 14190-094) for 15 minutes at 4°C, permeabilized with 0.15% Triton X-100 (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: 101895731) for 7 minutes at room temperature, blocked with 2% BSA (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: A7030-100G) in 1x sterile DPBS (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 14190-094) for 1 hour at room temperature, and incubated overnight with primary antibodies (anti-Vimentin, 1:1000; anti-MMP2, 1:1000; anti-α-SMA, 1:500; anti-COL1A1, 1:1000; anti-CTGF; antibodies details are reported in the Immunoblotting section). The following day cells were washed three times with 1x sterile DPBS (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 14190-094), incubated for 1 hour at room temperature with fluorophore-conjugated secondary antibodies for the detection of the antigens of interest and subjected to microscopy analysis. The following secondary antibodies were used: Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ Plus 488, 1:500 (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: A-32731TR); Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 488, 1:500 (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: A-11029). Images were collected and analyzed as described in the Cell lines immunofluorescence and confocal microscopy section.
4.6. Immunofluorescence of Cell Lines-Derived Spheroids
1 x 103 Caco-2, SW620, and HCEC-1CT cells were seeded into 3D low attachment 96-well cell culture plates (Corning, New York, New York, U.S.A.; Cat. No.: 4520) to obtain 3D cell-line spheroids with 100 µL of growth medium in each well. Cells were kept in culture in the above-described conditions. After seeding, all cell lines were treated with 0.5 μM iPolyP or with 10 μM TRPM8 inhibitor or both, while untreated cells were used as controls (untreated, UT). Spheroid cultures were observed at 24 and 96 hours and maintained in a humidified incubator set to 37°C and 5% CO2. After 96 hours, cells were treated with Dimethyl sulfoxide (DMSO, indicated as untreated, UT) (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: D8418-100 mL), or 0.5 μM iPolyP, or TRPM8 inhibitor 10 μM or iPolyP + TRPM8 inhibitor for 48 hours, and fixed with 4% paraformaldehyde solution (PFA) (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: P6148-500G) in 1x sterile DPBS (Thermo Fisher Scientific, Waltham, MA, USA; Cat. No.: 14190-094) for 15 minutes at 4°C, permeabilized with 0.15% Triton X-100 (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: 101895731) for 7 minutes at room temperature, blocked with 2% BSA (Sigma-Aldrich, St. Louis, MO, USA; Cat. No.: A7030-100G) in 1x sterile DPBS (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 14190-094) for 1 hour at room temperature, and incubated overnight with α-SMA antibody. The following day cells were washed three times with 1x sterile DPBS (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 14190-094), incubated for 1 hour at room temperature with fluorophore-conjugated secondary antibody: Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ Plus 488, 1:500 (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: A-32731TR). Nuclei were stained using PureBlu DAPI (Bio-Rad Laboratories, Hercules, California, U.S.A.; Cat. No.: 1351303). Spheroids were then subjected to microscopy analysis. Images were collected and analyzed as described in the Cell lines immunofluorescence and confocal microscopy section.
4.7. Cell Lines Immunofluorescence and Confocal Microscopy
2 x 105/mL HCEC-1CT, Caco-2, SW620 and THP-1 cells were grown in 35-mm petri dishes, no. 1.5 coverglass (MatTek, Ashland, Massachusetts, U.S.A.; Cat. No: P35G-1.5-14-C). HCEC-1CT, Caco-2, SW620 cells were treated with 0.5 μM iPolyP, or with 10 μM TRPM8 inhibitor or both for 48 hours. THP-1 cells were treated overnight with 300 ng/mL phorbol myristate acetate (PMA), (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: P8139-5MG). The following day, the priming step was performed by adding 1 μg/mL of lipopolysaccharides (LPS), (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: L4524-5MG), considered as positive control, or 0.5 μM iPolyP, to the cells for 4 hours. Pharmacological inhibition of the priming step of the NLRP3 inflammasome was performed by adding 10 μM of TRPM8 inhibitor for 4 hours. NLRP3 inflammasome activation was triggered by adding 5 mM of adenosine 5′-triphosphate (ATP) disodium salt hydrate (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: FLAAS-1VL), or 20 μM of Nigericin sodium salt (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: N7143). Pharmacological inhibition of the NLRP3 inflammasome was performed by adding 0.1 μM of the NLRP3 direct inhibitor MCC950 (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: 5.38120) 1 hour before the activation step. Fixative, permeabilization, and blocking buffers were prepared in 1x sterile DPBS (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 14190-094). Cells were fixed with 4% PFA for 30 minutes at 4°C and then washed twice using 1x sterile DPBS. Permeabilization was performed for 7 minutes at room temperature using 0.15% Triton X-100 (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: 101895731) diluted in 1x DPBS. Washing was performed to remove permeabilization buffer. Cells were then blocked for 1 hour at room temperature using blocking buffer (3% BSA), (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: A7030-100G)/1x sterile DPBS). Cells were incubated with primary (overnight) and secondary (1 hour) antibodies. Nuclei were stained using PureBlu DAPI (Bio-Rad Laboratories, Hercules, California, U.S.A.; Cat. No.: 1351303). In between, extensive washing steps were performed to remove unbound antibodies and stains. The following primary antibodies were employed: anti-Fibronectin, 1:1000 (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: MA5-11981); rhodamine phalloidin, for the F-actin detection, 1:250 (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: R415); anti-ASC (anti-ASC/TMS1 (E1E3I), 1:1000 (Cell Signaling Technology, Danvers, Massachusetts, U.S.A.; Cat. No.: 13833S). The following secondary antibodies were used: Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ Plus 488, 1:500 (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: A-32731TR); Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 488, 1:500 (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: A-11029). Images were collected with a Nikon Ti2 inverted laser-scanning confocal microscope equipped with Plan Apo 20x (0.75 numerical aperture) in bright-field and analyzed with NIS-Elements software (version 5.11.01) and Fiji software (version 2.16.0). Prior to acquisition, cells were placed in the microscope CO2 chamber. All images were collected with a confocal Yokogawa spinning-disk on a Nikon Ti inverted microscope equipped with Plan Fluor 40x (0.60 numerical aperture) lens. Images were acquired with a Hamamatsu ORCA ER cooled CCD camera controlled with NIS-Elements software (version 5.11.01). For time-lapse experiments, images were collected using an exposure time of 700 milliseconds. At each time point, z-series optical sections were collected with a step size of 1 μm at the indicated time intervals. Gamma, brightness, and contrast were adjusted on displayed images (identically for comparative image sets) using NIS-Elements software (version 5.11.01). The Perfect Focus System was kept running for continuous maintenance of focus. DMEM without phenol red (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 21063-045), or RPMI without phenol red (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 11835063) were used during image acquisition.
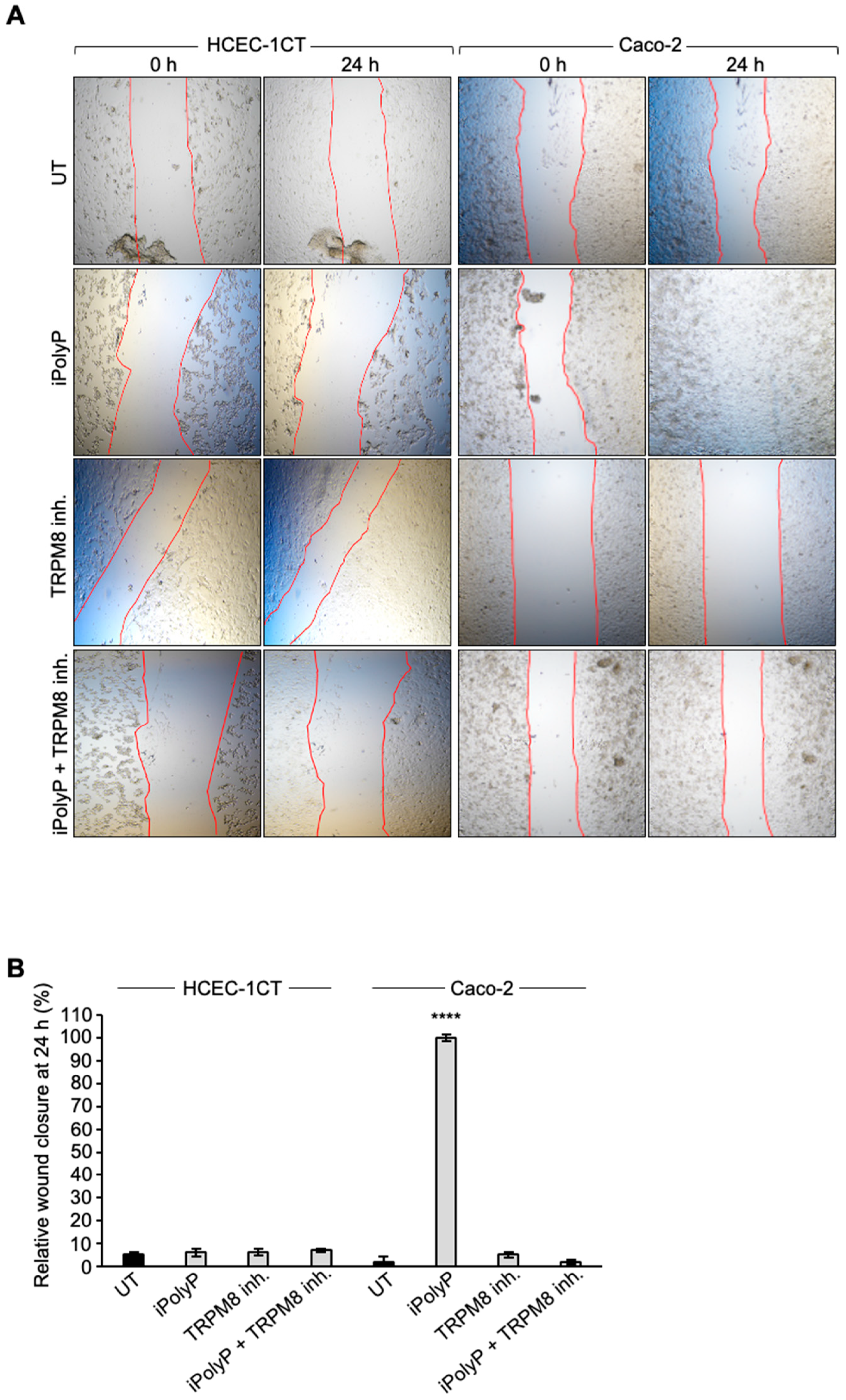
4.8. Wound Healing Assay
HCEC-1CC, Caco-2 and SW620 cells were seeded in a 12-well plate (Corning, New York, New York, U.S.A.; Cat. No.: 3513) in 10% FBS medium and grown to reach an 80–90% confluence. Then, a scratch was made on the cell monolayer using a p200 pipette tip. The plate was washed with 1x sterile DPBS to remove the debris and markings were created on the outer bottom of the plate with a tip marker, to be used as reference points. Each cell line was incubated with medium without FBS, containing either 0.5 μM iPolyP, 10 μM TRPM8 inhibitor or both. Images were acquired at 0 and 24 hours using a confocal microscope Nikon Eclipse Ti2 in bright field microscopy equipped with Plan Fluor 10x (0.25 numerical aperture) lens and Fiji software (version 2.16.0) was used to measure the scratched area. Cell migratory ability for wound-healing was assessed by applying the following formula: [(wound area at 0 hour) – (wound area at 24 hours)] / (wound area at 0 hour) [
66].
4.9. Detection of Cytokines by ELISA
The detection kit for human IL-1β (Abcam, Cambridge Biomedical Campus, Cambridge, U.K.; Cat. No.: ab214025) was used at the specified temperature and conditions according to the manufacturer’s instructions. Briefly, 50 μL plasma derived from Normal and Pathological individuals as well as supernatants derived from THP-1 cells at various conditions were placed into 96-well plates (Corning, New York, New York, U.S.A.; Cat. No. 3599), added with an additional 50 μL of antibody cocktail. The plates were then incubated for 2 hours at room temperature on a plate shaker set to 400 rpm. After extensive washing, 100 μL of TMB development solution were added to each well for 10 minutes in the dark on a plate shaker set to 400 rpm. Finally, 100 μL of stop solution were added to each well before recording the OD at 450 nm.
4.10. Propidium Iodide Fluorescence Cell Death Assay
After inflammasome priming by LPS or iPolyP for 4 hours and activation by LPS + nigericin, LPS + ATP, LPS + ATP + AMTB, iPolyP + ATP, iPolyP + ATP + AMTB, THP-1 cells were collected, alongside the untreated condition, and washed twice in 1x sterile DPBS. Pellets were then re-suspended in 2 μg/mL PI (ImmunoChemistry Technologies LLC, Davis, California, U.S.A.; Cat. No.: 638). The percentage of cells which took up propidium iodide (PI) was measured by flow cytometry (Navios), (Beckman Coulter, Brea, California, U.S.A.; Cat. No.: B83535) and the results were analyzed with FlowJo version 10 Software (FlowJo LLC, Ashland, Oregon, U.S.A.).
4.11. Cloning
Human ASC (a gift from Eicke Latz (Addgene plasmid # 41840;
http://n2t.net/addgene:41840; RRID:Addgene_41840) was subcloned into pLV-mScarlet (a gift from Pantelis Tsoulfas (Addgene plasmid # 159172;
http://n2t.net/addgene:159172; RRID:Addgene_159172)), plasmid between SalI-HF and EcoRV-HF (New England Biolabs Ipswich, Massachusetts, U.S.A.; Cat. No.: R0138M and R3195M respectively) restriction sites using the following primers (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.A.; Cat. No.: 10629186): forward primer of 5′ - ATACGTCGACGGATCTATGGGGCGC - 3′ and reverse primer of 5 ′- TTCAGATATCTAACTCGATGGTAGC - 3′.
4.12. Generation Stable Cell Lines
To generate stable cell lines, on day 0, lentivirus was produced using HEK293T cells by cotransfecting 1 mg of pLV plasmid containing the gene: 1. IL-1β: pLV-mTurquoise2-IL-1β-mNeonGreen was a gift from Hao Wu (Addgene plasmid # 166783;
http://n2t.net/addgene:166783; RRID:Addgene_166783) or 2. pLV mScarlet-I-ASC with 750 ng of psPAX2 packaging plasmid (a gift from Didier Trono (Addgene plasmid # 12260;
http://n2t.net/addgene:12260; RRID:Addgene_12260), and 250 ng of pMD2.G envelope plasmid (a gift from Didier Trono (Addgene plasmid # 12259;
http://n2t.net/addgene:12259; RRID:Addgene_12259)). The transfected cells were incubated overnight. The following day (day 1), the medium was removed and the cells were replenished with 1 mL of fresh medium and incubated for another day. On day 2, the supernatant containing the virus was filtered using a 0.45-mm filter (Biosigma, Cona, Italy; Cat. No.: 051230) and directly used to infect THP-1 with a spinfection protocol to increase the efficacy. Spinfection was performed at 2500 x g for 90 minutes at room temperature using 8 mg/mL polybrene (EMD Millipore Corp., Burlington, Massachusetts, U.S.A.; Cat. No.: TR-1003-G). Positive clones were selected by cell sorting and colonies were expanded from single clones. Positive clones were extensively validated by immunofluorescence microscopy.
4.13. Transmission Electron Microscopy
For transmission electron microscopy (TEM), THP-1 cells, following activation with ATP or nigericin activation, were processed for plastic embedding. Cells were first incubated in fixative for 1 hour at room temperature. To prevent cellular shock and facilitate gentle fixation, a 2X fixative mixture was added to the culture medium at a 1:1 ratio in the dish containing the cells. This approach ensured optimal preservation of cellular structures for subsequent TEM analysis. Fresh fixative was prepared using 1.25% PFA, 2.5% glutaraldehyde (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: 354400), and 0.03% picric acid (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No: P6744-1GA) in a 0.1 M sodium cacodylate buffer (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: 97068), pH 7.4. After fixation, the cells were washed three times in 0.1 M sodium cacodylate buffer. Subsequently, they were incubated with 1% osmium tetroxide/1.5% potassium ferrocyanide for 1 hour at room temperature. Following this step, the cells were washed three times with water and then incubated in aqueous solution with 1% uranyl acetate (Electron Microscopy Science, Hatfield, Pennsylvania, U.S.A.; Cat. No.: 541-09-3) for 30 minutes. This was followed by another three rounds of washing with water to ensure thorough removal of excess staining agents. Dehydration steps were performed twice in different grades of alcohol (70% ethanol for 15 minutes, 90% ethanol for 15 minutes, and 100% ethanol for 15 minutes). Samples were then placed in propyleneoxide (Sigma-Aldrich, St. Louis, Missouri, U.S.A.; Cat. No.: 110205) for 1 hour and infiltration was performed with Epon mixed 1 + 1 with propyleneoxide for 3 hours at room temperature. Samples were moved to the embedding mold filled with freshly mixed Epon and allowed to polymerize for 24 to 48 hours at 60°C. Ultrathin sections (~ 60 nm) were cut on a Reichert Ultracut-S microtome, placed on copper grids, and stained with lead citrate. The grids from the above-described electron microscopy procedures were examined using a JEOL Jem-1011 transmission electron microscope (JEOL U.S.A., Inc, Peabody, Massachusetts, U.S.A.) operating at an accelerating voltage of 100 kV. Images were acquired using an Olympus Quemesa Camera (11 Mpx) (Olympus, Shinjuku-ku, Tokyo, Japan) to ensure high-quality image capture.