Submitted:
29 October 2024
Posted:
30 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Metallation of Nuclear Fe-S Cluster Proteins
3. Fe-S Clusters Proteins Involved in Nuclear DNA Transactions
3.1. Fe-S Clusters and DNA Replication
3.1.1. DNA Polymerases
3.1.2. DNA Primase
3.1.3. Fe-S Clusters and DNA Replication Origins
3.1.4. Fe-S Clusters in Helicase Activity
3.1.5. Fe-S Clusters and Telomere Maintenance
3.2. Fe-S Clusters and DNA Repair
3.2.1. Fe-S Clusters, Transcription, and Nuclear RNA Transactions
3.2.2. Fe-S Cluster Binding Fe-Dependent Transcriptional Regulators
3.2.3. RNA Polymerase III
3.2.4. Transcription Factor IIH
3.2.5. RNA Modifying Proteins
4. Fe-S Cluster Proteins in Mitosis
5. Other Fe-S Cluster Proteins in the Nucleus
6. Conclusions and Open Questions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lill, R., Broderick, J. B. & Dean, D. R. Special issue on iron-sulfur proteins: Structure, function, biogenesis and diseases. Biochim Biophys Acta 1853, 1251-1252 (2015). [CrossRef]
- Braymer, J. J., Freibert, S. A., Rakwalska-Bange, M. & Lill, R. Mechanistic concepts of iron-sulfur protein biogenesis in Biology. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research 1868, 118863 (2021). [CrossRef]
- Bak, D. W. & Weerapana, E. Proteomic strategies to interrogate the Fe-S proteome. Biochim Biophys Acta Mol Cell Res 1871, 119791 (2024). [CrossRef]
- Rudolf, J., Makrantoni, V., Ingledew, W. J., Stark, M. J. & White, M. F. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Molecular cell 23, 801-808 (2006). [CrossRef]
- Thul, P. J. et al. A subcellular map of the human proteome. Science 356, eaal3321 (2017). [CrossRef]
- Consortium, T. U. UniProt: the universal protein knowledgebase in 2023. Nucleic acids research 51, D523-D531 (2023).
- Binder, J. X. et al. COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database 2014 (2014). [CrossRef]
- Klinge, S., Hirst, J., Maman, J. D., Krude, T. & Pellegrini, L. An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nature structural & molecular biology 14, 875-877 (2007). [CrossRef]
- Weiner, B. E. et al. An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. Journal of biological chemistry 282, 33444-33451 (2007). [CrossRef]
- Starokadomskyy, P. et al. DNA polymerase-α regulates the activation of type I interferons through cytosolic RNA: DNA synthesis. Nature immunology 17, 495-504 (2016). [CrossRef]
- Netz, D. J. et al. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nature chemical biology 8, 125-132 (2012).
- Chea, J. et al. Spatiotemporal recruitment of human DNA polymerase delta to sites of UV damage. Cell cycle 11, 2885-2895 (2012). [CrossRef]
- Roske, J. J. & Yeeles, J. T. Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol ε. Nature Structural & Molecular Biology, 1-11 (2024). [CrossRef]
- Ræder, S. B. et al. APIM-mediated REV3L–PCNA interaction important for error free TLS Over UV-induced DNA lesions in human cells. International Journal of Molecular Sciences 20, 100 (2018). [CrossRef]
- Cantor, S. B. et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 105, 149-160 (2001). [CrossRef]
- Seki, M., Takeda, Y., Iwai, K. & Tanaka, K. IOP1 protein is an external component of the human cytosolic iron-sulfur cluster assembly (CIA) machinery and functions in the MMS19 protein-dependent CIA pathway. Journal of Biological Chemistry 288, 16680-16689 (2013). [CrossRef]
- Wu, Y. et al. Fanconi anemia group J mutation abolishes its DNA repair function by uncoupling DNA translocation from helicase activity or disruption of protein-DNA complexes. Blood, The Journal of the American Society of Hematology 116, 3780-3791 (2010). [CrossRef]
- Parish, J. L. et al. The DNA helicase ChlR1 is required for sister chromatid cohesion in mammalian cells. Journal of cell science 119, 4857-4865 (2006). [CrossRef]
- Simon, A. K. et al. The iron–sulfur helicase DDX11 promotes the generation of single-stranded DNA for CHK1 activation. Life science alliance 3 (2020). [CrossRef]
- Landry, A. P. & Ding, H. The N-Terminal Domain of Human DNA Helicase Rtel1 Contains a Redox Active Iron-Sulfur Cluster. BioMed research international 2014, 285791 (2014). [CrossRef]
- Ito, S. et al. MMXD, a TFIIH-independent XPD-MMS19 protein complex involved in chromosome segregation. Molecular cell 39, 632-640 (2010). [CrossRef]
- Duxin, J. P. et al. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Molecular and cellular biology 29, 4274-4282 (2009). [CrossRef]
- Pokharel, S. & Campbell, J. L. Cross talk between the nuclease and helicase activities of Dna2: role of an essential iron–sulfur cluster domain. Nucleic acids research 40, 7821-7830 (2012). [CrossRef]
- Mariotti, L. et al. The iron–sulphur cluster in human DNA2 is required for all biochemical activities of DNA2. Communications biology 3, 322 (2020). [CrossRef]
- Sparks, J. L. et al. Human exonuclease 5 is a novel sliding exonuclease required for genome stability. Journal of Biological Chemistry 287, 42773-42783 (2012). [CrossRef]
- Komine, K. et al. Functional complementation assay for 47 MUTYH variants in a MutY-disrupted Escherichia coli strain. Human mutation 36, 704-711 (2015).
- Chepanoske, C. L., Golinelli, M.-P., Williams, S. D. & David, S. S. Positively Charged Residues within the Iron–Sulfur Cluster Loop of E. coli MutY Participate in Damage Recognition and Removal. Archives of biochemistry and biophysics 380, 11-19 (2000).
- Nuñez, N. N., Majumdar, C., Lay, K. T. & David, S. S. in Methods in enzymology Vol. 599 21-68 (Elsevier, 2018).
- Ikeda, S., Kohmoto, T., Tabata, R. & Seki, Y. Differential intracellular localization of the human and mouse endonuclease III homologs and analysis of the sorting signals. DNA repair 1, 847-854 (2002). [CrossRef]
- Kuo, C.-F. et al. Atomic structure of the DNA repair [4Fe-4S] enzyme endonuclease III. Science 258, 434-440 (1992). [CrossRef]
- Carroll, B. L. et al. Caught in motion: human NTHL1 undergoes interdomain rearrangement necessary for catalysis. Nucleic Acids Research 49, 13165-13178 (2021). [CrossRef]
- Kim, J.-H., Lane, W. S. & Reinberg, D. Human Elongator facilitates RNA polymerase II transcription through chromatin. Proceedings of the National Academy of Sciences 99, 1241-1246 (2002). [CrossRef]
- Paraskevopoulou, C., Fairhurst, S. A., Lowe, D. J., Brick, P. & Onesti, S. The elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Molecular microbiology 59, 795-806 (2006).
- Kimura, S. & Suzuki, T. Iron-sulfur proteins responsible for RNA modifications. Biochim Biophys Acta 1853, 1272-1283 (2015). [CrossRef]
- Ramsay, E. P. et al. Structure of human RNA polymerase III. Nature communications 11, 6409 (2020). [CrossRef]
- Blombach, F. et al. Archaeal TFEα/β is a hybrid of TFIIE and the RNA polymerase III subcomplex hRPC62/39. Elife 4, e08378 (2015).
- Li, L. et al. Structure of human RNA polymerase III elongation complex. Cell research 31, 791-800 (2021). [CrossRef]
- Chen, W. et al. CPSF4 activates telomerase reverse transcriptase and predicts poor prognosis in human lung adenocarcinomas. Molecular oncology 8, 704-716 (2014). [CrossRef]
- Shimberg, G. D. et al. Cleavage and polyadenylation specificity factor 30: An RNA-binding zinc-finger protein with an unexpected 2Fe-2S cluster. Proc Natl Acad Sci U S A 113, 4700-4705 (2016). [CrossRef]
- Pritts, J. D. et al. Unraveling the RNA Binding Properties of the Iron–Sulfur Zinc Finger Protein CPSF30. Biochemistry 59, 970-982 (2020). [CrossRef]
- Young, A. P. & Bandarian, V. in Methods in enzymology Vol. 606 119-153 (Elsevier, 2018).
- Reiter, V. et al. The CDK5 repressor CDK5RAP1 is a methylthiotransferase acting on nuclear and mitochondrial RNA. Nucleic Acids Res 40, 6235-6240 (2012). [CrossRef]
- Pierrel, F., Douki, T., Fontecave, M. & Atta, M. MiaB protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. Journal of Biological Chemistry 279, 47555-47563 (2004). [CrossRef]
- LEE, Y. M. et al. Human kinesin superfamily member 4 is dominantly localized in the nuclear matrix and is associated with chromosomes during mitosis. Biochemical Journal 360, 549-556 (2001).
- Ben-Shimon, L. et al. Fe-S cluster coordination of the chromokinesin KIF4A alters its subcellular localization during mitosis. Journal of Cell Science 131, jcs211433 (2018). [CrossRef]
- Bellelli, R. et al. NCOA4 transcriptional coactivator inhibits activation of DNA replication origins. Molecular cell 55, 123-137 (2014). [CrossRef]
- Kuno, S. & Iwai, K. Oxygen modulates iron homeostasis by switching iron sensing of NCOA4. Journal of Biological Chemistry 299 (2023). [CrossRef]
- Zhao, H. et al. NCOA4 requires a [3Fe-4S] to sense and maintain the iron homeostasis. Journal of Biological Chemistry 300 (2024). [CrossRef]
- Vinas-Castells, R. et al. Nuclear ubiquitination by FBXL5 modulates Snail1 DNA binding and stability. Nucleic acids research 42, 1079-1094 (2013). [CrossRef]
- Wang, H. et al. FBXL5 regulates IRP2 stability in iron homeostasis via an oxygen-responsive [2Fe2S] cluster. Molecular cell 78, 31-41. e35 (2020). [CrossRef]
- Bruening, W. et al. Expression of OVCA1, a candidate tumor suppressor, is reduced in tumors and inhibits growth of ovarian cancer cells. Cancer research 59, 4973-4983 (1999).
- Zhang, Y. et al. Diphthamide biosynthesis requires an organic radical generated by an iron–sulphur enzyme. Nature 465, 891-896 (2010). [CrossRef]
- Kwon, Y. T., Kashina, A. S. & Varshavsky, A. Alternative splicing results in differential expression, activity, and localization of the two forms of arginyl-tRNA-protein transferase, a component of the N-end rule pathway. Molecular and cellular biology 19, 182-193 (1999).
- Van, V. et al. Iron-sulfur clusters are involved in post-translational arginylation. Nature Communications 14, 458 (2023). [CrossRef]
- Du, Z. et al. Structure of the human respiratory complex II. Proceedings of the National Academy of Sciences 120, e2216713120 (2023).
- Hao, Z. et al. Subcellular localization of CIAPIN1. Journal of Histochemistry & Cytochemistry 54, 1437-1444 (2006).
- Banci, L. et al. Anamorsin is a [2Fe-2S] cluster-containing substrate of the Mia40-dependent mitochondrial protein trapping machinery. Chemistry & biology 18, 794-804 (2011). [CrossRef]
- Zhang, Y., Yang, C., Dancis, A. & Nakamaru-Ogiso, E. EPR studies of wild type and mutant Dre2 identify essential [2Fe–-2S] and [4Fe–-4S] clusters and their cysteine ligands. The journal of biochemistry, mvw054 (2016). [CrossRef]
- Willems, P. et al. BOLA1 is an aerobic protein that prevents mitochondrial morphology changes induced by glutathione depletion. Antioxidants & redox signaling 18, 129-138 (2013). [CrossRef]
- Li, H., Mapolelo, D. T., Randeniya, S., Johnson, M. K. & Outten, C. E. Human glutaredoxin 3 forms [2Fe-2S]-bridged complexes with human BolA2. Biochemistry 51, 1687-1696 (2012). [CrossRef]
- Frey, A. G., Palenchar, D. J., Wildemann, J. D. & Philpott, C. C. A Glutaredoxin· BolA Complex Serves as an Iron-Sulfur Cluster Chaperone for the Cytosolic Cluster Assembly Machinery*♦. Journal of Biological Chemistry 291, 22344-22356 (2016).
- Pandya, P., Braiman, A. & Isakov, N. PICOT (GLRX3) is a positive regulator of stress-induced DNA-damage response. Cellular signalling 62, 109340 (2019). [CrossRef]
- Okuno, T., Yamabayashi, H. & Kogure, K. Comparison of intracellular localization of Nubp1 and Nubp2 using GFP fusion proteins. Molecular biology reports 37, 1165-1168 (2010). [CrossRef]
- Netz, D. J., Pierik, A. J., Stümpfig, M., Mühlenhoff, U. & Lill, R. The Cfd1–Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol. Nature chemical biology 3, 278-286 (2007). [CrossRef]
- Stehling, O. et al. Human Nbp35 is essential for both cytosolic iron-sulfur protein assembly and iron homeostasis. Molecular and cellular biology 28, 5517-5528 (2008). [CrossRef]
- Tong, W.-H., Jameson, G. N., Huynh, B. H. & Rouault, T. A. Subcellular compartmentalization of human Nfu, an iron–sulfur cluster scaffold protein, and its ability to assemble a [4Fe–4S] cluster. Proceedings of the National Academy of Sciences 100, 9762-9767 (2003).
- Cai, K. et al. Structural/functional properties of human NFU1, an intermediate [4Fe-4S] carrier in human mitochondrial iron-sulfur cluster biogenesis. Structure 24, 2080-2091 (2016). [CrossRef]
- Land, T. & Rouault, T. A. Targeting of a human iron–sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Molecular cell 2, 807-815 (1998). [CrossRef]
- Fox, N. G. et al. Structure of the human frataxin-bound iron-sulfur cluster assembly complex provides insight into its activation mechanism. Nature communications 10, 2210 (2019). [CrossRef]
- Marelja, Z., Stöcklein, W., Nimtz, M. & Leimkühler, S. A novel role for human Nfs1 in the cytoplasm: Nfs1 acts as a sulfur donor for MOCS3, a protein involved in molybdenum cofactor biosynthesis. Journal of Biological Chemistry 283, 25178-25185 (2008).
- Lundberg, M. et al. Cloning and expression of a novel human glutaredoxin (Grx2) with mitochondrial and nuclear isoforms. Journal of Biological Chemistry 276, 26269-26275 (2001). [CrossRef]
- Lillig, C. H. et al. Characterization of human glutaredoxin 2 as iron–sulfur protein: a possible role as redox sensor. Proceedings of the National Academy of Sciences 102, 8168-8173 (2005). [CrossRef]
- Kispal, G., Csere, P., Prohl, C. & Lill, R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. The EMBO journal 18, 3981-3989 (1999). [CrossRef]
- Netz, D. J., Mascarenhas, J., Stehling, O., Pierik, A. J. & Lill, R. Maturation of cytosolic and nuclear iron–sulfur proteins. Trends in cell biology 24, 303-312 (2014). [CrossRef]
- Song, D. & Lee, F. S. A role for IOP1 in mammalian cytosolic iron-sulfur protein biogenesis. Journal of Biological Chemistry 283, 9231-9238 (2008). [CrossRef]
- Netz, D. J. et al. A bridging [4Fe-4S] cluster and nucleotide binding are essential for function of the Cfd1-Nbp35 complex as a scaffold in iron-sulfur protein maturation. Journal of Biological Chemistry 287, 12365-12378 (2012). [CrossRef]
- Stehling, O. et al. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science 337, 195-199 (2012). [CrossRef]
- Gari, K. et al. MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science 337, 243-245 (2012). [CrossRef]
- Balk, J., Pierik, A. J., Netz, D. J. A., Mühlenhoff, U. & Lill, R. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron–sulphur proteins. The EMBO journal 23, 2105-2115 (2004). [CrossRef]
- SantaMaria, A. M. & Rouault, T. A. Regulatory and Sensing Iron–Sulfur Clusters: New Insights and Unanswered Questions. Inorganics 12, 101 (2024). [CrossRef]
- Querci, L., Piccioli, M., Ciofi-Baffoni, S. & Banci, L. Structural aspects of iron-sulfur protein biogenesis: An NMR view. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 119786 (2024). [CrossRef]
- Maio, N. et al. Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell metabolism 19, 445-457 (2014). [CrossRef]
- Li, K., Tong, W.-H., Hughes, R. M. & Rouault, T. A. Roles of the mammalian cytosolic cysteine desulfurase, ISCS, and scaffold protein, ISCU, in iron-sulfur cluster assembly. Journal of Biological Chemistry 281, 12344-12351 (2006). [CrossRef]
- Kim, K. S., Maio, N., Singh, A. & Rouault, T. A. Cytosolic HSC20 integrates de novo iron–sulfur cluster biogenesis with the CIAO1-mediated transfer to recipients. Human molecular genetics 27, 837-852 (2018). [CrossRef]
- Shi, H., Bencze, K. Z., Stemmler, T. L. & Philpott, C. C. A cytosolic iron chaperone that delivers iron to ferritin. Science 320, 1207-1210 (2008). [CrossRef]
- Philpott, C. C. Coming into view: eukaryotic iron chaperones and intracellular iron delivery. Journal of Biological Chemistry 287, 13518-13523 (2012). [CrossRef]
- Philpott, C. C., Ryu, M.-S., Frey, A. & Patel, S. Cytosolic iron chaperones: proteins delivering iron cofactors in the cytosol of mammalian cells. Journal of Biological Chemistry 292, 12764-12771 (2017). [CrossRef]
- Philpott, C. C., Patel, S. J. & Protchenko, O. Management versus miscues in the cytosolic labile iron pool: The varied functions of iron chaperones. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research 1867, 118830 (2020). [CrossRef]
- Patel, S. J. et al. A PCBP1–BolA2 chaperone complex delivers iron for cytosolic [2Fe–2S] cluster assembly. Nature chemical biology 15, 872-881 (2019).
- Adamec, J. et al. Iron-dependent self-assembly of recombinant yeast frataxin: implications for Friedreich ataxia. The American Journal of Human Genetics 67, 549-562 (2000). [CrossRef]
- Bulteau, A.-L. et al. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science 305, 242-245 (2004). [CrossRef]
- Cavadini, P., O’Neill, H. A., Benada, O. & Isaya, G. Assembly and iron-binding properties of human frataxin, the protein deficient in Friedreich ataxia. Human molecular genetics 11, 217-227 (2002). [CrossRef]
- Bencze, K. Z. et al. The structure and function of frataxin. Critical reviews in biochemistry and molecular biology 41, 269-291 (2006).
- Parent, A. et al. Mammalian frataxin directly enhances sulfur transfer of NFS1 persulfide to both ISCU and free thiols. Nature communications 6, 5686 (2015). [CrossRef]
- Bridwell-Rabb, J., Fox, N. G., Tsai, C.-L., Winn, A. M. & Barondeau, D. P. Human frataxin activates Fe–S cluster biosynthesis by facilitating sulfur transfer chemistry. Biochemistry 53, 4904-4913 (2014). [CrossRef]
- Balk, J., Aguilar Netz, D. J., Tepper, K., Pierik, A. J. & Lill, R. The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron-sulfur protein assembly. Molecular and cellular biology 25, 10833-10841 (2005). [CrossRef]
- Stehling, O. et al. Human CIA2A-FAM96A and CIA2B-FAM96B integrate iron homeostasis and maturation of different subsets of cytosolic-nuclear iron-sulfur proteins. Cell metabolism 18, 187-198 (2013). [CrossRef]
- Marquez, M. D. et al. Cytosolic iron–sulfur protein assembly system identifies clients by a C-terminal tripeptide. Proceedings of the National Academy of Sciences 120, e2311057120 (2023). [CrossRef]
- Paul, V. D. et al. The deca-GX3 proteins Yae1-Lto1 function as adaptors recruiting the ABC protein Rli1 for iron-sulfur cluster insertion. Elife 4, e08231 (2015). [CrossRef]
- Odermatt, D. & Gari, K. (2017).
- Kassube, S. A. & Thomä, N. H. Structural insights into Fe–S protein biogenesis by the CIA targeting complex. Nature structural & molecular biology 27, 735-742 (2020). [CrossRef]
- Alhebshi, A., Sideri, T. C., Holland, S. L. & Avery, S. V. The essential iron-sulfur protein Rli1 is an important target accounting for inhibition of cell growth by reactive oxygen species. Molecular biology of the cell 23, 3582-3590 (2012). [CrossRef]
- Honarmand Ebrahimi, K. et al. Iron–sulfur clusters as inhibitors and catalysts of viral replication. Nature Chemistry 14, 253-266 (2022). [CrossRef]
- Jang, S. & Imlay, J. A. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. Journal of Biological Chemistry 282, 929-937 (2007). [CrossRef]
- Mettert, E. L. & Kiley, P. J. Fe–S proteins that regulate gene expression. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research 1853, 1284-1293 (2015).
- Imlay, J. A. Iron-sulphur clusters and the problem with oxygen. Molecular microbiology 59, 1073-1082 (2006). [CrossRef]
- Mettert, E. L. & Kiley, P. J. How is Fe-S cluster formation regulated? Annual review of microbiology 69, 505-526 (2015).
- Lauble, H., Kennedy, M., Beinert, H. & Stout, C. Crystal structures of aconitase with isocitrate and nitroisocitrate bound. Biochemistry 31, 2735-2748 (1992). [CrossRef]
- Federico, G. et al. NCOA4 links iron bioavailability to DNA metabolism. Cell Reports 40 (2022). [CrossRef]
- van Wietmarschen, N., Moradian, A., Morin, G. B., Lansdorp, P. M. & Uringa, E.-J. The mammalian proteins MMS19, MIP18, and ANT2 are involved in cytoplasmic iron-sulfur cluster protein assembly. Journal of Biological Chemistry 287, 43351-43358 (2012). [CrossRef]
- Puig, S., Ramos-Alonso, L., Romero, A. & Martínez-Pastor, M. (2017).
- Troadec, M.-B., Loréal, O. & Brissot, P. The interaction of iron and the genome: For better and for worse. Mutation Research/Reviews in Mutation Research 774, 25-32 (2017). [CrossRef]
- Veatch, J. R., McMurray, M. A., Nelson, Z. W. & Gottschling, D. E. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell 137, 1247-1258 (2009). [CrossRef]
- Shi, R., Hou, W., Wang, Z.-Q. & Xu, X. Biogenesis of iron–sulfur clusters and their role in DNA metabolism. Frontiers in Cell and Developmental Biology 9, 735678 (2021). [CrossRef]
- Loeb, L. A. & Monnat Jr, R. J. DNA polymerases and human disease. Nature Reviews Genetics 9, 594-604 (2008).
- Ter Beek, J. et al. Structural evidence for an essential Fe–S cluster in the catalytic core domain of DNA polymerase ϵ. Nucleic acids research 47, 5712-5722 (2019). [CrossRef]
- Jain, R. et al. An iron–sulfur cluster in the polymerase domain of yeast DNA polymerase ε. Journal of molecular biology 426, 301-308 (2014).
- Baranovskiy, A. G. et al. DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ. Journal of Biological Chemistry 287, 17281-17287 (2012).
- Lisova, A. E. et al. The iron-sulfur cluster is essential for DNA binding by human DNA polymerase ε. Scientific reports 12, 17436 (2022). [CrossRef]
- Bartels, P. L., Stodola, J. L., Burgers, P. M. & Barton, J. K. A redox role for the [4Fe4S] cluster of yeast DNA polymerase δ. Journal of the American Chemical Society 139, 18339-18348 (2017). [CrossRef]
- Baranovskiy, A. G. et al. Mechanism of concerted RNA-DNA primer synthesis by the human primosome. Journal of Biological Chemistry 291, 10006-10020 (2016). [CrossRef]
- Suwa, Y. et al. Crystal structure of the human Pol α B subunit in complex with the C-terminal domain of the catalytic subunit. Journal of Biological Chemistry 290, 14328-14337 (2015). [CrossRef]
- Klinge, S., Núñez-Ramírez, R., Llorca, O. & Pellegrini, L. 3D architecture of DNA Pol α reveals the functional core of multi-subunit replicative polymerases. The EMBO journal 28, 1978-1987 (2009). [CrossRef]
- Zhang, Y., Baranovskiy, A. G., Tahirov, T. H. & Pavlov, Y. I. The C-terminal domain of the DNA polymerase catalytic subunit regulates the primase and polymerase activities of the human DNA polymerase α-primase complex. Journal of Biological Chemistry 289, 22021-22034 (2014). [CrossRef]
- Baranovskiy, A. G. & Tahirov, T. H. Elaborated action of the human primosome. Genes 8, 62 (2017). [CrossRef]
- Pritts, J. D. & Michel, S. L. Fe-S clusters masquerading as zinc finger proteins. Journal of Inorganic Biochemistry 230, 111756 (2022). [CrossRef]
- Maio, N. et al. An iron–sulfur cluster in the zinc-binding domain of the SARS-CoV-2 helicase modulates its RNA-binding and-unwinding activities. Proceedings of the National Academy of Sciences 120, e2303860120 (2023).
- Maio, N. et al. Fe-S cofactors in the SARS-CoV-2 RNA-dependent RNA polymerase are potential antiviral targets. Science 373, 236-241 (2021).
- Shimberg, G. D. et al. Cleavage and polyadenylation specificity factor 30: An RNA-binding zinc-finger protein with an unexpected 2Fe–2S cluster. Proceedings of the National Academy of Sciences 113, 4700-4705 (2016). [CrossRef]
- Conlan, A. R. et al. Crystal structure of Miner1: The redox-active 2Fe-2S protein causative in Wolfram Syndrome 2. Journal of molecular biology 392, 143-153 (2009). [CrossRef]
- Cutone, A. et al. Pichia pastoris Fep1 is a [2Fe-2S] protein with a Zn finger that displays an unusual oxygen-dependent role in cluster binding. Scientific Reports 6, 31872 (2016). [CrossRef]
- Liu, L. & Huang, M. Essential role of the iron-sulfur cluster binding domain of the primase regulatory subunit Pri2 in DNA replication initiation. Protein & Cell 6, 194-210 (2015). [CrossRef]
- Baranovskiy, A. G. et al. Crystal structure of the human primase. Journal of Biological Chemistry 290, 5635-5646 (2015). [CrossRef]
- Sauguet, L., Klinge, S., Perera, R. L., Maman, J. D. & Pellegrini, L. Shared active site architecture between the large subunit of eukaryotic primase and DNA photolyase. PLoS One 5, e10083 (2010). [CrossRef]
- Agarkar, V. B., Babayeva, N. D., Pavlov, Y. I. & Tahirov, T. H. Crystal structure of the C-terminal domain of human DNA primase large subunit: Implications for the mechanism of the primase-polymerase α switch. Cell Cycle 10, 926-931 (2011).
- O’Brien, E. et al. The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport. Science 355, eaag1789 (2017). [CrossRef]
- O’Brien, E. et al. Yeast require redox switching in DNA primase. Proceedings of the National Academy of Sciences 115, 13186-13191 (2018).
- Amin, M. & Brooks, B. R. The oxidation of the [4Fe-4S] cluster of DNA primase alters the binding energies with DNA and RNA primers. Biophysical Journal (2024). [CrossRef]
- O’Brien, E., Holt, M. E., Salay, L. E., Chazin, W. J. & Barton, J. K. Substrate binding regulates redox signaling in human DNA primase. Journal of the American Chemical Society 140, 17153-17162 (2018). [CrossRef]
- Mancias, J. D., Wang, X., Gygi, S. P., Harper, J. W. & Kimmelman, A. C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509, 105-109 (2014). [CrossRef]
- Dowdle, W. E. et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nature cell biology 16, 1069-1079 (2014). [CrossRef]
- Singleton, M. R., Dillingham, M. S. & Wigley, D. B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76, 23-50 (2007). [CrossRef]
- Wu, Y. & Brosh Jr, R. M. DNA helicase and helicase–nuclease enzymes with a conserved iron–sulfur cluster. Nucleic acids research 40, 4247-4260 (2012).
- Yeeles, J. T., Cammack, R. & Dillingham, M. S. An iron-sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases. Journal of Biological Chemistry 284, 7746-7755 (2009).
- Zheng, L. et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Molecular cell 32, 325-336 (2008). [CrossRef]
- Estep, K. N. & Brosh Jr, R. M. RecQ and Fe–S helicases have unique roles in DNA metabolism dictated by their unwinding directionality, substrate specificity, and protein interactions. Biochemical Society Transactions 46, 77-95 (2018).
- Matsuzaki, K., Borel, V., Adelman, C. A., Schindler, D. & Boulton, S. J. FANCJ suppresses microsatellite instability and lymphomagenesis independent of the Fanconi anemia pathway. Genes & Development 29, 2532-2546 (2015). [CrossRef]
- Capo-Chichi, J. M. et al. Identification and biochemical characterization of a novel mutation in DDX11 causing W arsaw breakage syndrome. Human mutation 34, 103-107 (2013).
- Deng, Z. et al. Inherited mutations in the helicase RTEL1 cause telomere dysfunction and Hoyeraal–Hreidarsson syndrome. Proceedings of the National Academy of Sciences 110, E3408-E3416 (2013). [CrossRef]
- Falquet, B. et al. Disease-associated DNA2 nuclease–helicase protects cells from lethal chromosome under-replication. Nucleic acids research 48, 7265-7278 (2020). [CrossRef]
- Sommers, J. A. et al. FANCJ uses its motor ATPase to destabilize protein-DNA complexes, unwind triplexes, and inhibit RAD51 strand exchange. Journal of Biological Chemistry 284, 7505-7517 (2009). [CrossRef]
- Levran, O. et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nature genetics 37, 931-933 (2005). [CrossRef]
- Odermatt, D. C. et al. Cancer-associated mutations in the iron-sulfur domain of FANCJ affect G-quadruplex metabolism. PLoS genetics 16, e1008740 (2020). [CrossRef]
- Bharti, S. K. et al. Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability. Journal of Biological Chemistry 288, 28217-28229 (2013). [CrossRef]
- Wulfridge, P. & Sarma, K. Intertwining roles of R-loops and G-quadruplexes in DNA repair, transcription and genome organization. Nature Cell Biology, 1-12 (2024). [CrossRef]
- Hänsel-Hertsch, R., Di Antonio, M. & Balasubramanian, S. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nature reviews Molecular cell biology 18, 279-284 (2017). [CrossRef]
- Wu, Y., Shin-ya, K. & Brosh Jr, R. M. FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Molecular and cellular biology 28, 4116-4128 (2008).
- London, T. B. et al. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. Journal of Biological Chemistry 283, 36132-36139 (2008). [CrossRef]
- Wu, C. G. & Spies, M. G-quadruplex recognition and remodeling by the FANCJ helicase. Nucleic acids research 44, 8742-8753 (2016). [CrossRef]
- Vannier, J.-B. et al. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science 342, 239-242 (2013). [CrossRef]
- Uringa, E.-J., Youds, J. L., Lisaingo, K., Lansdorp, P. M. & Boulton, S. J. RTEL1: an essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic acids research 39, 1647-1655 (2011). [CrossRef]
- Wu, W. et al. RTEL1 suppresses G-quadruplex-associated R-loops at difficult-to-replicate loci in the human genome. Nature structural & molecular biology 27, 424-437 (2020). [CrossRef]
- Ghisays, F. et al. RTEL1 influences the abundance and localization of TERRA RNA. Nature communications 12, 3016 (2021). [CrossRef]
- Wu, Y., Sommers, J. A., Khan, I., de Winter, J. P. & Brosh, R. M. Biochemical characterization of Warsaw breakage syndrome helicase. Journal of Biological Chemistry 287, 1007-1021 (2012). [CrossRef]
- van Schie, J. J. et al. Warsaw Breakage Syndrome associated DDX11 helicase resolves G-quadruplex structures to support sister chromatid cohesion. Nature communications 11, 4287 (2020).
- Gray, L. T., Vallur, A. C., Eddy, J. & Maizels, N. G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nature chemical biology 10, 313-318 (2014). [CrossRef]
- Kou, H., Zhou, Y., Gorospe, R. C. & Wang, Z. Mms19 protein functions in nucleotide excision repair by sustaining an adequate cellular concentration of the TFIIH component Rad3. Proceedings of the National Academy of Sciences 105, 15714-15719 (2008). [CrossRef]
- Seroz, T. et al. Cloning of a human homolog of the yeast nucleotide excision repair gene MMS19 and interaction with transcription repair factor TFIIH via the XPB and XPD helicases. Nucleic acids research 28, 4506-4513 (2000). [CrossRef]
- Ding, H. et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell 117, 873-886 (2004).
- Askree, S. H. et al. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proceedings of the National Academy of Sciences 101, 8658-8663 (2004). [CrossRef]
- Friedberg, E. C., Walker, G. C., Siede, W. & Wood, R. D. DNA repair and mutagenesis. (American Society for Microbiology Press, 2005).
- Knijnenburg, T. A. et al. Genomic and molecular landscape of DNA damage repair deficiency across the cancer genome atlas. Cell reports 23, 239-254. e236 (2018). [CrossRef]
- Fuss, J. O., Tsai, C.-L., Ishida, J. P. & Tainer, J. A. Emerging critical roles of Fe–S clusters in DNA replication and repair. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research 1853, 1253-1271 (2015). [CrossRef]
- Cunningham, R. P. et al. Endonuclease III is an iron-sulfur protein. Biochemistry 28, 4450-4455 (1989). [CrossRef]
- Hinks, J. A. et al. An iron-sulfur cluster in the family 4 uracil-DNA glycosylases. Journal of Biological Chemistry 277, 16936-16940 (2002). [CrossRef]
- Parker, A., Gu, Y. & Lu, A.-L. Purification and characterization of a mammalian homolog of Escherichia coli MutY mismatch repair protein from calf liver mitochondria. Nucleic acids research 28, 3206-3215 (2000). [CrossRef]
- Jacobs, A. L. & Schär, P. DNA glycosylases: in DNA repair and beyond. Chromosoma 121, 1-20 (2012). [CrossRef]
- McGoldrick, J. P., Yeh, Y.-C., Solomon, M., Essigmann, J. M. & Lu, A.-L. Characterization of a mammalian homolog of the Escherichia coli MutY mismatch repair protein. Molecular and cellular biology 15, 989-996 (1995). [CrossRef]
- Aspinwall, R. et al. Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proceedings of the National Academy of Sciences 94, 109-114 (1997). [CrossRef]
- Lukianova, O. A. & David, S. S. A role for iron–sulfur clusters in DNA repair. Current opinion in chemical biology 9, 145-151 (2005).
- Trasviña-Arenas, C. H., Lopez-Castillo, L. M., Sanchez-Sandoval, E. & Brieba, L. G. Dispensability of the [4Fe-4S] cluster in novel homologues of adenine glycosylase MutY. The FEBS journal 283, 521-540 (2016). [CrossRef]
- Guan, Y. et al. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nature structural biology 5, 1058-1064 (1998). [CrossRef]
- Boal, A. K. et al. DNA-bound redox activity of DNA repair glycosylases containing [4Fe-4S] clusters. Biochemistry 44, 8397-8407 (2005). [CrossRef]
- Romano, C. A., Sontz, P. A. & Barton, J. K. Mutants of the base excision repair glycosylase, endonuclease III: DNA charge transport as a first step in lesion detection. Biochemistry 50, 6133-6145 (2011). [CrossRef]
- Porello, S. L., Cannon, M. J. & David, S. S. A substrate recognition role for the [4Fe-4S] 2+ cluster of the DNA repair glycosylase MutY. Biochemistry 37, 6465-6475 (1998). [CrossRef]
- Bartels, P. L. et al. Electrochemistry of the [4Fe4S] cluster in base excision repair proteins: tuning the redox potential with DNA. Langmuir 33, 2523-2530 (2017). [CrossRef]
- Tse, E. C., Zwang, T. J. & Barton, J. K. The oxidation state of [4Fe4S] clusters modulates the DNA-binding affinity of DNA repair proteins. Journal of the American Chemical Society 139, 12784-12792 (2017). [CrossRef]
- Bruner, S. D., Norman, D. P. & Verdine, G. L. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. nature 403, 859-866 (2000). [CrossRef]
- Poor, C. B. et al. Molecular mechanism and structure of the Saccharomyces cerevisiae iron regulator Aft2. Proc Natl Acad Sci U S A 111, 4043-4048 (2014). [CrossRef]
- Ueta, R., Fujiwara, N., Iwai, K. & Yamaguchi-Iwai, Y. Iron-induced dissociation of the Aft1p transcriptional regulator from target gene promoters is an initial event in iron-dependent gene suppression. Mol Cell Biol 32, 4998-5008 (2012). [CrossRef]
- Li, H. et al. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry 48, 9569-9581 (2009).
- Mercier, A., Watt, S., Bähler, J. r. & Labbé, S. Key function for the CCAAT-binding factor Php4 to regulate gene expression in response to iron deficiency in fission yeast. Eukaryotic cell 7, 493-508 (2008). [CrossRef]
- Vachon, P., Mercier, A., Jbel, M. & Labbé, S. The monothiol glutaredoxin Grx4 exerts an iron-dependent inhibitory effect on Php4 function. Eukaryotic Cell 11, 806-819 (2012). [CrossRef]
- Dlouhy, A. C., Beaudoin, J., Labbé, S. & Outten, C. E. Schizosaccharomyces pombe Grx4 regulates the transcriptional repressor Php4 via [2Fe–2S] cluster binding. Metallomics 9, 1096-1105 (2017). [CrossRef]
- Hati, D. et al. Iron homeostasis proteins Grx4 and Fra2 control activity of the Schizosaccharomyces pombe iron repressor Fep1 by facilitating [2Fe-2S] cluster removal. J Biol Chem 299, 105419 (2023). [CrossRef]
- Jacques, J. F., Mercier, A., Brault, A., Mourer, T. & Labbe, S. Fra2 is a co-regulator of Fep1 inhibition in response to iron starvation. PLoS One 9, e98959 (2014). [CrossRef]
- Pelletier, B., Trott, A., Morano, K. A. & Labbé, S. Functional characterization of the iron-regulatory transcription factor Fep1 from Schizosaccharomyces pombe. Journal of Biological Chemistry 280, 25146-25161 (2005). [CrossRef]
- Ogunjimi, B. et al. Inborn errors in RNA polymerase III underlie severe varicella zoster virus infections. The Journal of clinical investigation 127, 3543-3556 (2017). [CrossRef]
- Girbig, M. et al. Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states. Nature structural & molecular biology 28, 210-219 (2021). [CrossRef]
- Wang, Q. et al. Structural insights into transcriptional regulation of human RNA polymerase III. Nature structural & molecular biology 28, 220-227 (2021). [CrossRef]
- Wang, Z. & Roeder, R. G. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes & development 11, 1315-1326 (1997). [CrossRef]
- Chiu, Y.-H., MacMillan, J. B. & Chen, Z. J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138, 576-591 (2009).
- Tian, K., Wang, R., Huang, J., Wang, H. & Ji, X. Subcellular localization shapes the fate of RNA polymerase III. Cell Reports 42 (2023). [CrossRef]
- Zurita, M. & Merino, C. The transcriptional complexity of the TFIIH complex. TRENDS in Genetics 19, 578-584 (2003). [CrossRef]
- Giglia-Mari, G. et al. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nature genetics 36, 714-719 (2004). [CrossRef]
- Keriel, A., Stary, A., Sarasin, A., Rochette-Egly, C. & Egly, J.-M. XPD mutations prevent TFIIH-dependent transactivation by nuclear receptors and phosphorylation of RARα. Cell 109, 125-135 (2002). [CrossRef]
- Dubaele, S. et al. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Molecular cell 11, 1635-1646 (2003). [CrossRef]
- Compe, E. et al. Dysregulation of the peroxisome proliferator-activated receptor target genes by XPD mutations. Molecular and cellular biology 25, 6065-6076 (2005). [CrossRef]
- Pritts, J. D. & Michel, S. L. J. Fe-S clusters masquerading as zinc finger proteins. J Inorg Biochem 230, 111756 (2022). [CrossRef]
- Abbassi, N.-e.-H. et al. Cryo-EM structures of the human Elongator complex at work. Nature Communications 15, 4094 (2024). [CrossRef]
- Selvadurai, K., Wang, P., Seimetz, J. & Huang, R. H. Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nature chemical biology 10, 810-812 (2014). [CrossRef]
- Dalwadi, U. & Yip, C. K. Structural insights into the function of Elongator. Cellular and Molecular Life Sciences 75, 1613-1622 (2018). [CrossRef]
- Maio, N. et al. CIAO1 loss of function causes a neuromuscular disorder with compromise of nucleocytoplasmic Fe-S enzymes. The Journal of Clinical Investigation 134 (2024). [CrossRef]
- Romero, A. M., Martinez-Pastor, M. T. & Puig, S. Iron in Translation: From the Beginning to the End. Microorganisms 9 (2021). [CrossRef]
- Sun, C. et al. Wybutosine hypomodification of tRNAphe activates HERVK and impairs neuronal differentiation. Iscience 27 (2024). [CrossRef]
- Kessler, A. C., Silveira d'Almeida, G. & Alfonzo, J. D. The role of intracellular compartmentalization on tRNA processing and modification. RNA biology 15, 554-566 (2018).
- Wei, F. Y. et al. Cdk5rap1-mediated 2-methylthio modification of mitochondrial tRNAs governs protein translation and contributes to myopathy in mice and humans. Cell Metab 21, 428-442 (2015). [CrossRef]
- Sheng, L., Hao, S.-L., Yang, W.-X. & Sun, Y. The multiple functions of kinesin-4 family motor protein KIF4 and its clinical potential. Gene 678, 90-99 (2018). [CrossRef]
- Lee, Y. M. & Kim, W. Kinesin superfamily protein member 4 (KIF4) is localized to midzone and midbody in dividing cells. Experimental & Molecular Medicine 36, 93-97 (2004). [CrossRef]
- Mazumdar, M., Sundareshan, S. & Misteli, T. Human chromokinesin KIF4A functions in chromosome condensation and segregation. The Journal of cell biology 166, 613-620 (2004). [CrossRef]
- Zhu, C. & Jiang, W. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proceedings of the National Academy of Sciences 102, 343-348 (2005). [CrossRef]
- Kurasawa, Y., Earnshaw, W. C., Mochizuki, Y., Dohmae, N. & Todokoro, K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. The EMBO journal 23, 3237-3248 (2004). [CrossRef]
- Mazumdar, M., Sung, M.-H. & Misteli, T. Chromatin maintenance by a molecular motor protein. Nucleus 2, 591-600 (2011). [CrossRef]
- Wu, G. et al. A novel role of the chromokinesin Kif4A in DNA damage response. Cell Cycle 7, 2013-2020 (2008). [CrossRef]
- Kalantari, S. et al. Expanding the KIF4A-associated phenotype. American Journal of Medical Genetics Part A 185, 3728-3739 (2021).
- Gad, S. A. et al. The kinesin KIF4 mediates HBV/HDV entry through the regulation of surface NTCP localization and can be targeted by RXR agonists in vitro. PLoS Pathogens 18, e1009983 (2022). [CrossRef]
- Mazumdar, M. & Misteli, T. Chromokinesins: multitalented players in mitosis. Trends in cell biology 15, 349-355 (2005). [CrossRef]
- Wu, G. & Chen, P.-L. Structural requirements of chromokinesin Kif4A for its proper function in mitosis. Biochemical and biophysical research communications 372, 454-458 (2008). [CrossRef]
- Barton, J. K., Silva, R. M. & O'Brien, E. Redox chemistry in the genome: emergence of the [4Fe4S] cofactor in repair and replication. Annual review of biochemistry 88, 163-190 (2019). [CrossRef]
- Ueda, C., Langton, M., Chen, J. & Pandelia, M.-E. The HBx protein from hepatitis B virus coordinates a redox-active Fe-S cluster. Journal of Biological Chemistry 298 (2022). [CrossRef]
- Henkler, F. et al. Intracellular localization of the hepatitis B virus HBx protein. Journal of General Virology 82, 871-882 (2001). [CrossRef]
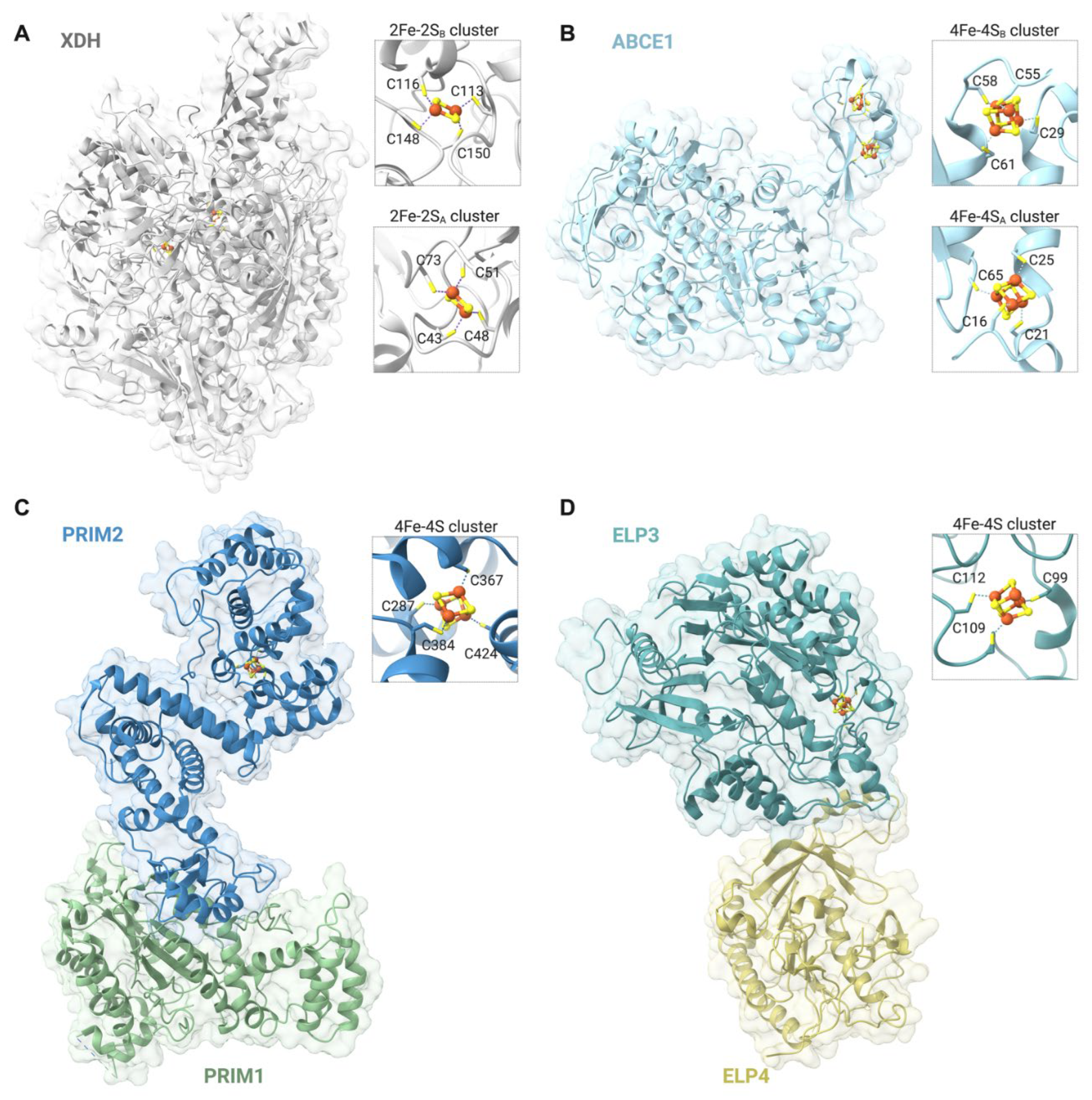
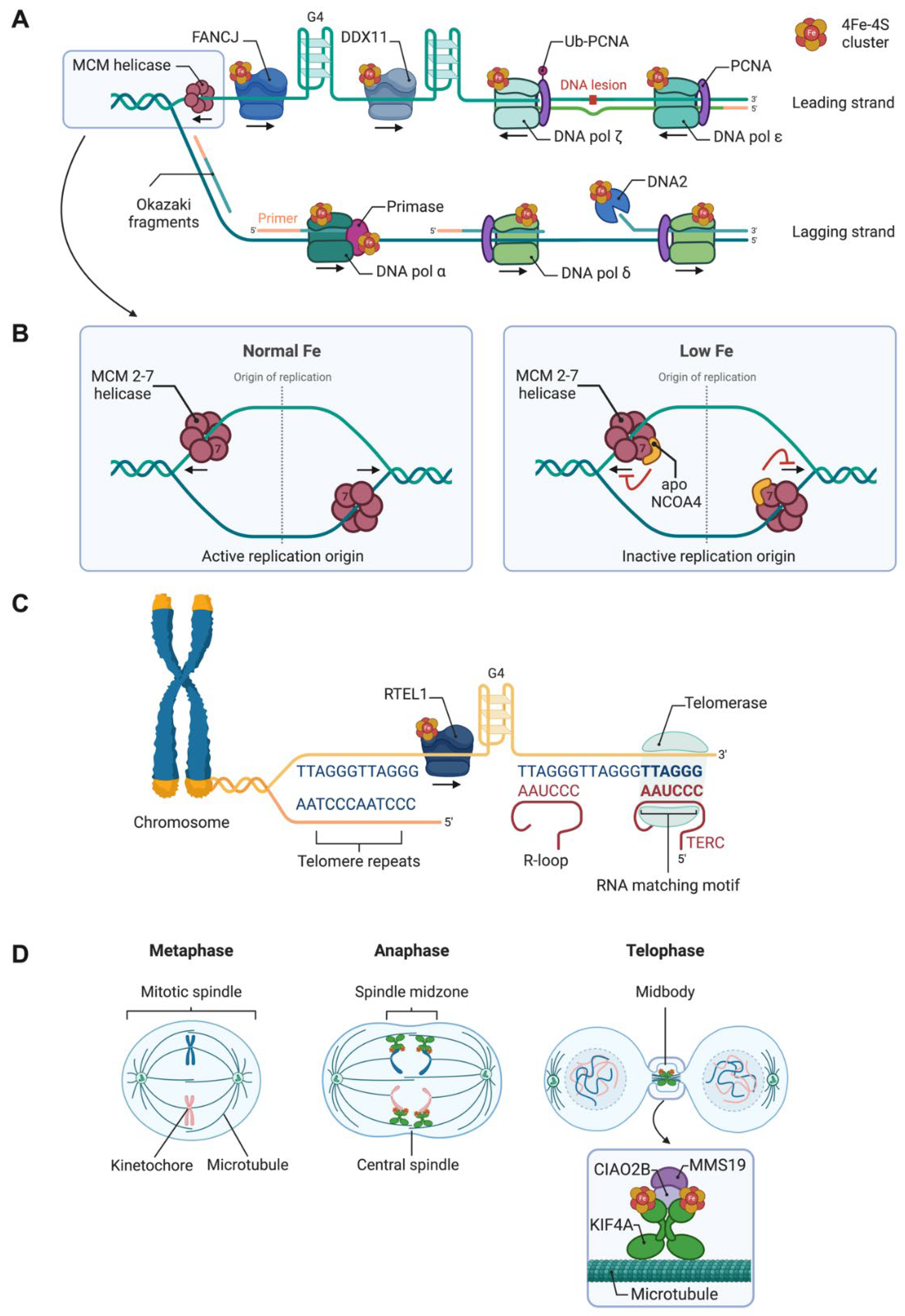
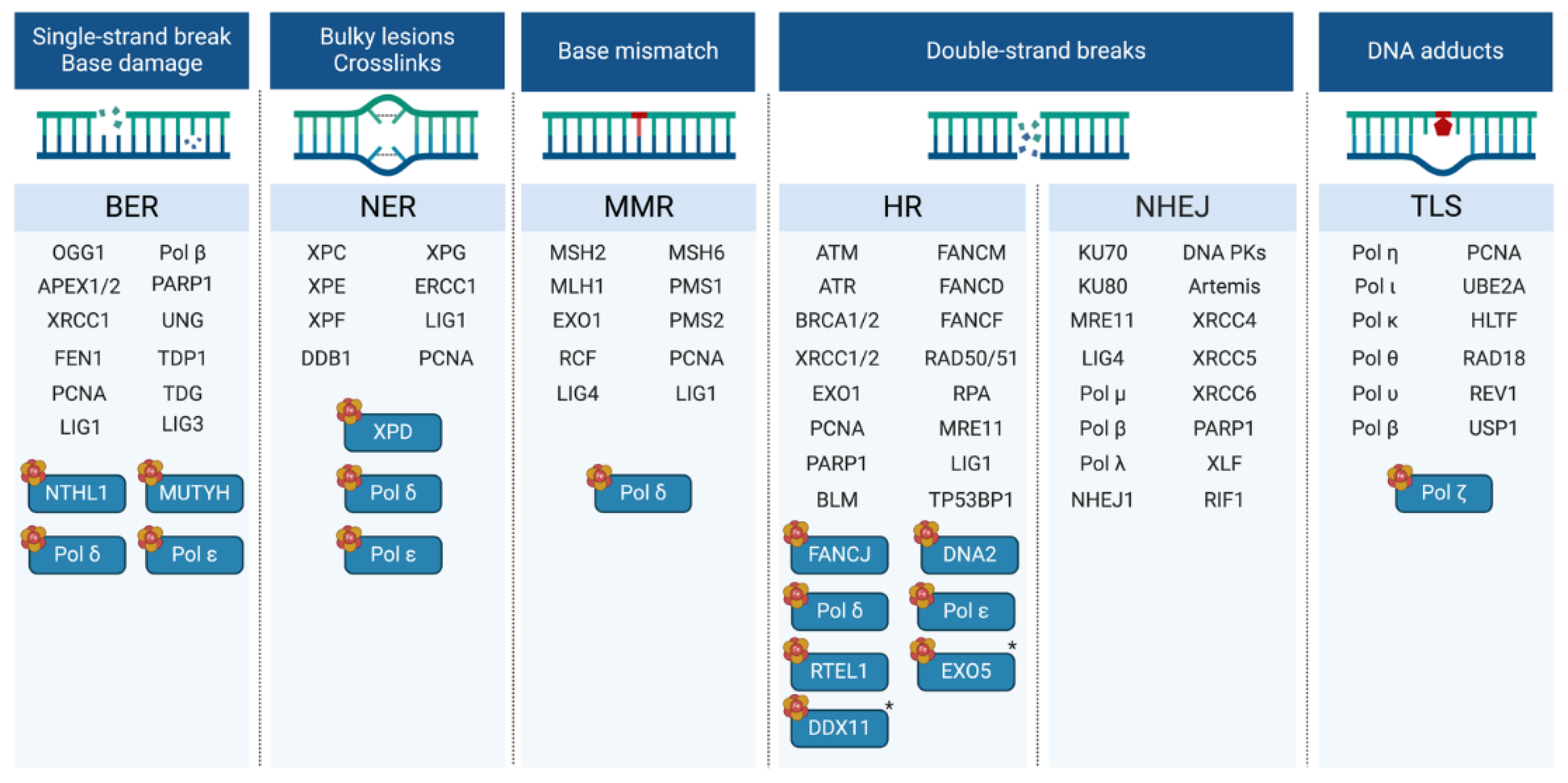
| Participates in | Name | Consolidated Localization | Fe-S cluster type |
|---|---|---|---|
| DNA replication DNA repair | PRIM2 | Nucleus | [4Fe-4S] (Cys)4 [8,9] |
| POLA1 | Nucleus, Cytosol [10] | [4Fe-4S] (Cys)4 [11] | |
| POLD1 | Nucleus, Cytosol [12] | [4Fe-4S] (Cys)4 [11] | |
| POLE | Nucleus | [4Fe-4S] (Cys)4 [11,13] | |
| REV3L | Nucleus, Cytosol [14] | [4Fe-4S] (Cys)4 [11] | |
| FANCJ | Nucleus, Cytosol [15,16] | [4Fe-4S] (Cys)4 [4,17] | |
| DDX11 | Nucleus [18] | [4Fe-4S] (Cys)4 [19] | |
| RTEL1 | Nucleus [16] | [4Fe-4S] (Cys)4 [20] | |
| XPD | Nucleus, Cytosol [16,21] | [4Fe-4S] (Cys)4 [4] | |
| DNA2 | Nucleus, Mitochondria [22] | [4Fe-4S] (Cys)4 [23,24] | |
| EXO5 | Nucleus, Cytosol [25] | [4Fe-4S] (Cys)4 [25] | |
| MUTYH | Nucleus, Mitochondria [26] | [4Fe-4S] (Cys)4 [27,28] | |
| NTHL1 | Nucleus, Mitochondria [29] | [4Fe-4S] (Cys)4 [30,31] | |
| RNA transactions | ELP3 | Nucleus, Cytosol [32] | [4Fe-4S] (Cys)3 SAM [33,34] |
| RPC6 | Nucleus [35] | [4Fe-4S] (Cys)4 [36,37] | |
| CPSF4 | Nucleus [38] | [2Fe-2S] (Cys)3 (His)? [39,40] | |
| TYW1 | Nucleus, Cytosol | [4Fe-4S] (Cys)3 SAM [34,41] | |
| TYW1B | Nucleus | [4Fe-4S] (Cys)3 SAM [34,41] | |
| CDK5RAP1 | Nucleus, Mitochondria, Cytosol [42] | [4Fe-4S] (Cys)4, [4Fe-4S] (Cys)3 SAM [34,43] | |
| Mitosis | KIF4A | Nucleus [44] | [4Fe-4S] (Cys)4? [45] |
| KIF4B | Nucleus [44] | [4Fe-4S] (Cys)4? [45] | |
| Iron metabolism | NCOA4 | Nucleus, Cytosol [46] | [3Fe-4S] (Cys)4? [47,48] |
| FBXL5 | Nucleus, Perinuclear region [49] | [2Fe-2S] (Cys)4 [50] | |
| Post-translational modifications | DPH1 | Nucleus, Cytosol [51] | [4Fe-4S] (Cys)3 SAM [52] |
| DPH2 | Nucleus, Cytosol | [4Fe-4S] (Cys)3 SAM [52] | |
| ATE1 | Nucleus, Cytosol [53] | [4Fe-4S] (Cys)4 [54] | |
| Respiration | SDHB | Mitochondria, Nucleus [55] | [2Fe-2S] (Cys)4, [3Fe-4S] (Cys)3, [4Fe-4S] (Cys)4 [55] |
| Unknown | RFESD | Nucleus | [2Fe-2S] (Cys)2 (His)2 |
| CIA | CIAPIN1 | Nucleus, Mitochondria, Cytosol [56] | [2Fe-2S] (Cys)4, [4Fe-4S] (Cys)4? [57,58] |
| BOLA2 | Nucleus, Cytosol [59] | [2Fe-2S]* Ligands shared with GLRX3 [60,61] | |
| GLRX3 | Nucleus, Cytosol [62] | [2Fe-2S]* Ligands shared with BOLA2 [60,61] | |
| NUBP2 | Nucleus, Cytosol [63] | [4Fe-4S]* Ligands shared with NUBP1 [64,65] | |
| ISC | NFU1 | Nucleus, Mitochondria, Cytosol [66] | [4Fe-4S]* Ligands shared between dimers [66,67] |
| NFS1 | Nucleus, Mitochondria, Cytosol [68] | [2Fe-2S]* Ligands shared with ISCU [69,70] | |
| GLRX2 | Nucleus, Mitochondria [71] | [2Fe-2S]* Ligands shared between dimers [72] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





