Submitted:
30 October 2024
Posted:
31 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
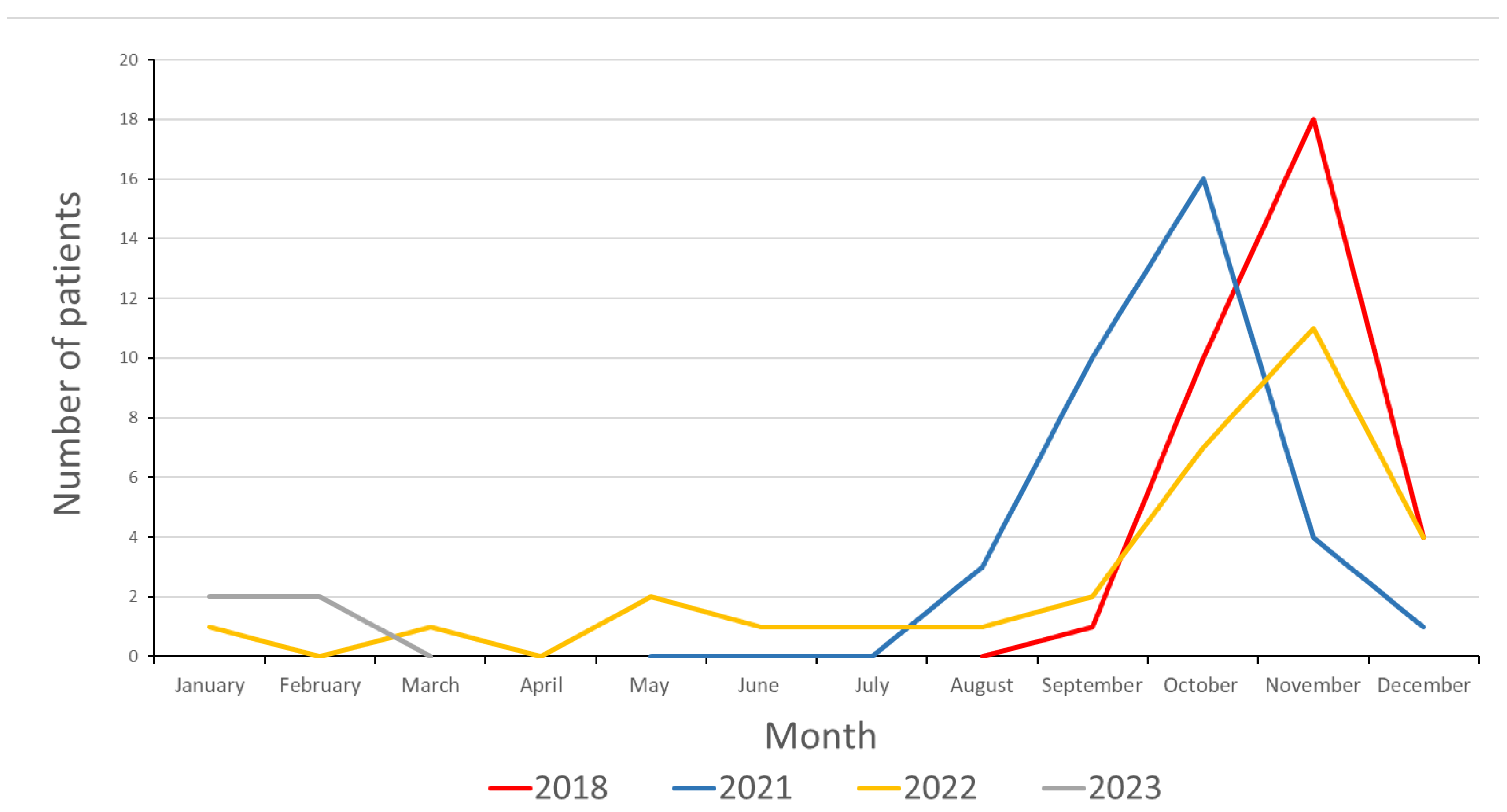
3.1. Epidemiology
3.2. Clinical and Laboratory Characteristics
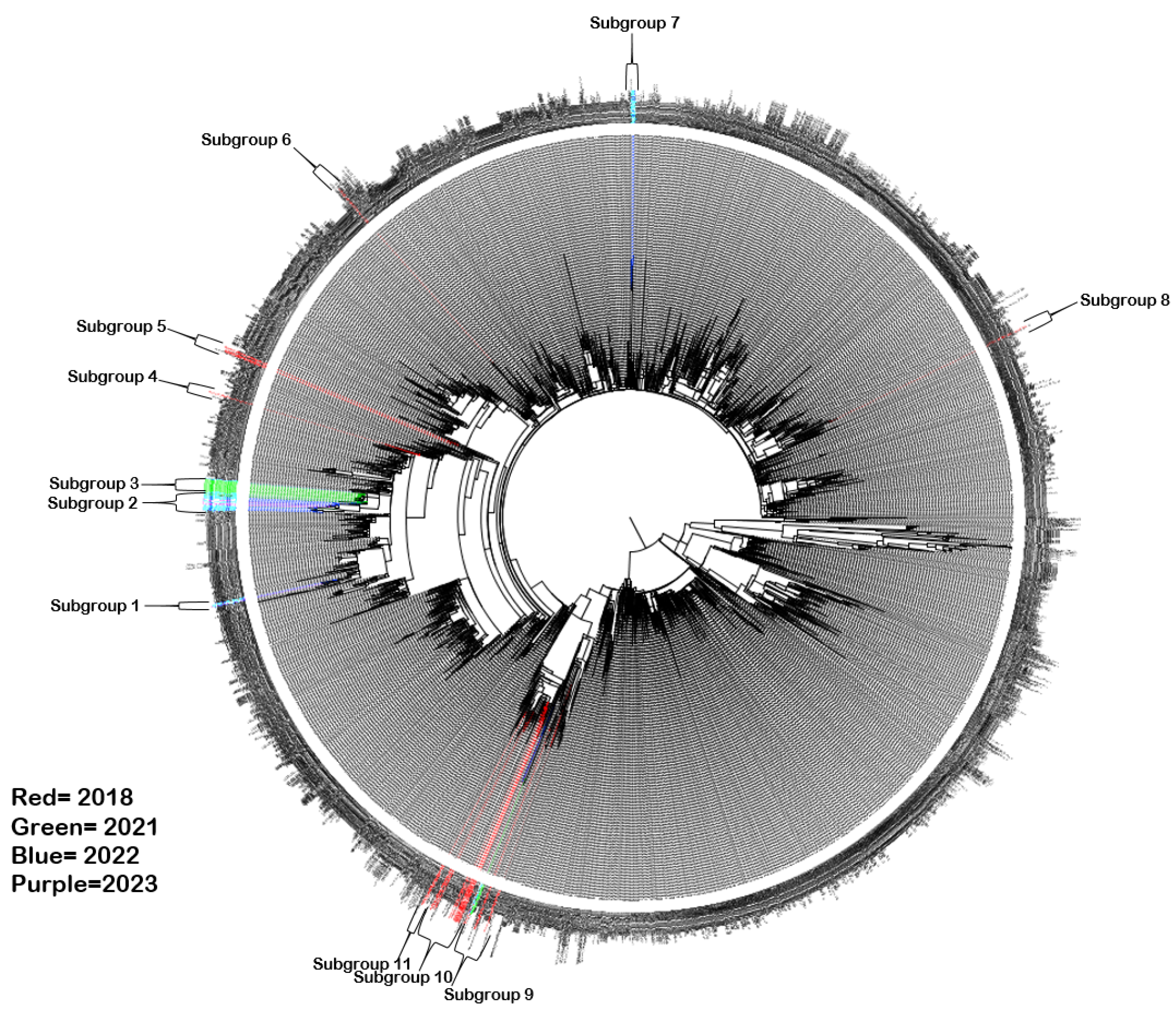
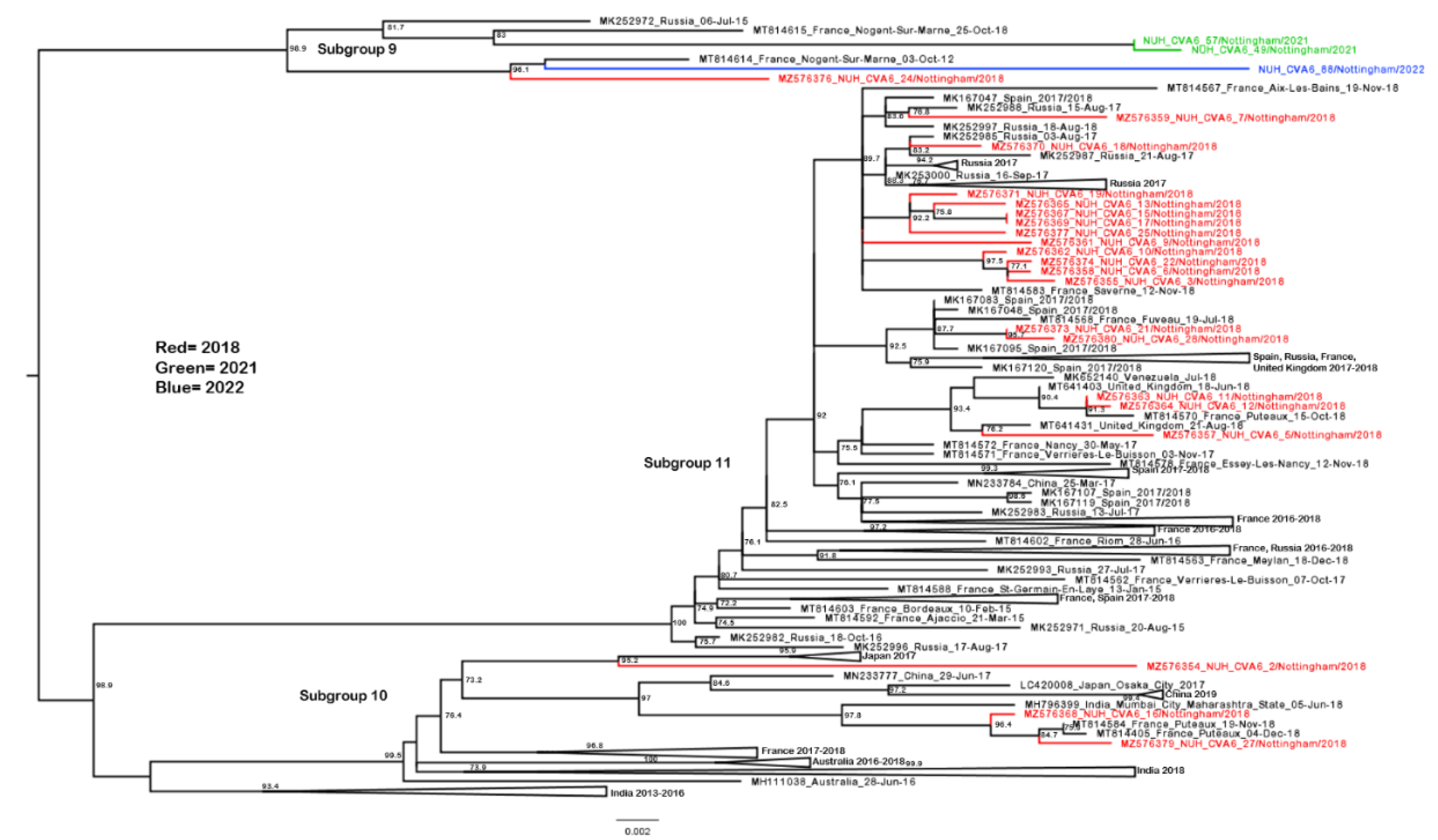
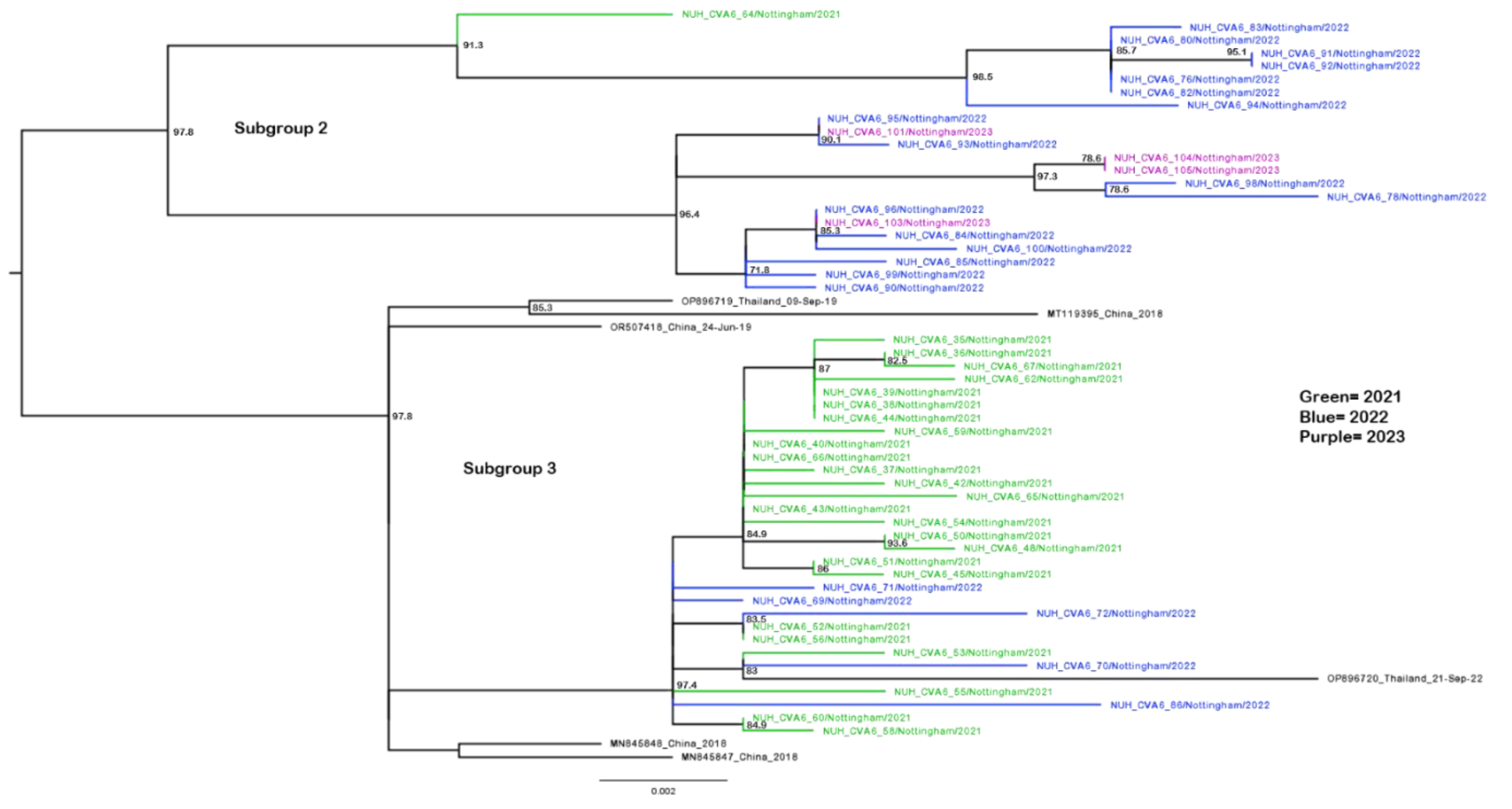
3.3. Phylogenetic Analysis of CVA6 VP1 Sequences
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abzug MJ. Presentation, Diagnosis, and Management of Enterovirus Infections in Neonates. Pediatric Drugs 2004; 6:1 - 10.
- Glaser C, Wilson MR. Enteroviruses: the elephants in the room. Lancet Infect Dis 2020; 20:153-5.
- Lugo D, Krogstad P. Enteroviruses in the early 21st century: new manifestations and challenges. Curr Opin Pediatr 2016; 28:107-13.
- Bubba L, Broberg EK, Jasir A, Simmonds P, Harvala H, Enterovirus study c. Circulation of non-polio enteroviruses in 24 EU and EEA countries between 2015 and 2017: a retrospective surveillance study. Lancet Infect Dis 2020; 20:350-61.
- Esposito S, Principi N. Hand, foot and mouth disease: current knowledge on clinical manifestations, epidemiology, aetiology and prevention. Eur J Clin Microbiol Infect Dis 2018; 37:391-8.
- Gonzalez G, Carr MJ, Kobayashi M, Hanaoka N, Fujimoto T. Enterovirus-Associated Hand-Foot and Mouth Disease and Neurological Complications in Japan and the Rest of the World. Int J Mol Sci 2019; 20.
- Anh NT, Nhu LNT, Van HMT, et al. Emerging Coxsackievirus A6 Causing Hand, Foot and Mouth Disease, Vietnam. Emerg Infect Dis 2018; 24:654-62.
- Li J, Zhu R, Huo D, et al. An outbreak of Coxsackievirus A6-associated hand, foot, and mouth disease in a kindergarten in Beijing in 2015. BMC Pediatr 2018; 18:277.
- Luchs A, Azevedo LS, Souza EV, et al. Coxsackievirus A6 strains causing an outbreak of hand-foot-and-mouth disease in Northeastern Brazil in 2018. Rev Inst Med Trop Sao Paulo 2022; 64:e16.
- Sinclair C, Gaunt E, Simmonds P, et al. Atypical hand, foot, and mouth disease associated with coxsackievirus A6 infection, Edinburgh, United Kingdom, January to February 2014. Euro Surveill 2014; 19:20745.
- Osterback R, Vuorinen T, Linna M, Susi P, Hyypia T, Waris M. Coxsackievirus A6 and hand, foot, and mouth disease, Finland. Emerg Infect Dis 2009; 15:1485-8.
- Huang WC, Huang LM, Lu CY, Cheng AL, Chang LY. Atypical hand-foot-mouth disease in children: a hospital-based prospective cohort study. Virol J 2013; 10:209.
- Andres C, Guasch E, Pinana M, et al. Recombinant CV-A6 strains related to hand-foot-mouth disease and herpangina at primary care centers (Barcelona, Spain). Future Microbiol 2019; 14:499-507.
- Li W, Gao HH, Zhang Q, et al. Large outbreak of herpangina in children caused by enterovirus in summer of 2015 in Hangzhou, China. Sci Rep 2016; 6:35388.
- Zhao TS, Du J, Li HJ, et al. Molecular epidemiology and clinical characteristics of herpangina children in Beijing, China: a surveillance study. PeerJ 2020; 8:e9991.
- Drago F, Ciccarese G, Broccolo F, Rebora A, Parodi A. Atypical hand, foot, and mouth disease in adults. J Am Acad Dermatol 2017; 77:e51-e6.
- Broccolo F, Drago F, Ciccarese G, et al. Severe atypical hand-foot-and-mouth disease in adults due to coxsackievirus A6: Clinical presentation and phylogenesis of CV-A6 strains. J Clin Virol 2019; 110:1-6.
- Blomqvist S, Klemola P, Kaijalainen S, et al. Co-circulation of coxsackieviruses A6 and A10 in hand, foot and mouth disease outbreak in Finland. J Clin Virol 2010; 48:49-54.
- Wei SH, Huang YP, Liu MC, et al. An outbreak of coxsackievirus A6 hand, foot, and mouth disease associated with onychomadesis in Taiwan, 2010. BMC Infect Dis 2011; 11:346.
- Miyamoto A, Hirata R, Ishimoto K, et al. An outbreak of hand-foot-and-mouth disease mimicking chicken pox, with a frequent association of onychomadesis in Japan in 2009: a new phenotype caused by coxsackievirus A6. Eur J Dermatol 2014; 24:103-4.
- Fujimoto T, Iizuka S, Enomoto M, et al. Hand, foot, and mouth disease caused by coxsackievirus A6, Japan, 2011. Emerg Infect Dis 2012; 18:337-9.
- Yang F, Yuan J, Wang X, et al. Severe hand, foot, and mouth disease and coxsackievirus A6-Shenzhen, China. Clin Infect Dis 2014; 59:1504-5.
- Yang X, Li Y, Zhang C, et al. Clinical features and phylogenetic analysis of severe hand-foot-and-mouth disease caused by Coxsackievirus A6. Infect Genet Evol 2020; 77:104054.
- S A, Sabeena S, Bhat KG, Bharani KC, Ramachandran S, Arunkumar G. Coxsackievirus A6 (CV-A6) Encephalomyelitis in an Immunocompromised Child: a Case Report and Brief Review of the Literature. Jpn J Infect Dis 2018; 71:388-9.
- Qian SS, Wei ZN, Jin WP, et al. Efficacy of a coxsackievirus A6 vaccine candidate in an actively immunized mouse model. Emerg Microbes Infect 2021; 10:763-73.
- Oberste MS, Maher K, Flemister MR, Marchetti G, Kilpatrick DR, Pallansch MA. Comparison of classic and molecular approaches for the identification of untypeable enteroviruses. J Clin Microbiol 2000; 38:1170-4.
- Gaunt E, Harvala H, Osterback R, et al. Genetic characterization of human coxsackievirus A6 variants associated with atypical hand, foot and mouth disease: a potential role of recombination in emergence and pathogenicity. J Gen Virol 2015; 96:1067-79.
- Simmonds P, Welch J. Frequency and dynamics of recombination within different species of human enteroviruses. J Virol 2006; 80:483-93.
- Nix WA, Oberste MS, Pallansch MA. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol 2006; 44:2698-704.
- Howson-Wells HC, Winckles S, Aliker C, et al. Enterovirus subtyping in a routine UK laboratory setting between 2013 and 2017. J Clin Virol 2020; 132:104646.
- Hassel C, Mirand A, Lukashev A, et al. Transmission patterns of human enterovirus 71 to, from and among European countries, 2003 to 2013. Euro Surveill 2015; 20:30005.
- Lott JP, Liu K, Landry ML, et al. Atypical hand-foot-and-mouth disease associated with coxsackievirus A6 infection. J Am Acad Dermatol 2013; 69:736-41.
- Wang J, Zhang S. Epidemiological characteristics and trends of hand-foot-mouth disease in Shanghai, China from 2011 to 2021. Front Public Health 2023; 11:1162209.
- Liu H, Zhang M, Feng C, et al. Characterization of Coxsackievirus A6 Strains Isolated From Children With Hand, Foot, and Mouth Disease. Front Cell Infect Microbiol 2021; 11:700191.
- Song Y, Zhang Y, Ji T, et al. Persistent circulation of Coxsackievirus A6 of genotype D3 in mainland of China between 2008 and 2015. Sci Rep 2017; 7:5491.
- Mirand A, Cohen R, Bisseux M, et al. A large-scale outbreak of hand, foot and mouth disease, France, as at 28 September 2021. Euro Surveill 2021; 26.
- Tomba Ngangas S, Bisseux M, Jugie G, et al. Coxsackievirus A6 Recombinant Subclades D3/A and D3/H Were Predominant in Hand-Foot-And-Mouth Disease Outbreaks in the Paediatric Population, France, 2010-2018. Viruses 2022; 14.
- Forero EL, Knoester M, Gard L, et al. Changes in enterovirus epidemiology after easing of lockdown measures. J Clin Virol 2023; 169:105617.
- Howson-Wells HC, Tsoleridis T, Zainuddin I, et al. Enterovirus D68 epidemic, UK, 2018, was caused by subclades B3 and D1, predominantly in children and adults, respectively, with both subclades exhibiting extensive genetic diversity. Microb Genom 2022; 8.
- Bian L, Wang Y, Yao X, Mao Q, Xu M, Liang Z. Coxsackievirus A6: a new emerging pathogen causing hand, foot and mouth disease outbreaks worldwide. Expert Rev Anti Infect Ther 2015; 13:1061-71.
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 2016; 33:1870-4.
- Kroneman A, Vennema H, Deforche K, et al. An automated genotyping tool for enteroviruses and noroviruses. J Clin Virol 2011; 51:121-5.
- Minh BQ, Schmidt HA, Chernomor O, et al. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol 2020; 37:1530-4.
- Francis RV, Billam H, Clarke M, et al. The Impact of Real-Time Whole-Genome Sequencing in Controlling Healthcare-Associated SARS-CoV-2 Outbreaks. J Infect Dis 2022; 225:10-8.
- Bagasi AA, Howson-Wells HC, Clark G, et al. Human Bocavirus infection and respiratory tract disease identified in a UK patient cohort. J Clin Virol 2020; 129:104453.
- Majumdar M, Celma C, Pegg E, Polra K, Dunning J, Martin J. Detection and Typing of Human Enteroviruses from Clinical Samples by Entire-Capsid Next Generation Sequencing. Viruses 2021; 13.
- Yoshitomi H, Ashizuka Y, Ichihara S, et al. Molecular epidemiology of coxsackievirus A6 derived from hand, foot, and mouth disease in Fukuoka between 2013 and 2017. J Med Virol 2018; 90:1712-9.
- Cobbin JCA, Britton PN, Burrell R, et al. A complex mosaic of enteroviruses shapes community-acquired hand, foot and mouth disease transmission and evolution within a single hospital. Virus Evol 2018; 4:vey020.
- Simoes MP, Hodcroft EB, Simmonds P, et al. Epidemiological and clinical insights into the enterovirus D68 upsurge in Europe 2021/22 and the emergence of novel B3-derived lineages, ENPEN multicentre study. J Infect Dis 2024.
- Abedi GR, Watson JT, Pham H, Nix WA, Oberste MS, Gerber SI. Enterovirus and Human Parechovirus Surveillance - United States, 2009-2013. MMWR Morb Mortal Wkly Rep 2015; 64:940-3.
- Moriyama M, Hugentobler WJ, Iwasaki A. Seasonality of Respiratory Viral Infections. Annu Rev Virol 2020; 7:83-101.
- Kamau E, Nguyen D, Celma C, et al. Seroprevalence and Virologic Surveillance of Enterovirus 71 and Coxsackievirus A6, United Kingdom, 2006-2017. Emerg Infect Dis 2021; 27:2261-8.
- Messacar K, Baker RE, Park SW, Nguyen-Tran H, Cataldi JR, Grenfell B. Preparing for uncertainty: endemic paediatric viral illnesses after COVID-19 pandemic disruption. Lancet 2022; 400:1663-5.
- Cai W, Kondgen S, Tolksdorf K, et al. Atypical age distribution and high disease severity in children with RSV infections during two irregular epidemic seasons throughout the COVID-19 pandemic, Germany, 2021 to 2023. Euro Surveill 2024; 29.
- Honemann M, Thiem S, Bergs S, et al. In-Depth Analysis of the Re-Emergence of Respiratory Syncytial Virus at a Tertiary Care Hospital in Germany in the Summer of 2021 after the Alleviation of Non-Pharmaceutical Interventions Due to the SARS-CoV-2 Pandemic. Viruses 2023; 15.
- Fricke LM, Glockner S, Dreier M, Lange B. Impact of non-pharmaceutical interventions targeted at COVID-19 pandemic on influenza burden - a systematic review. J Infect 2021; 82:1-35.
- Paget J, Caini S, Del Riccio M, van Waarden W, Meijer A. Has influenza B/Yamagata become extinct and what implications might this have for quadrivalent influenza vaccines? Euro Surveill 2022; 27.
- Davis WW, Mott JA, Olsen SJ. The role of non-pharmaceutical interventions on influenza circulation during the COVID-19 pandemic in nine tropical Asian countries. Influenza Other Respir Viruses 2022; 16:568-76.
- Liu B, Luo L, Yan S, et al. Clinical Features for Mild Hand, Foot and Mouth Disease in China. PLoS One 2015; 10:e0135503.
- Lau SKP, Zhao PSH, Sridhar S, et al. Molecular epidemiology of coxsackievirus A6 circulating in Hong Kong reveals common neurological manifestations and emergence of novel recombinant groups. J Clin Virol 2018; 108:43-9.
- Messacar K, Asturias EJ, Hixon AM, et al. Enterovirus D68 and acute flaccid myelitis-evaluating the evidence for causality. Lancet Infect Dis 2018; 18:e239-e47.
- de Sousa IP, Jr. , Giamberardino HI, Raboni SM, et al. Simultaneous enterovirus EV-D68 and CVA6 infections causing acute respiratory distress syndrome and hand, foot and mouth disease. Virol J 2021; 18:88.
- Yu F, Zhu R, Jia L, et al. Sub-genotype change and recombination of coxsackievirus A6s may be the cause of it being the predominant pathogen for HFMD in children in Beijing, as revealed by analysis of complete genome sequences. Int J Infect Dis 2020; 99:156-62.
- Chappell JG, Tsoleridis T, Clark G, et al. Retrospective screening of routine respiratory samples revealed undetected community transmission and missed intervention opportunities for SARS-CoV-2 in the United Kingdom. J Gen Virol 2021; 102.
- Gomez GB, Mahe C, Chaves SS. Uncertain effects of the pandemic on respiratory viruses. Science 2021; 372:1043-4.
- Fischer TK, Simmonds P, Harvala H. The importance of enterovirus surveillance in a post-polio world. Lancet Infect Dis 2022; 22:e35-e40.
| Pandemic era | |||
| Pre- | Post- | ||
| Cases | 33 | 69 | |
| Clinical feature | N (%) | N (%) | p value |
| Male gender | 20 (60.6) | 43 (62.3) | 0.866 |
| Hospital admission | 14 (42.4) | 24 (34.8) | 0.4552 |
| Comorbidities | 3 (9.1) | 26 (37.7) | 0.0018 |
| Neurological symptoms | 4 (12.1) | 0 (0.0) | 0.0039 |
| Fever | 13 (39.4) | 21 (30.4) | 0.4543 |
| Gastro | 4 (12.1) | 7 (10.1) | 0.8211 |
| Rash | 23 (69.7) | 62 (89.9) | 0.0106 |
| Respiratory symptoms | 15 (45.5) | 13 (18.8) | 0.073 |
| HFMD primary diagnosis | 22 (66.7) | 59 (85.5) | 0.0057 |
| Sample site | |||
| Skin swab | 15 (45.5) | 59 (85.5) | <0.0001 |
| Oral swab | 13 (39.4) | 6 (8.7) | |
| Other sample | 5 (15.2) | 4 (5.8) | |
| Age group | 0.5569 | ||
| Under 1 year | 9 (27.3) | 13 (18.8) | |
| 1 year | 15 (45.5) | 27 (39.1) | |
| 2 years | 3 (9.1) | 11 (15.9) | |
| 3 years | 0 (0.0) | 6 (8.7) | |
| 4 to 17 years of age | 1 (3.0) | 5 (7.2) | |
| Adult | 5 (15.2) | 7 (10.1) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





