Submitted:
30 October 2024
Posted:
30 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Identification
2.2. Susceptibility Testing
2.3. Calculation of Provisional Lactococcus garvieae Epidemiological Cut-Off Values
2.4. Data Analysis
3. Results
3.1. Bacterial Identification
3.2. Quality Control
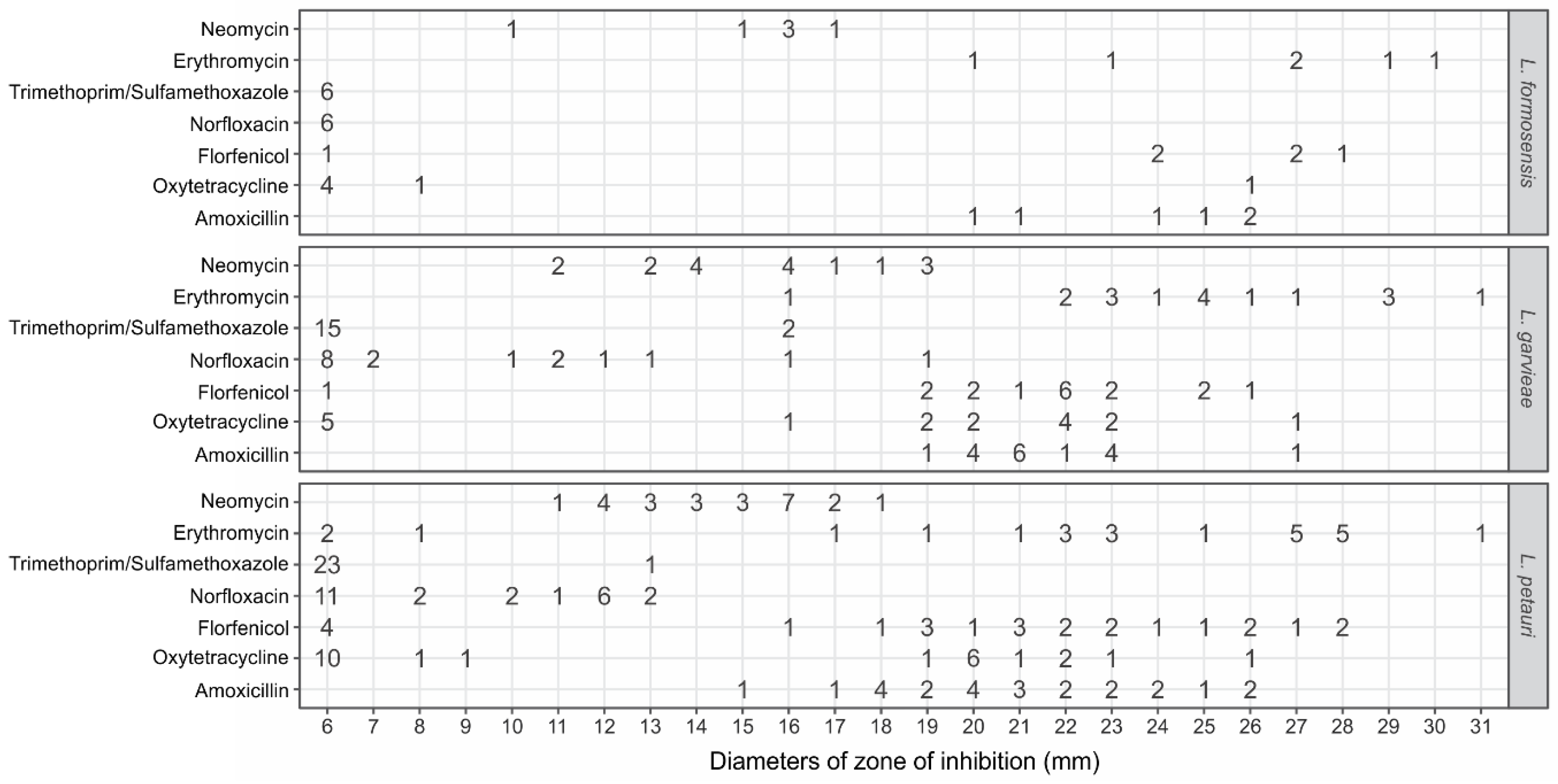
3.3. Antimicrobial Susceptibility for Lactococcus formosensis
3.4. Antimicrobial Susceptibility for Lactococcus garvieae
3.5. Antimicrobial Susceptibility for Lactococcus petauri
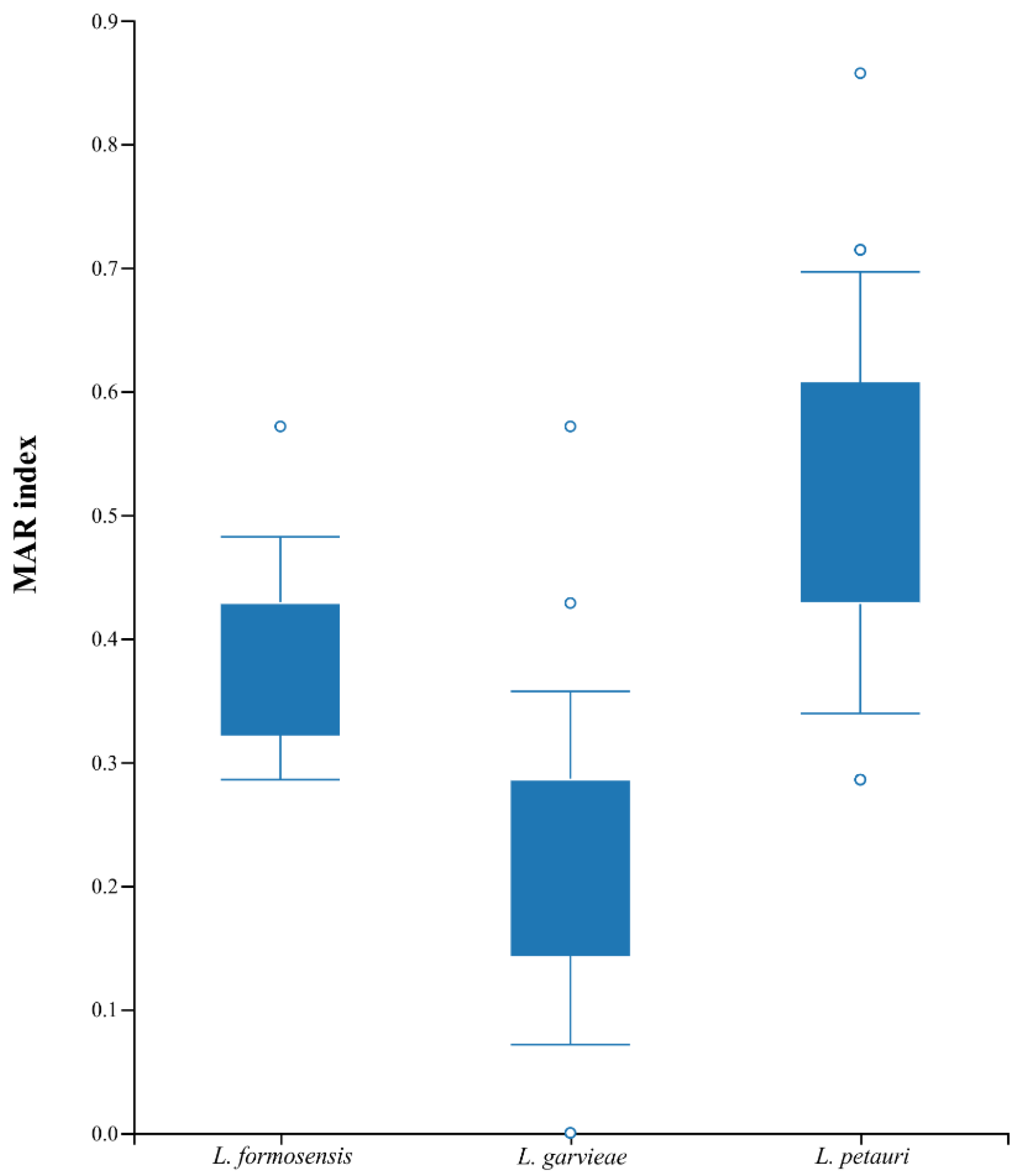
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Ethical Approval
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Altinok, I.; Ozturk, R.C.; Ture, M. NGS Analysis revealed that Lactococcus garvieae Lg-Per was Lactococcus petauri in Türkiye. J. Fish Dis. 2022, 45, 1839–1843. [Google Scholar] [CrossRef] [PubMed]
- Barbanti, A.C.C.; do Rosário, A.E.C.; da Silva Maia, C.R.M.; Rocha, V.P.; Costa, H.L.; Trindade, J.M.; Nogueira, L.F.F.; Rosa, J.C.C.; Ranzani-Paiva, M.J.T.; Pilarski, F.; et al. Genetic characterization of lactococcosis-causing bacteria isolated from Brazilian native fish species. Aquaculture 2024, 593, 741305. [Google Scholar] [CrossRef]
- Bondavalli, F.; Colussi, S.; Pastorino, P.; Zanoli, A.; Bezzo Llufrio, T.; Fernández-Garayzábal, J.F.; Acutis, P.L.; Prearo, M. First report of Lactococcus petauri in the pumpkinseed (Lepomis gibbosus) from Candia Lake (Northwestern Italy). Fishes 2024, 9. [Google Scholar] [CrossRef]
- Khoo, L.H.; Austin, F.W.; Quiniou, S.M.A.; Gaunt, P.S.; Riecke, D.K.; Jacobs, A.M.; Meals, K.O.; Dunn, A.W.; Griffin, M.J. Lactococcosis in silver carp. J. Aquat. Anim. Health 2014, 26, 1–8. [Google Scholar] [CrossRef]
- Abraham, T.; Yazdi, Z.; Littman, E.; Shahin, K.; Heckman, T.I.; Quijano Cardé, E.M.; Nguyen, D.T.; Hu, R.; Adkison, M.; Veek, T.; et al. Detection and virulence of Lactococcus garvieae and L. petauri from four lakes in Southern California. J. Aquat. Anim. Health 2023, 35, 187–198. [Google Scholar] [CrossRef]
- Salogni, C.; Bertasio, C.; Accini, A.; Gibelli, L.R.; Pigoli, C.; Susini, F.; Podavini, E.; Scali, F.; Varisco, G.; Alborali, G.L. The characterisation of Lactococcus garvieae isolated in an outbreak of septicaemic disease in farmed sea bass (Dicentrarchus labrax, Linnaues 1758) in Italy. Pathogens 2024, 13. [Google Scholar] [CrossRef]
- Neupane, S.; Rao, S.; Yan, W.-X.; Wang, P.-C.; Chen, S.-C. First identification, molecular characterization, and pathogenicity assessment of Lactococcus garvieae isolated from cultured pompano in Taiwan. J. Fish Dis. 2023, 46, 1295–1309. [Google Scholar] [CrossRef]
- Shahin, K.; Veek, T.; Heckman, T.I.; Littman, E.; Mukkatira, K.; Adkison, M.; Welch, T.J.; Imai, D.M.; Pastenkos, G.; Camus, A.; et al. Isolation and characterization of Lactococcus garvieae from rainbow trout, Onchorhyncus mykiss, from California, USA. Transbound. Emerg. Dis. 2022, 69, 2326–2343. [Google Scholar] [CrossRef]
- Ortega, C.; Irgang, R.; Valladares-Carranza, B.; Collarte, C.; Avendaño-Herrera, R. First identification and characterization of Lactococcus garvieae isolated from rainbow trout (Oncorhynchus mykiss) cultured in Mexico. Animals 2020, 10. [Google Scholar] [CrossRef]
- Egger, R.C.; Rosa, J.C.C.; Resende, L.F.L.; de Pádua, S.B.; de Oliveira Barbosa, F.; Zerbini, M.T.; Tavares, G.C.; Figueiredo, H.C.P. Emerging fish pathogens Lactococcus petauri and L. garvieae in Nile tilapia (Oreochromis niloticus) farmed in Brazil. Aquaculture 2023, 565, 739093. [Google Scholar] [CrossRef]
- Gazal, L.E. de S.; Brito, K.C.T. de; Kobayashi, R.K.T.; Nakazato, G.; Cavalli, L.S.; Otutumi, L.K.; Brito, B.G. de Antimicrobials and resistant bacteria in global fish farming and the possible risk for public health. Arq. Inst. Biol. (Sao. Paulo). 2020, 87. [Google Scholar]
- Duman, M.; Buyukekiz, A.G.; Saticioglu, I.B.; Cengiz, M.; Sahinturk, P.; Altun, S. Epidemiology, genotypic diversity, and antimicrobial resistance of Lactococcus garvieae in farmed rainbow trout (Oncorhynchus mykiss). IFRO 2020, 19, 1–18. [Google Scholar]
- Raissy, M.; Moumeni, M. Detection of antibiotic resistance genes in some Lactococcus garvieae strains isolated from infected rainbow trout. Iran. J. Fish. Sci. 2016, 15, 221–229. [Google Scholar]
- Monteiro, S.H.; Francisco, J.G.; Andrade, G.C.R.M.; Botelho, R.G.; Figueiredo, L.A.; Tornisielo, V.L. Study of spatial and temporal distribution of antimicrobial in water and sediments from caging fish farms by on-line SPE-LC-MS/MS. J. Environ. Sci. Heal. Part B 2016, 51, 634–643. [Google Scholar] [CrossRef] [PubMed]
- Leal, C.A.G. Resistência a antimicrobianos na piscicultura. Panor. da Aquicultura 2022, 31, 38–47. [Google Scholar]
- Öztürk, R.Ç.; Ustaoglu, D.; Ture, M.; Bondavalli, F.; Colussi, S.; Pastorino, P.; Vela, A.I.; Kotzamanidis, C.; Fernandez-Garayzábal, J.F.; Bitchava, K.; et al. Epidemiological cutoff values and genetic antimicrobial resistance of Lactococcus garvieae and L. petauri. Aquaculture 2024, 593, 741340. [Google Scholar] [CrossRef]
- Carvalho, G.M.; Silva, B.A.; Xavier, R.G.C.; Zanon, I.P.; Vilela, E.G.; Nicolino, R.R.; Tavares, G.C.; Silva, R.O.S. Evaluation of disk diffusion method for testing the rifampicin, erythromycin, and tetracycline susceptibility of Clostridioides (Prev. Clostridium) difficile. Anaerobe 2023, 80, 102720. [Google Scholar] [CrossRef]
- Sebastião, F.A.; Furlan, L.R.; Hashimoto, D.T.; Pilarski, F. Identification of bacterial fish pathogens in Brazil by direct colony PCR and 16S rRNA gene sequencing. Adv. Microbiol. 2015, 05, 409–424. [Google Scholar] [CrossRef]
- Fukushima, H.C.S.; Leal, C.A.G.; Cavalcante, R.B.; Figueiredo, H.C.P.; Arijo, S.; Moriñigo, M.A.; Ishikawa, M.; Borra, R.C.; Ranzani-Paiva, M.J.T. Lactococcus garvieae outbreaks in Brazilian farms: Lactococcosis in Pseudoplatystoma sp. – Development of an autogenous vaccine as a control strategy. J. Fish Dis. 2017, 40, 263–272. [Google Scholar] [CrossRef]
- Assis, G.B.N.; Pereira, F.L.; Zegarra, A.U.; Tavares, G.C.; Leal, C.A.; Figueiredo, H.C.P. Use of MALDI-TOF mass spectrometry for the fast identification of Gram-positive fish pathogens. Front. Microbiol. 2017, 8, 1492. [Google Scholar] [CrossRef]
- Tavares, G.C.; de Queiroz, G.A.; Assis, G.B.N.; Leibowitz, M.P.; Teixeira, J.P.; Figueiredo, H.C.P.; Leal, C.A.G. Disease outbreaks in farmed Amazon catfish (Leiarius marmoratus x Pseudoplatystoma corruscans) caused by Streptococcus agalactiae, S. iniae, and S. dysgalactiae. Aquaculture 2018, 495, 384–392. [Google Scholar] [CrossRef]
- Rosário, A.E.C. do; Henrique, M.C.; Costa, É.J.C. da; Barbanti, A.C.C.; Tavares, G.C. Surtos de bacteriose em juvenis de pirarucu (Arapaima gigas) provevientes de pisciculturas amazônicas e avaliação da sensibilidade de Aeromonas jandaei a antimicrobianos. In Enfermidades parasitárias e bacterianas na piscicultura brasileira: insights e perspectivas; Cruz, M.G. da, Castro, J. da S., Jerônimo, G.T., Eds.; i-EDUCAM, 2023; pp. 33–46. [Google Scholar]
- Ferrario, C.; Ricci, G.; Milani, C.; Lugli, G.A.; Ventura, M.; Eraclio, G.; Borgo, F.; Fortina, M.G. Lactococcus garvieae: where is it from? A first approach to explore the evolutionary history of this emerging pathogen. PLoS One 2013, 8, e84796. [Google Scholar] [CrossRef] [PubMed]
- CLSI VET03: Methods for Antimicrobial Broth Dilution and Disk Diffusion Susceptibility Testing of Bacteria Isolated from Aquatic Animals; Clinical and Laboratory Standards Institute: Wayne, USA, 2020.
- Smith, P.; Finnegan, W.; Ngo, T.; Kronvall, G. Influence of incubation temperature and time on the precision of MIC and disc diffusion antimicrobial susceptibility test data. Aquaculture 2018, 490, 19–24. [Google Scholar] [CrossRef]
- Kronvall, G.; Kahlmeter, G.; Myhre, E.; Galas, M.F. A new method for normalized interpretation of antimicrobial resistance from disk test results for comparative purposes. Clin. Microbiol. Infect. 2003, 9, 120–132. [Google Scholar] [CrossRef]
- Silley, P. Susceptibility testing methods, resistance and breakpoints: what do these terms really mean? Rev. Sci. Tech. - Off. Int. des Épizooties 2012, 31, 33–41. [Google Scholar] [CrossRef]
- Smith, P. Eight rules for improving the quality of papers on the antimicrobial susceptibility of bacteria isolated from aquatic animals. Dis. Aquat. Organ. 2020, 139, 87–92. [Google Scholar] [CrossRef]
- R Core Team R: A language and environment for statistical computing 2024.
- Mauri, M.; Elli, T.; Caviglia, G.; Uboldi, G.; Azzi, M. RAWGraphs: A visualisation platform to create open outputs. In Proceedings of the Proceedings of the 12th Biannual Conference on Italian SIGCHI Chapter; Association for Computing Machinery: New York, NY, USA, 2017. [Google Scholar]
- Schwarz, S.; Silley, P.; Simjee, S.; Woodford, N.; van Duijkeren, E.; Johnson, A.P.; Gaastra, W. Assessing the antimicrobial susceptibility of bacteria obtained from animals. Vet. Microbiol. 2010, 141, 1–4. [Google Scholar] [CrossRef]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- McEwen, S.A.; Collignon, P.J. Antimicrobial resistance: a One Health perspective. Microbiol. Spectr. 2018, 6, 10–1128. [Google Scholar] [CrossRef]
- Torres-Corral, Y.; Santos, Y. Predicting antimicrobial resistance of Lactococcus garvieae: PCR detection of resistance genes versus MALDI-TOF protein profiling. Aquaculture 2022, 553, 738098. [Google Scholar] [CrossRef]
- Altan, E.; Korun, J. Lactococcus garvieae isolates from rainbow trout (Oncorhynchus mykiss, W.) compared by PLG and SA1B10 PCR primer pairs. J. Ilm. Platax 2021, 9, 18–28. [Google Scholar] [CrossRef]
- Kawanishi, M.; Kojima, A.; Ishihara, K.; Esaki, H.; Kijima, M.; Takahashi, T.; Suzuki, S.; Tamura, Y. Drug resistance and pulsed-field gel electrophoresis patterns of Lactococcus garvieae isolates from cultured Seriola (Yellowtail, Amberjack and Kingfish) in Japan. Lett. Appl. Microbiol. 2005, 40, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Lim, F.H.; Jenkins, D.R. Native valve endocarditis caused by Lactococcus garvieae: an emerging human pathogen. BMJ Case Rep. 2017, 2017, bcr–2017. [Google Scholar] [CrossRef] [PubMed]
- Korun, J.; Altan, E.; Teker, S.; Ulutaş, A. A study on the antimicrobial resistance of Lactococcus garvieae. Acta Aquat. 2021, 8, 98–102. [Google Scholar] [CrossRef]
- Sezgin, S.S.; Yılmaz, M.; Arslan, T.; Kubilay, A. Current antibiotic sensitivity of Lactococcus garvieae in rainbow trout (Oncorhynchus mykiss) farms from Southwestern Turkey. J. Agric. Sci. 2023, 29, 630–642. [Google Scholar] [CrossRef]
- Chang, P.H.; Lin, C.W.; Lee, Y.C. Lactococcus garvieae infection of cultured rainbow trout, Oncorhynchus mykiss, in Taiwan and associated biophysical characteristics and histopathology. Bull. Eur. Assoc. Fish Pathol. 2002, 22, 319–327. [Google Scholar]
- Majeed, S.; De Silva, L.A.D.S.; Kumarage, P.M.; Heo, G.J. Characterization of pathogenic Lactococcus garvieae isolated from farmed mullet (Mugil cephalus). Vet. Integr. Sci. 2025, 23, 1–17. [Google Scholar]
- Ture, M.; Boran, H. Phenotypic and genotypic antimicrobial resistance of Lactococcus sp. strains isolated from rainbow trout (Oncorhynchus mykiss). Bull. Vet. Inst. Pulawy 2015, 59, 37–42. [Google Scholar] [CrossRef]
- Vela, A.I.; del Mar Blanco, M.; Colussi, S.; Kotzamanidis, C.; Prearo, M.; Altinok, I.; Acutis, P.L.; Volpatti, D.; Alba, P.; Feltrin, F.; et al. The association of Lactococcus petauri with lactococcosis is older than expected. Aquaculture 2024, 578, 740057. [Google Scholar] [CrossRef]
- Chan, Y.-X.; Cao, H.; Jiang, S.; Li, X.; Fung, K.-K.; Lee, C.-H.; Sridhar, S.; Chen, J.H.-K.; Ho, P.-L. Genomic investigation of Lactococcus formosensis, Lactococcus garvieae, and Lactococcus petauri reveals differences in species distribution by human and animal sources. Microbiol. Spectr. 2024, 12, e00541–24. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Han, J.; Barkema, H.W.; Wang, Y.; Gao, J.; Kastelic, J.P.; Han, B.; Qin, S.; Deng, Z. Comparative genomic analyses of Lactococcus garvieae isolated from bovine mastitis in China. Microbiol. Spectr. 2023, 11, e02995–22. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, T.F.; Leibowitz, M.P.; Leal, C.A.G. Local epidemiological cutoff values and antimicrobial susceptibility profile for Brazilian Francisella orientalis isolates. Aquaculture 2022, 553, 738054. [Google Scholar] [CrossRef]
- Sun, K.; Wang, H.; Zhang, M.; Xiao, Z.; Sun, L. Genetic mechanisms of multi-antimicrobial resistance in a pathogenic Edwardsiella tarda strain. Aquaculture 2009, 289, 134–139. [Google Scholar] [CrossRef]
- Bondad-Reantaso, M.G.; MacKinnon, B.; Karunasagar, I.; Fridman, S.; Alday-Sanz, V.; Brun, E.; Le Groumellec, M.; Li, A.; Surachetpong, W.; Karunasagar, I.; et al. Review of alternatives to antibiotic use in aquaculture. Rev. Aquac. 2023, 15, 1421–1451. [Google Scholar] [CrossRef]
- Leal, C.A.G.; Silva, B.A.; Colombo, S.A. Susceptibility profile and epidemiological cut-off values are influenced by serotype in fish pathogenic Streptococcus agalactiae. Antibiotics 2023, 12. [Google Scholar] [CrossRef]
- Savvidis, G.K.; Anatoliotis, C.; Kanaki, Z.; Vafeas, G. Epizootic outbreaks of lactococcosis disease in rainbow trout, Oncorhynchus mykiss (Walbaum), culture in Greece. Bull. Eur. Assoc. Fish Pathol. 2007, 27, 223–228. [Google Scholar]
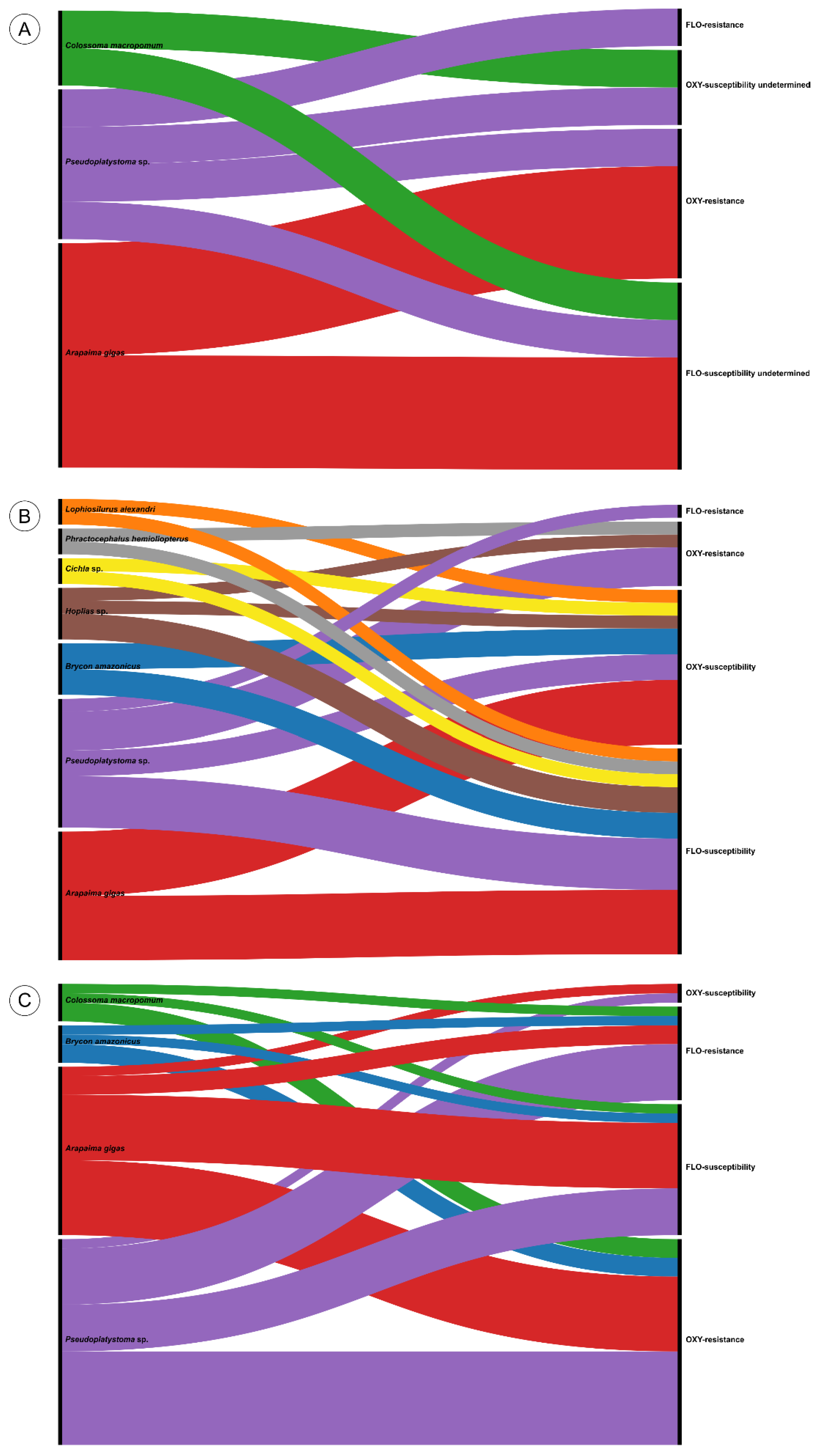
| Isolate | Species | Host | Origin | Tissue | Year | State | Reference |
|---|---|---|---|---|---|---|---|
| 167/23-02 | L. formosensis | Arapaima gigas | Farmed | Brain | 2023 | BA | [2] |
| 167/23-06 | L. formosensis | Arapaima gigas | Farmed | Brain | 2023 | BA | [2] |
| 167/23-09 | L. formosensis | Arapaima gigas | Farmed | Kidney | 2023 | BA | This study |
| AM-LG05 | L. formosensis | Colossoma macropomum | Farmed | Intestine | 2022 | AM | [2] |
| 52MS | L. formosensis | Pseudoplatystoma fasciatum | Farmed | Brain | 2012 | MS | [18] |
| LG91-23 | L. formosensis | Pseudoplatystoma sp. | Farmed | Brain | 2023 | MG | [2] |
| CRBP53 | L. garvieae | Arapaima gigas | Farmed | Intestine | 2023 | AM | [2] |
| CRBP54 | L. garvieae | Arapaima gigas | Farmed | Intestine | 2023 | AM | [2] |
| CRBP138 | L. garvieae | Arapaima gigas | Farmed | Intestine | 2023 | AM | [2] |
| CRBP144 | L. garvieae | Arapaima gigas | Farmed | Intestine | 2023 | AM | [2] |
| PA-LG01 | L. garvieae | Arapaima gigas | Farmed | Brain | 2018 | PA | [22] |
| LG88-23 | L. garvieae | Brycon amazonicus | Farmed | Brain | 2023 | MG | [2] |
| LG89-23 | L. garvieae | Brycon amazonicus | Farmed | Kidney | 2023 | MG | [2] |
| LG116-23 | L. garvieae | Cichla sp. | Wild | Brain | 2023 | MG | This study |
| LG63-21 | L. garvieae | Hoplias macrophtalmus | Farmed | Kidney | 2021 | MG | [2] |
| LG114-23 | L. garvieae | Hoplias malabaricus | Wild | Brain | 2023 | AM | This study |
| LG10-14 | L. garvieae | Lophiosilurus alexandri | Farmed | Brain | 2014 | MG | [20] |
| LG66-22 | L. garvieae | Phractocephalus hemioliopterus | Farmed | Kidney | 2022 | MG | [2] |
| LG09-14 | L. garvieae | Pseudoplatystoma corruscans | Farmed | Kidney | 2014 | SP | [20] |
| LG23-16 | L. garvieae | Pseudoplatystoma corruscans | Farmed | Brain | 2016 | SP | [21] |
| 177 | L. garvieae | Pseudoplatystoma fasciatum | Farmed | Brain | 2012 | MS | [19] |
| 31MS | L. garvieae | Pseudoplatystoma fasciatum | Farmed | Kidney | 2012 | MS | [18] |
| LG119-24 | L. garvieae | Pseudoplatystoma sp. | Farmed | Brain | 2024 | MG | This study |
| 167/23-03 | L. petauri | Arapaima gigas | Farmed | Kidney | 2023 | BA | This study |
| 167/23-04 | L. petauri | Arapaima gigas | Farmed | Kidney | 2023 | BA | This study |
| 167/23-05 | L. petauri | Arapaima gigas | Farmed | Kidney | 2023 | BA | This study |
| 167/23-07 | L. petauri | Arapaima gigas | Farmed | Kidney | 2023 | BA | This study |
| 167/23-08 | L. petauri | Arapaima gigas | Farmed | Kidney | 2023 | BA | This study |
| 167/23-10 | L. petauri | Arapaima gigas | Farmed | Spleen | 2023 | BA | This study |
| CRBT89 | L. petauri | Arapaima gigas | Farmed | Intestine | 2023 | AM | [2] |
| CRBT98 | L. petauri | Arapaima gigas | Farmed | Intestine | 2023 | AM | [2] |
| CRBP146 | L. petauri | Arapaima gigas | Farmed | Intestine | 2023 | AM | [2] |
| AM-LG07 | L. petauri | Brycon amazonicus | Farmed | Brain | 2022 | AM | [2] |
| AM-LG08 | L. petauri | Brycon amazonicus | Farmed | Brain | 2022 | AM | [2] |
| AM-LG02 | L. petauri | Colossoma macropomum | Farmed | Intestine | 2020 | AM | [2] |
| AM-LG03 | L. petauri | Colossoma macropomum | Farmed | Intestine | 2022 | AM | [2] |
| LG03-18 | L. petauri | Pseudoplatystoma corruscans | Farmed | Brain | 2018 | MG | [2] |
| 14MS | L. petauri | Pseudoplatystoma fasciatum | Farmed | Kidney | 2012 | MS | [18] |
| 176 | L. petauri | Pseudoplatystoma fasciatum | Farmed | Brain | 2012 | MS | [19] |
| 86 | L. petauri | Pseudoplatystoma sp. | Farmed | Brain | 2012 | MS | [19] |
| 89/2 | L. petauri | Pseudoplatystoma sp. | Farmed | Brain | 2012 | MS | [19] |
| 93 | L. petauri | Pseudoplatystoma sp. | Farmed | Brain | 2012 | MS | [19] |
| LG86-23 | L. petauri | Pseudoplatystoma sp. | Farmed | Kidney | 2023 | MG | [2] |
| LG94-23 | L. petauri | Pseudoplatystoma sp. | Farmed | Brain | 2023 | MG | [2] |
| LG104-23 | L. petauri | Pseudoplatystoma sp. | Farmed | Brain | 2023 | MG | [2] |
| LG106-23 | L. petauri | Pseudoplatystoma sp. | Farmed | Kidney | 2023 | MG | [2] |
| LG117-23 | L. petauri | Pseudoplatystoma sp. | Farmed | Kidney | 2023 | MG | This study |
| Antimicrobials | Minimum Value | Maximum Value | Mean ± SD | ECV (mm) | WT (%) | NWT* (%) |
|---|---|---|---|---|---|---|
| Lactococcus formosensisa | ||||||
| Amoxicillin | 19 | 27 | 23.2 ± 2.7 | - | - | - |
| Erythromycin | 20 | 30 | 25.7 ± 3.7 | - | - | - |
| Florfenicol | 6 | 28 | 22.3 ± 7.8 | - | - | 16.7 |
| Neomycin | 10 | 17 | 14.9 ± 2.4 | - | - | - |
| Norfloxacin | 6 | 6 | 6.0 ± 0.0 | - | - | 100 |
| Oxytetracycline | 6 | 27 | 9.5 ± 7.5 | - | - | 66.7 |
| Trimethoprim-sulfametoxazole | 6 | 6 | 6.0 ± 0.0 | - | - | 100 |
| Lactococcus garvieaeb | ||||||
| Amoxicillin | 18 | 28 | 21.4 ± 2.2 | ≥ 11 | 100 | 0 |
| Erythromycin | 16 | 31 | 24.7 ± 3.7 | ≥ 16 | 100 | 0 |
| Florfenicol | 6 | 29 | 20.9 ± 4.4 | ≥ 12 | 94.4 | 5.6 |
| Neomycin | 10 | 19 | 15.1 ± 2.7 | ≥ 7 | 100 | 0 |
| Norfloxacin | 6 | 19 | 9.0 ± 4.0 | - | - | 47 |
| Oxytetracycline | 6 | 27 | 16.5 ± 7.3 | ≥ 10 | 72.2 | 27.8 |
| Trimethoprim-sulfametoxazole | 6 | 19 | 7.2 ± 3.4 | - | - | 88.2 |
| Lactococcus petauric | ||||||
| Amoxicillin | 15 | 26 | 20.5 ± 3.0 | ≥ 16 | 95.8 | 4.2 |
| Erythromycin | 6 | 31 | 22.4 ± 7.1 | ≥ 23 | 66.7 | 33.3 |
| Florfenicol | 6 | 29 | 19.5 ± 6.9 | ≥ 21 | 62.5 | 37.5 |
| Neomycin | 10 | 19 | 14.2 ± 2.2 | ≥ 9 | 100 | 0 |
| Norfloxacin | 6 | 14 | 8.6 ± 2.9 | ≥ 13 | 16.7 | 83.3 |
| Oxytetracycline | 6 | 26 | 13.6 ± 7.5 | ≥ 23 | 16.7 | 83.3 |
| Trimethoprim-sulfametoxazole | 6 | 14 | 6.3 ± 1.4 | - | - | 95.8 |
| Escherichia coliATCC 25922d | ||||||
| Amoxicillin | 14 | 19 | 15.8 ± 2.4 | - | - | - |
| Erythromycin | 12 | 18 | 14.6 ± 2.8 | - | - | - |
| Florfenicol | 19 | 28 | 23.5 ± 4.4 | - | - | - |
| Neomycin | 16 | 20 | 18.0 ± 2.0 | - | - | - |
| Norfloxacin | 24 | 34 | 30.6 ± 5.7 | - | - | - |
| Oxytetracycline | 19 | 27 | 23.2 ± 3.3 | - | - | - |
| Trimethoprim-sulfametoxazole | 25 | 26 | 25.5 ± 0.7 | - | - | - |
| Aeromonas salmonicida subsp. salmonicidaATCC 33658d | ||||||
| Amoxicillin | 24 | 30 | 27.4 ± 3.1 | - | - | - |
| Erythromycin | 19 | 22 | 20.7 ± 1.5 | - | - | - |
| Florfenicol | 32 | 36 | 34.2 ± 1.7 | - | - | - |
| Neomycin | 12 | 20 | 17.3 ± 4.6 | - | - | - |
| Norfloxacin | 21 | 37 | 29.6 ± 8.0 | - | - | - |
| Oxytetracycline | 29 | 32 | 29.7 ± 1.5 | - | - | - |
| Trimethoprim-sulfametoxazole | 24 | 26 | 25.0 ± 1.4 | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





