Submitted:
30 October 2024
Posted:
31 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
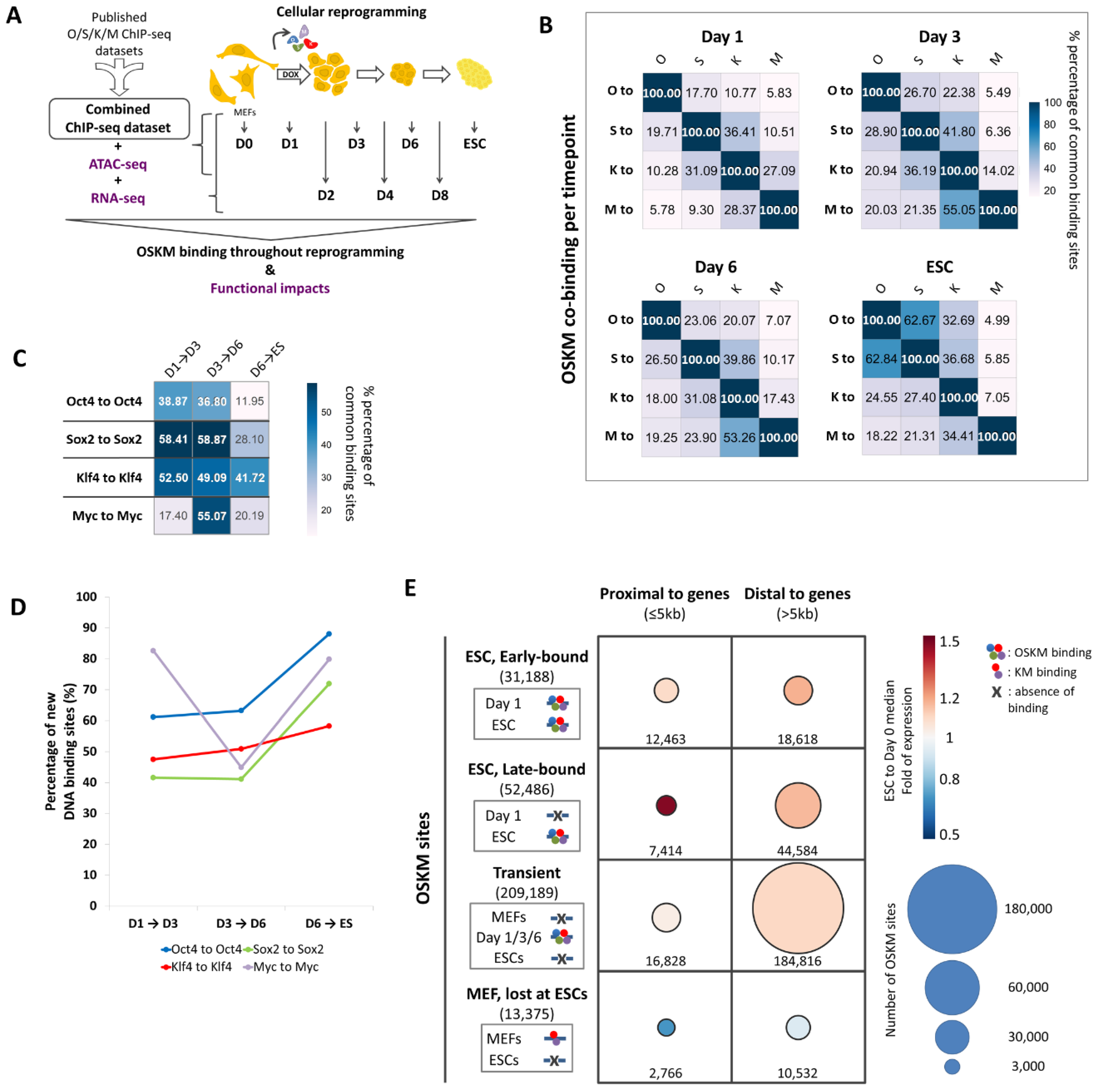
2.1. Creation of an Integrated ChIP-Seq Dataset to Monitor Global Binding of OSKM During Cellular Reprogramming
2.2. An Unprecedented Highly Dynamic OSKM Binding During Cellular Reprogramming
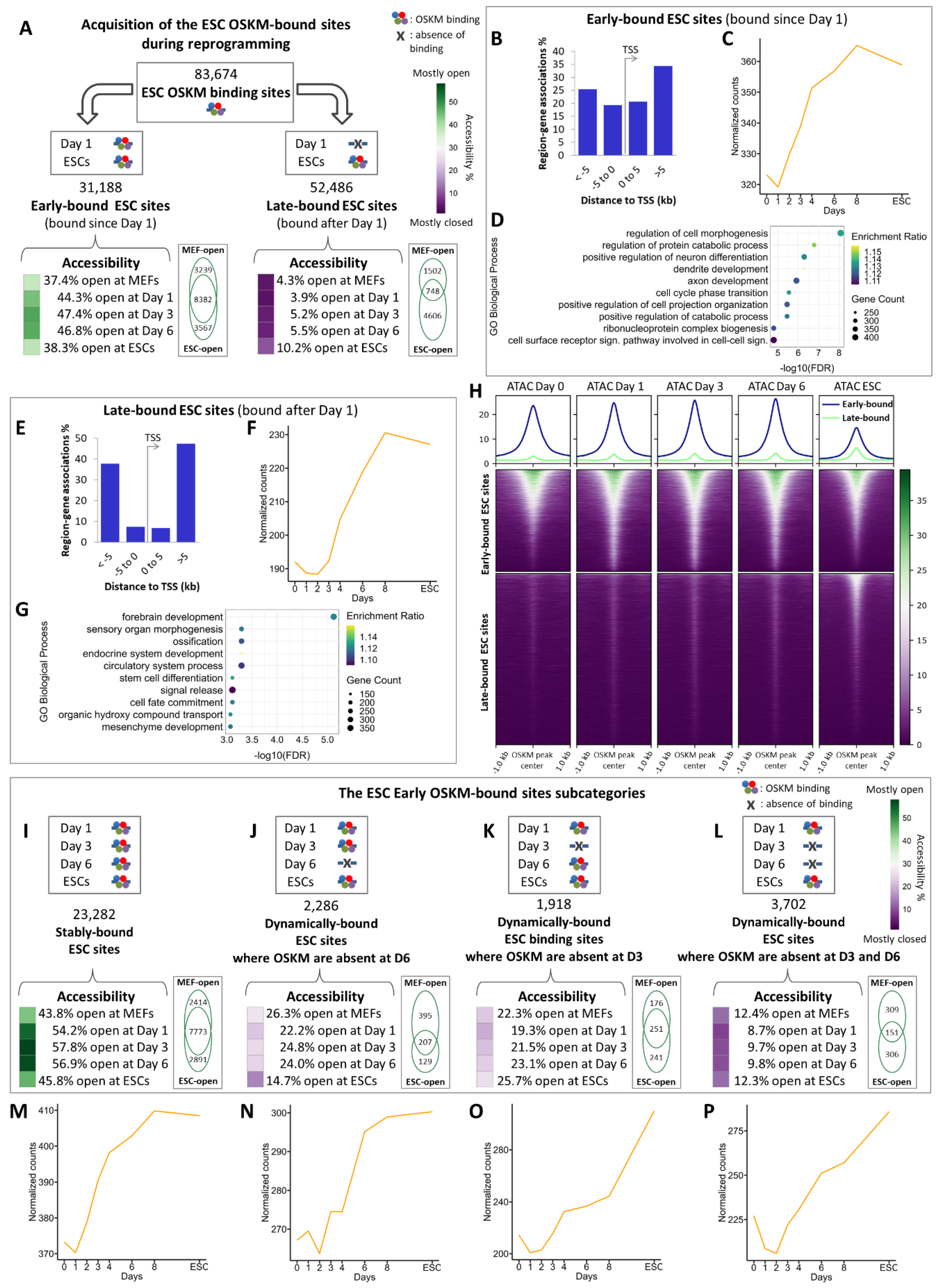
2.3. The Combinatorial Binding of OSKM to Early and Late Elements Prefigures the Induction of Pluripotency-Related Genes During Reprogramming
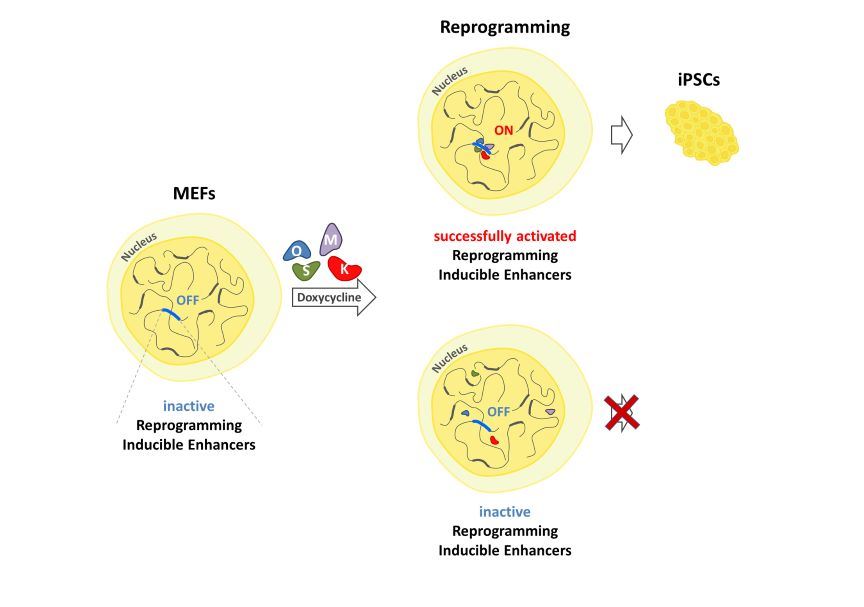
2.4. Identification of Reprogramming-Inducible Enhancers in the Mouse Genome
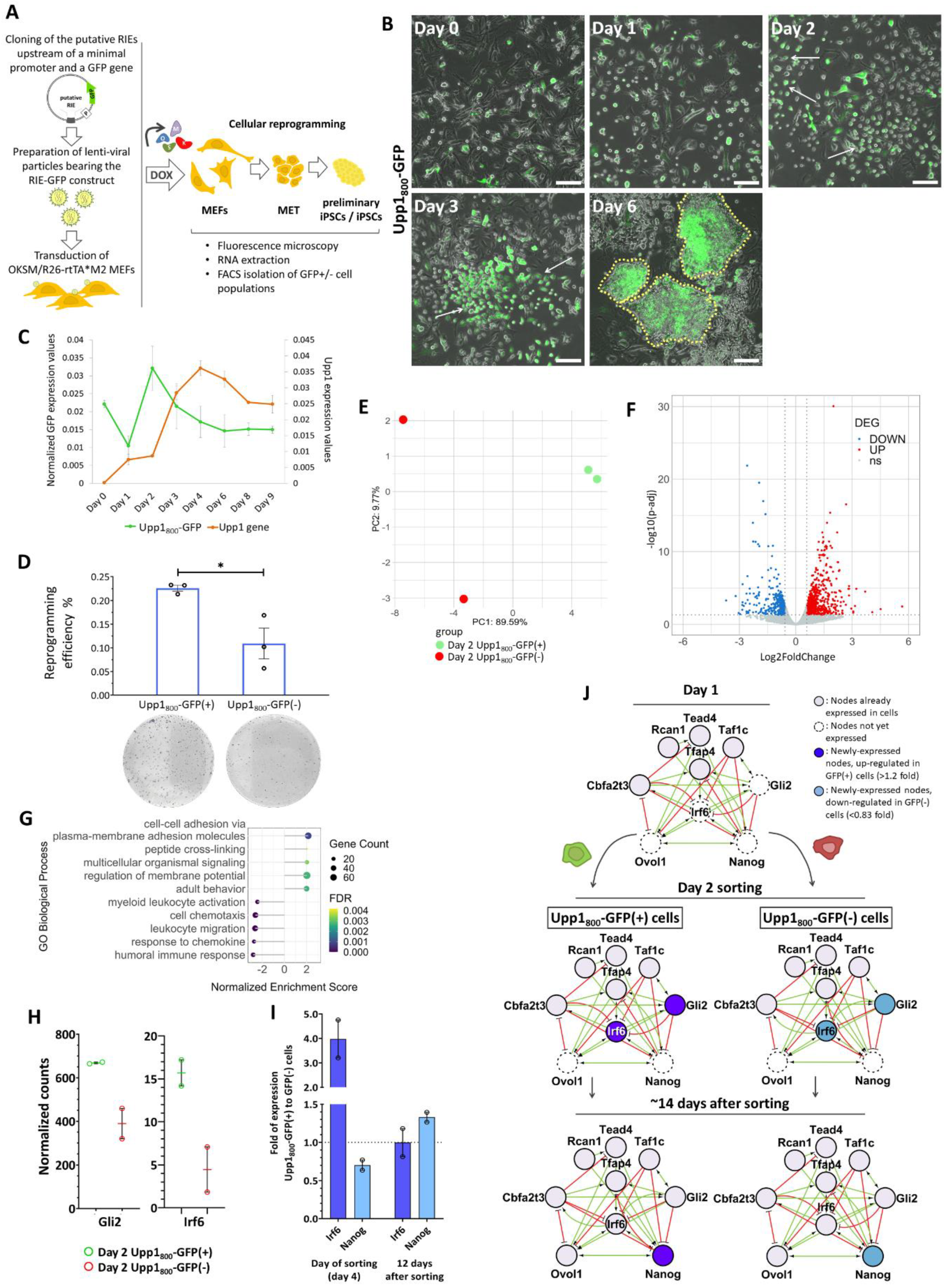
2.5. The Upp1800 Element Functions as a Reprogramming Inducible Enhancer Marking Cells Undergoing Reprogramming to Pluripotency
2.6. The Cells “Marked” by the Reprogramming Inducible Enhancer Upp1800 Element Achieve Earlier and More Robust Induction of the 9TR Network Leading to Efficient Reprogramming
3. Discussion
4. Materials and Methods
4.1. Experimental Protocols
4.1.1. MEFs Isolation from Mouse Embryos
4.1.2. Cellular Reprogramming Protocol
4.1.3. Nanog Chromatin Immunoprecipitation Followed by High-Throughput Sequencing
4.1.4. ATAC-Seq
4.1.5. Construction of Enhancer Reporters and Production of Lenti-Viral Particles
4.1.6. Generation of Lenti-Viral Particles
4.1.7. Reporter Assays
- MEFs transduction and cellular reprogramming
- 2.
- Microscopy
- 3.
- Reporter assay for the comparison of exogenous-GFP and endogenous gene expression pattern (Figure 4C, Figure S5C-D)
- 4.
- Sorting of Upp1800-GFP(+) and GFP(-) cells for transcriptome analysis and reprogramming efficiency calculation
- 5.
- Alkaline Phosphatase (AP) staining for reprogramming efficiency calculation
- 6.
- RNA isolation and real-time PCR
- 7.
- RNA sequencing
- Reprogramming time-course
- Upp1800-GFP sorted cells
4.2. Bioinformatics Analyses
4.2.1. ChIP-Seq Data Analysis
- 8.
- Selection and primary analysis of published O/S/K/M and Nanog ChIP-seq datasets
- 9.
- Merging of datasets
- 10.
- ATAC-seq analysis
4.2.2. Investigation of OSKM Binding Sites
- 11.
- Calculation of common binding sites between Oct4, Sox2, Klf4 and Myc
- 12.
- Identification of ESC, transient and MEF sites
- 13.
- Overlaps of OSKM sites with ESC Super-enhancers
- 14.
- Epigenetic characterization of OSKM binding sites
- 15.
- Gene assignment to OSKM binding sites
- 16.
- Motif analysis
4.3. Method for the Identification of Putative Reprogramming Inducible Enhancers
4.4. RNA-Seq Analysis
4.5. Functional Enrichment of Gene Sets
4.6. Network Analysis
4.7. Other Graphical Representations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–72. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, S. Elite and stochastic models for induced pluripotent stem cell generation. Nature 2009, 460, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013, 14, 427–39. [Google Scholar] [CrossRef]
- Takahashi K, Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol. 2016, 17, 183–93. [Google Scholar] [CrossRef]
- David L, Polo JM. Phases of reprogramming. Stem Cell Res. 2014, 12, 754–61. [Google Scholar] [CrossRef]
- Samavarchi-Tehrani P, Golipour A, David L, Sung H, Beyer TA, Datti A, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010, 7, 64–77. [Google Scholar] [CrossRef]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010, 7, 51–63. [Google Scholar] [CrossRef]
- Shu X, Pei D. The function and regulation of mesenchymal-to-epithelial transition in somatic cell reprogramming. Curr Opin Genet Dev. 2014, 28, 32–7. [Google Scholar] [CrossRef]
- Papathanasiou M, Tsiftsoglou SA, Polyzos AP, Papadopoulou D, Valakos D, Klagkou E, et al. Identification of a dynamic gene regulatory network required for pluripotency factor-induced reprogramming of mouse fibroblasts and hepatocytes. EMBO J. 2021, 40, e102236. [Google Scholar] [CrossRef]
- Deng W, Jacobson EC, Collier AJ, Plath K. The transcription factor code in iPSC reprogramming. Curr Opin Genet Dev. 2021, 70, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell 2009, 136, 364–77. [Google Scholar] [CrossRef] [PubMed]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 2012, 151, 1617–32. [Google Scholar] [CrossRef] [PubMed]
- Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 2012, 151, 994–1004. [Google Scholar] [CrossRef]
- Cacchiarelli D, Trapnell C, Ziller MJ, Soumillon M, Cesana M, Karnik R, et al. Integrative Analyses of Human Reprogramming Reveal Dynamic Nature of Induced Pluripotency. Cell 2015, 162, 412–424. [Google Scholar] [CrossRef]
- Chen J, Chen X, Li M, Liu X, Gao Y, Kou X, et al. Hierarchical Oct4 Binding in Concert with Primed Epigenetic Rearrangements during Somatic Cell Reprogramming. Cell Rep. 2016, 14, 1540–1554. [Google Scholar] [CrossRef]
- Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, et al. Cooperative Binding of Transcription Factors Orchestrates Reprogramming. Cell. 2017, 168, 442–459e20. [Google Scholar] [CrossRef]
- Knaupp AS, Buckberry S, Pflueger J, Lim SM, Ford E, Larcombe MR, et al. Transient and Permanent Reconfiguration of Chromatin and Transcription Factor Occupancy Drive Reprogramming. Cell Stem Cell. 2017, 21, 834–845e6. [Google Scholar] [CrossRef]
- Li D, Liu J, Yang X, Zhou C, Guo J, Wu C, et al. Chromatin Accessibility Dynamics during iPSC Reprogramming. Cell Stem Cell. 2017, 21, 819–833e6. [Google Scholar] [CrossRef]
- Schwarz BA, Cetinbas M, Clement K, Walsh RM, Cheloufi S, Gu H, et al. Prospective Isolation of Poised iPSC Intermediates Reveals Principles of Cellular Reprogramming. Cell Stem Cell. 2018, 23, 289–305e5. [Google Scholar] [CrossRef]
- Zviran A, Mor N, Rais Y, Gingold H, Peles S, Chomsky E, et al. Deterministic Somatic Cell Reprogramming Involves Continuous Transcriptional Changes Governed by Myc and Epigenetic-Driven Modules. Cell Stem Cell. 2019, 24, 328–341e9. [Google Scholar] [CrossRef] [PubMed]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 2015, 161, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Roberts GA, Ozkan B, Gachulincová I, O’Dwyer MR, Hall-Ponsele E, Saxena M, et al. Dissecting OCT4 defines the role of nucleosome binding in pluripotency. Nat Cell Biol. 2021, 23, 834–845. [Google Scholar] [CrossRef] [PubMed]
- Li D, Shu X, Zhu P, Pei D. Chromatin accessibility dynamics during cell fate reprogramming. EMBO Rep. 2021, 22, e51644. [Google Scholar] [CrossRef]
- King HW, Klose RJ. The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. Elife 2017, 6, 1–24. [Google Scholar] [CrossRef]
- Valakos D, Klagkou E, Kokkalis A, Polyzos A, Kyrilis FL, Banos A, et al. Combinatorial targeting of a specific EMT/MET network by macroH2A variants safeguards mesenchymal identity. PLoS One 2023, 18, e0288005. [Google Scholar] [CrossRef]
- Theunissen TW, Jaenisch R. Molecular control of induced pluripotency. Cell Stem Cell 2014, 14, 720–34. [Google Scholar] [CrossRef]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 2007, 1, 55–70. [Google Scholar] [CrossRef]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 2007, 448, 318–24. [Google Scholar] [CrossRef]
- Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, Zaidi S, Gokhale S, et al. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell 2007, 1, 403–15. [Google Scholar] [CrossRef]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008, 2, 230–40. [Google Scholar] [CrossRef] [PubMed]
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008, 2, 151–9. [Google Scholar] [CrossRef] [PubMed]
- Hotta A, Cheung AYLL, Farra N, Vijayaragavan K, Séguin CA, Draper JS, et al. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat Methods 2009, 6, 370–6. [Google Scholar] [CrossRef] [PubMed]
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature 2013, 502, 65–70. [Google Scholar] [CrossRef]
- Chen J, Liu J, Chen Y, Yang J, Chen J, Liu H, et al. Rational optimization of reprogramming culture conditions for the generation of induced pluripotent stem cells with ultra-high efficiency and fast kinetics. Cell Res. 2011, 21, 884–94. [Google Scholar] [CrossRef]
- Bar-Nur O, Brumbaugh J, Verheul C, Apostolou E, Pruteanu-Malinici I, Walsh RM, et al. Small molecules facilitate rapid and synchronous iPSC generation. Nat Methods 2014, 11, 1170–6. [Google Scholar] [CrossRef]
- Vidal SE, Amlani B, Chen T, Tsirigos A, Stadtfeld M. Combinatorial modulation of signaling pathways reveals cell-type-specific requirements for highly efficient and synchronous iPSC reprogramming. Stem cell reports. 2014, 3, 574–84. [Google Scholar] [CrossRef]
- Di Giammartino DC, Kloetgen A, Polyzos A, Liu Y, Kim D, Murphy D, et al. KLF4 is involved in the organization and regulation of pluripotency-associated three-dimensional enhancer networks. Nat Cell Biol. 2019, 21, 1179–1190. [Google Scholar] [CrossRef]
- Huyghe A, Trajkova A, Lavial F. Cellular plasticity in reprogramming, rejuvenation and tumorigenesis: a pioneer TF perspective. Trends Cell Biol. 2024, 34, 255–267. [Google Scholar] [CrossRef]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014, 15, 178–96. [Google Scholar] [CrossRef]
- Young, RA. Control of the embryonic stem cell state. Cell. 2011, 144, 940–54. [Google Scholar] [CrossRef] [PubMed]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013, 153, 307–19. [Google Scholar] [CrossRef] [PubMed]
- Meno C, Saijoh Y, Fujii H, Ikeda M, Yokoyama T, Yokoyama M, et al. Left-right asymmetric expression of the TGF beta-family member lefty in mouse embryos. Nature 1996, 381, 151–5. [Google Scholar] [CrossRef] [PubMed]
- Tabibzadeh S, Hemmati-Brivanlou A. Lefty at the crossroads of “stemness” and differentiative events. Stem Cells. 2006, 24, 1998–2006. [Google Scholar] [CrossRef]
- Jerabek S, Merino F, Schöler HR, Cojocaru V. OCT4: dynamic DNA binding pioneers stem cell pluripotency. Biochim Biophys Acta. 2014, 1839, 138–54. [Google Scholar] [CrossRef]
- Watanabe S-I, Hino A, Wada K, Eliason JF, Uchida T. Purification, cloning, and expression of murine uridine phosphorylase. J Biol Chem. 1995, 270, 12191–6. [Google Scholar] [CrossRef]
- Li Y, Jiang M, Aye L, Luo L, Zhang Y, Xu F, et al. UPP1 promotes lung adenocarcinoma progression through the induction of an immunosuppressive microenvironment. Nat Commun. 2024, 15, 1200. [Google Scholar] [CrossRef]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008, 133, 1106–17. [Google Scholar] [CrossRef]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005, 121, 465–77. [Google Scholar] [CrossRef]
- Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods. 2010, 7, 53–5. [Google Scholar] [CrossRef]
- Bader GD, Hogue CW V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003, 4, 2. [Google Scholar] [CrossRef]
- Chen J, Liu H, Liu J, Qi J, Wei B, Yang J, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet. 2013, 45, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Zeng S, Zhang Y, Ma J, Deng G, Qu Y, Guo C, et al. BMP4 promotes metastasis of hepatocellular carcinoma by an induction of epithelial-mesenchymal transition via upregulating ID2. Cancer Lett. 2017, 390, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Huang J, Wang F, Okuka M, Liu N, Ji G, Ye X, et al. Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res. 2011, 21, 779–92. [Google Scholar] [CrossRef]
- Wang F, Yin Y, Ye X, Liu K, Zhu H, Wang L, et al. Molecular insights into the heterogeneity of telomere reprogramming in induced pluripotent stem cells. Cell Res. 2012, 22, 757–68. [Google Scholar] [CrossRef]
- Kinoshita T, Nagamatsu G, Saito S, Takubo K, Horimoto K, Suda T. Telomerase reverse transcriptase has an extratelomeric function in somatic cell reprogramming. J Biol Chem. 2014, 289, 15776–87. [Google Scholar] [CrossRef]
- Hidema S, Fukuda T, Date S, Tokitake Y, Matsui Y, Sasaki H, et al. Transgenic expression of Telomerase reverse transcriptase (Tert) improves cell proliferation of primary cells and enhances reprogramming efficiency into the induced pluripotent stem cell. Biosci Biotechnol Biochem. 2016, 80, 1925–33. [Google Scholar] [CrossRef]
- Tejera AM, Stagno d’Alcontres M, Thanasoula M, Marion RM, Martinez P, Liao C, et al. TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming, and normal skin development in mice. Dev Cell. 2010, 18, 775–89. [Google Scholar] [CrossRef]
- Nagamatsu G, Saito S, Kosaka T, Takubo K, Kinoshita T, Oya M, et al. Optimal ratio of transcription factors for somatic cell reprogramming. J Biol Chem. 2012, 287, 36273–36282. [Google Scholar] [CrossRef]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008, 26, 101–6. [Google Scholar] [CrossRef]
- Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008, 2, 10–2. [Google Scholar] [CrossRef] [PubMed]
- Cao D, Nimmakayalu MA, Wang F, Zhang D, Handschumacher RE, Bray-Ward P, et al. Genomic structure, chromosomal mapping, and promoter region analysis of murine uridine phosphorylase gene. Cancer Res. 1999, 59, 4997–5001. Available online: http://www.ncbi.nlm.nih.gov/pubmed/10519414.
- Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J Biol Chem. 2005, 280, 5307–17. [Google Scholar] [CrossRef] [PubMed]
- Nakatake Y, Fukui N, Iwamatsu Y, Masui S, Takahashi K, Yagi R, et al. Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol Cell Biol. 2006, 26, 7772–82. [Google Scholar] [CrossRef]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013, 10, 1213–8. [Google Scholar] [CrossRef]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015, 109, 21.29.1–21299. [Google Scholar] [CrossRef]
- Ackermann AM, Wang Z, Schug J, Naji A, Kaestner KH. Integration of ATAC-seq and RNA-seq identifies human alpha cell and beta cell signature genes. Mol Metab. 2016, 5, 233–244. [Google Scholar] [CrossRef]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017, 14, 959–962. [Google Scholar] [CrossRef]
- Weber K, Bartsch U, Stocking C, Fehse B. A multicolor panel of novel lentiviral “gene ontology” (LeGO) vectors for functional gene analysis. Mol Ther. 2008, 16, 698–706. [Google Scholar] [CrossRef]
- Juven-Gershon T, Cheng S, Kadonaga JT. Rational design of a super core promoter that enhances gene expression. Nat Methods. 2006, 3, 917–22. [Google Scholar] [CrossRef]
- Arnold CD, Gerlach D, Stelzer C, Boryń ŁM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 2013, 339, 1074–7. [Google Scholar] [CrossRef] [PubMed]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012, 9, 676–82. [Google Scholar] [CrossRef] [PubMed]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012, 9, 357–9. [Google Scholar] [CrossRef] [PubMed]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010, 26, 841–2. [Google Scholar] [CrossRef]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009, 25, 2078–9. [Google Scholar] [CrossRef]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–5. [Google Scholar] [CrossRef]
- Andrews S (Babraham, B. FastQC: a quality control tool for high throughput sequence data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011, 29, 24–6. [Google Scholar] [CrossRef]
- Zerbino DR, Johnson N, Juettemann T, Wilder SP, Flicek P. WiggleTools: parallel processing of large collections of genome-wide datasets for visualization and statistical analysis. Bioinformatics. 2014, 30, 1008–9. [Google Scholar] [CrossRef]
- Galaxy Community. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022, 50, W345–W351. [Google Scholar] [CrossRef]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014, 30, 2114–20. [Google Scholar] [CrossRef] [PubMed]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014, 42, W187–91. [Google Scholar] [CrossRef] [PubMed]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010, 28, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Tanigawa Y, Dyer ES, Bejerano G. WhichTF is functionally important in your open chromatin data? Ay F, editor. PLoS Comput Biol. 2022, 18, e1010378. [Google Scholar] [CrossRef]
- Oliveros, JC. VENNY. An interactive tool for comparing lists with Venn Diagrams. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010, 38, 576–89. [Google Scholar] [CrossRef]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015, 31, 166–9. [Google Scholar] [CrossRef]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013, 14, 128. [Google Scholar] [CrossRef]
- Kuleshov M V., Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–7. [Google Scholar] [CrossRef] [PubMed]
- Xie Z, Bailey A, Kuleshov M V., Clarke DJB, Evangelista JE, Jenkins SL, et al. Gene Set Knowledge Discovery with Enrichr. Curr Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000, 25, 25–9. [Google Scholar] [CrossRef]
- Gene Ontology Consortium, Aleksander SA, Balhoff J, Carbon S, Cherry JM, Drabkin HJ, et al. The Gene Ontology knowledgebase in 2023. Baryshnikova A, editor. Genetics. 2023, 224, 1–14. [Google Scholar] [CrossRef]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011, 27, 1739–40. [Google Scholar] [CrossRef] [PubMed]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Franzén O, Gan L-M, Björkegren JLM. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database (Oxford). 2019, 2019, 1–9. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, 2016. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




