Submitted:
31 October 2024
Posted:
01 November 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Thermodynamic Analysis
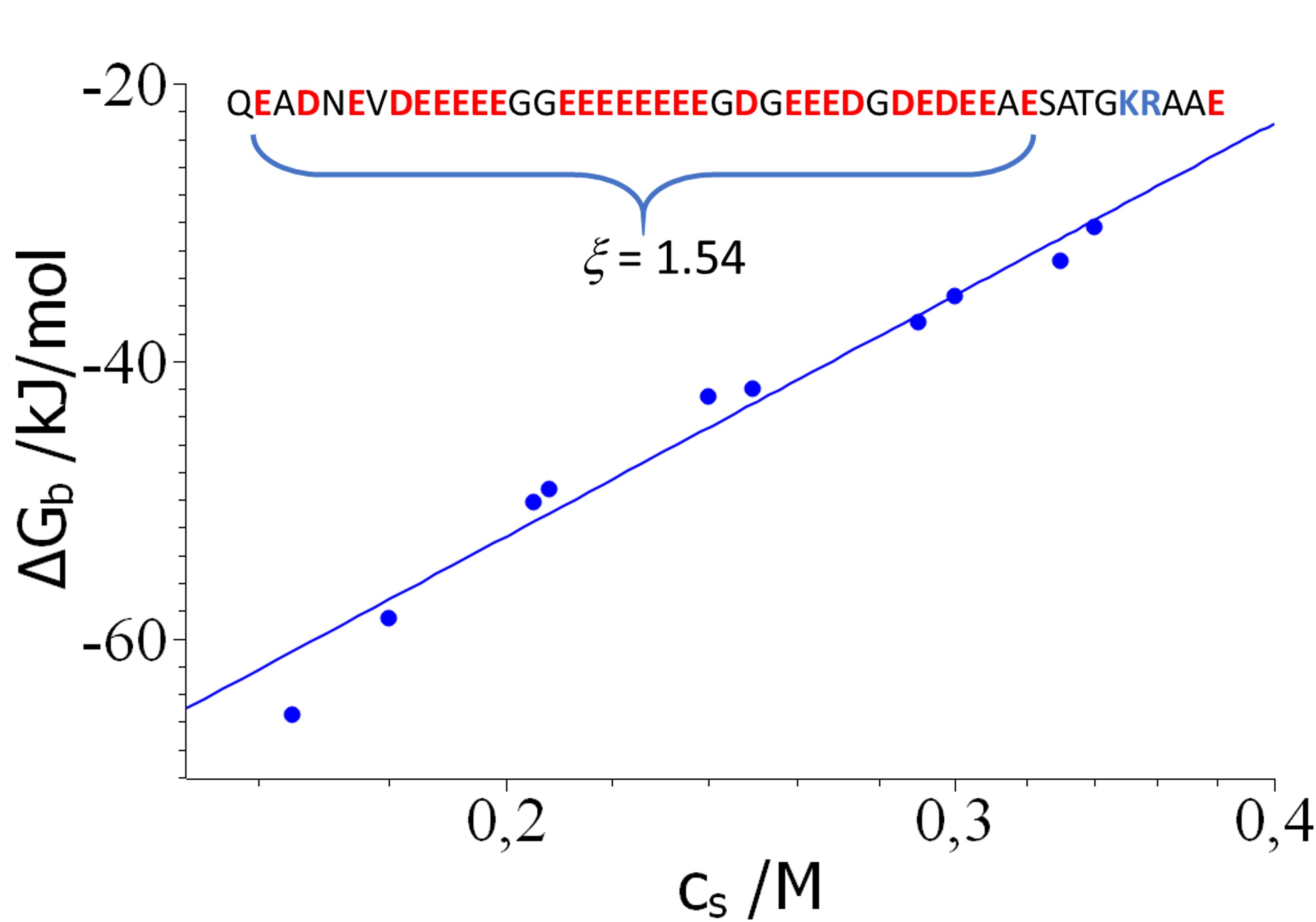
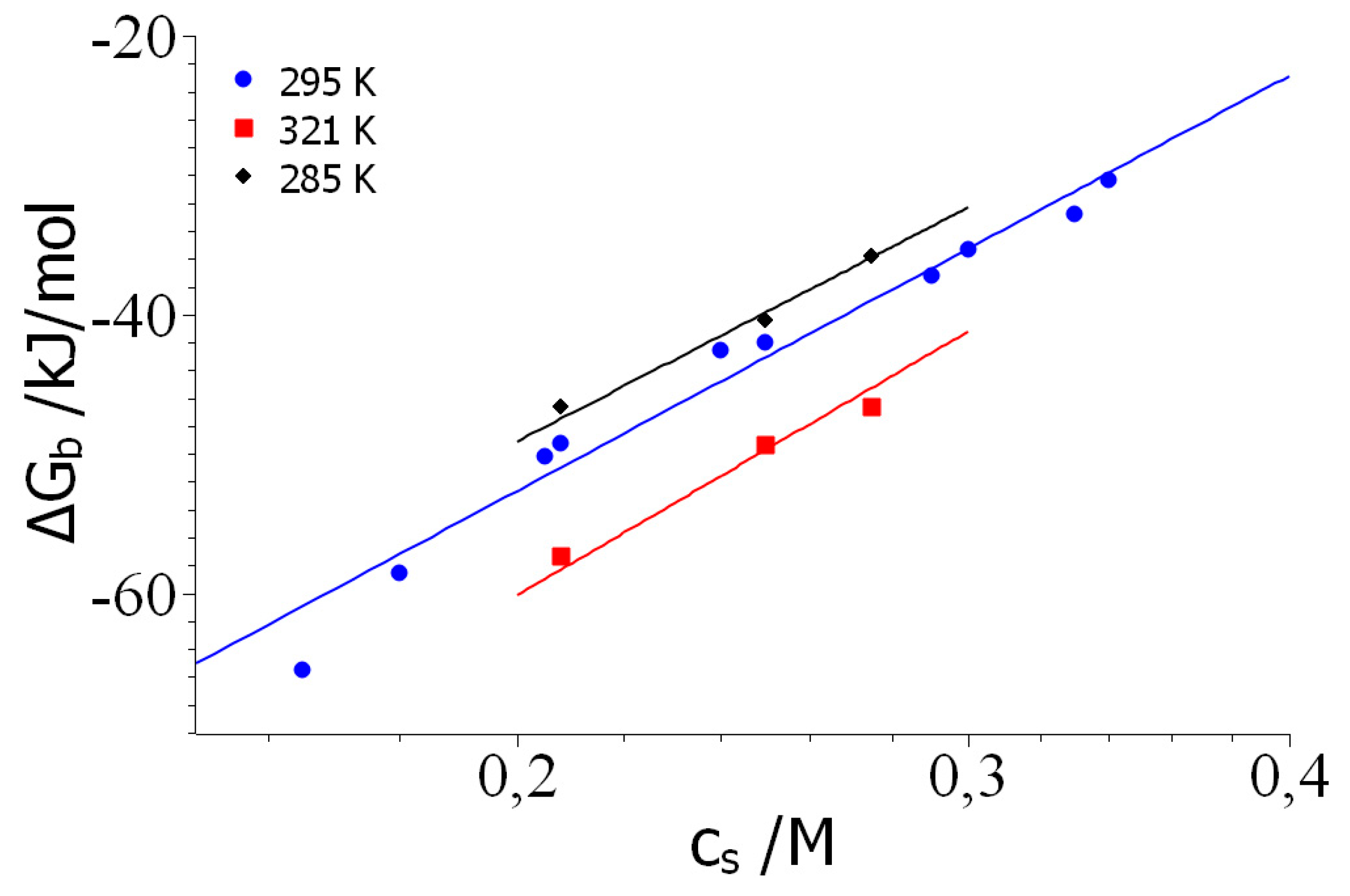
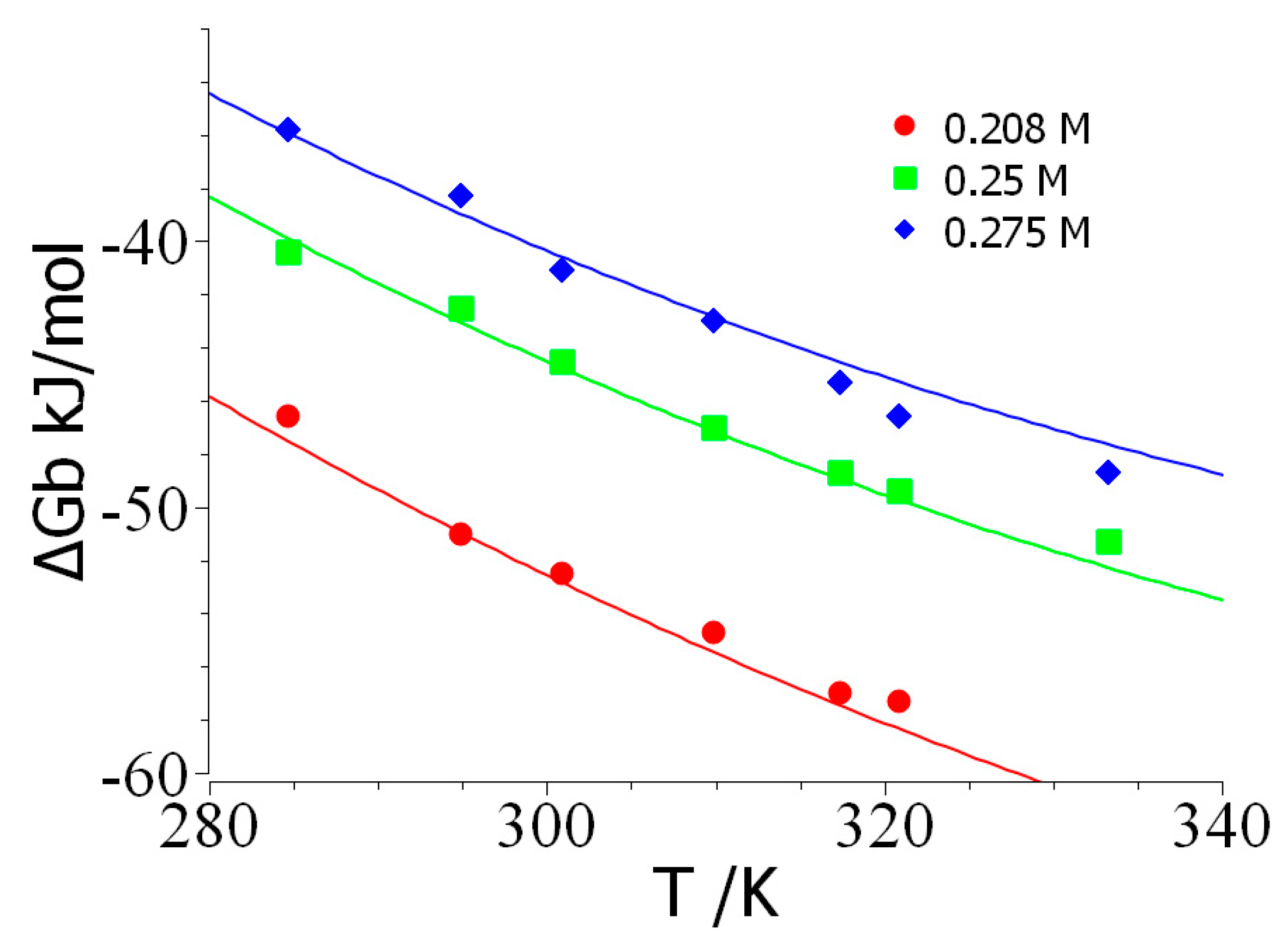
Results and Discussion
Conclusions
Acknowledgments
Conflicts of Interest
References
- F. Weinbreck, R.H. Tromp, C.G. de Kruif. Composition and structure of whey protein/gum arabic coacervates. Biomacromolecules 2004, 5, 1437–1445. [Google Scholar] [CrossRef]
- C.G. de Kruif, F. Weinbreck, R. de Vries. C.G. de Kruif, F. Weinbreck, R. de Vries. Curr Opin Colloid In 2004, 9, 340–349. [Google Scholar]
- E. Kizilay, A.B. Kayitmazer, P.L. Dubin. Complexation and coacervation of polyelectrolytes with oppositely charged colloids. Adv Colloid Interfac 2011, 167, 24–37. [Google Scholar] [CrossRef]
- A.B. Kayitmazer, D. Seeman, B.B. Minsky. Protein–polyelectrolyte interactions. Soft Matter 2013, 9, 2553–2583. [Google Scholar] [CrossRef]
- S.F. Banani, H.O. Lee, A.A. Hyman. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Bio 2017, 18, 285–298. [Google Scholar] [CrossRef]
- A. Klosin, F. Oltsch, T. Harmon. Phase separation provides a mechanism to reduce noise in cells. Science 2020, 367, 464. [Google Scholar] [CrossRef]
- C.E. Sing, S.L. Perry. Recent progress in the science of complex coacervation. Soft Matter 2020, 16, 2885–2914. [Google Scholar] [CrossRef]
- E., Dolgin. The shape-shifting blobs that shook up cell biology. Nature 2022, 611, 24–27. [Google Scholar]
- A. Abyzov, M. Blackledge, M. Zweckstetter. Conformational Dynamics of Intrinsically Disordered Proteins Regulate Biomolecular Condensate Chemistry. Chem Rev 2022, 122, 6719–6748. [Google Scholar] [CrossRef]
- B. Schuler, A. Borgia, M.B. Borgia. Binding without folding - the biomolecular function of disordered polyelectrolyte complexes. Curr Opin Struc Biol 2020, 60, 66–76. [Google Scholar] [CrossRef]
- N. Galvanetto, M.T. Ivanovic, A. Chowdhury. Extreme dynamics in a biomolecular condensate. Nature 2023, 619, 876–883. [Google Scholar] [CrossRef] [PubMed]
- Y.F. Dai, M. Farag, D. Lee. Programmable synthetic biomolecular condensates for cellular control. Nature Chemical Biology 2023, 19, 518. [Google Scholar]
- D. Priftis, N. Laugel, M. Tirrell. Thermodynamic Characterization of Polypeptide Complex Coacervation. Langmuir 2012, 28, 15947–15957. [Google Scholar] [CrossRef]
- D. Priftis, K. Megley, N. Laugel. Complex coacervation of poly(ethylene-imine)/polypeptide aqueous solutions: Thermodynamic and rheological characterization. J Colloid Interf Sci 2013, 398, 39–50. [Google Scholar] [CrossRef] [PubMed]
- S.L. Perry, Y. Li, D. Priftis. The Effect of Salt on the Complex Coacervation of Vinyl Polyelectrolytes. Polymers 2014, 6, 1756–1772. [Google Scholar] [CrossRef]
- L. Li, S. Srivastava, M. Andreev. Phase Behavior and Salt Partitioning in Polyelectrolyte Complex Coacervates. Macromolecules 2018, 51, 2988–2995. [Google Scholar] [CrossRef]
- Z.Y. Ou, M. Muthukumar, Entropy and enthalpy of polyelectrolyte complexation: Langevin dynamics simulations, J Chem Phys 124(15) (2006).
- T. Lytle, L.W. Chang, J. Madinya, S. Perry, C. Sing, Tuning complex coacervation using sequence-defined polyelectrolytes: A molecular understanding, Abstr Pap Am Chem S 254 (2017).
- T.K. Lytle, L.W. Chang, N. Markiewicz. Designing Electrostatic Interactions via Polyelectrolyte Monomer Sequence. Acs Central Sci 2019, 5, 709–718. [Google Scholar] [CrossRef]
- J.J. Zhou, M. Barz, F. Schmid, Complex formation between polyelectrolytes and oppositely charged oligoelectrolytes, J Chem Phys 144(16) (2016).
- M.T. Record Jr, C.F. Anderson, T.M. Lohman. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q. Rev. Biophys 1978, 11, 103–178. [Google Scholar] [CrossRef]
- X. Xu, S. Angioletti-Uberti, Y. Lu. Interaction of Proteins with Polyelectrolytes: Comparison of Theory to Experiment. Langmuir 2019, 35, 5373–5391. [Google Scholar] [CrossRef]
- S.S. Chen, Z.G. Wang, Driving force and pathway in polyelectrolyte complex coacervation, P Natl Acad Sci USA 119(36) (2022).
- K. Henzler, B. Haupt, K. Lauterbach. Adsorption of beta-lactoglobulin on spherical polyelectrolyte brushes: direct proof of counterion release by isothermal titration calorimetry. J Am Chem Soc 2010, 132, 3159–63. [Google Scholar] [CrossRef]
- M.T. Record, J.H. Ha, M.A. Fisher. Analysis of Equilibrium and Kinetic Measurements to Determine Thermodynamic Origins of Stability and Specificity and Mechanism of Formation of Site-Specific Complexes between Proteins and Helical DNA. Methods in Enzymology 1991, 208, 291–343. [Google Scholar]
- D.P. Mascotti, T.M. Lohman. Thermodynamic Extent of Counterion Release Upon Binding Oligolysines to Single-Stranded Nucleic-Acids. P Natl Acad Sci USA 1990, 87, 3142–3146. [Google Scholar] [CrossRef] [PubMed]
- D.P. Mascotti, T.M. Lohman. Thermodynamics of Single-Stranded Rna-Binding to Oligolysines Containing Tryptophan. Biochemistry 1992, 31, 8932–8946. [Google Scholar] [CrossRef] [PubMed]
- D.P. Mascotti, T.M. Lohman. Thermodynamics of charged oligopeptide-heparin interactions. Biochemistry 1995, 34, 2908–2915. [Google Scholar] [CrossRef] [PubMed]
- D.P. Mascotti, T.M. Lohman. Thermodynamics of oligoarginines binding to RNA and DNA. Biochemistry 1997, 36, 7272–7279. [Google Scholar] [CrossRef]
- X. Xu, Q.D. Ran, P. Dey. Counterion-Release Entropy Governs the Inhibition of Serum Proteins by Polyelectrolyte Drugs. Biomacromolecules 2018, 19, 409–416. [Google Scholar] [CrossRef]
- M.K. Hazra, Y. Levy, Affinity of disordered protein complexes is modulated by entropy-energy reinforcement, P Natl Acad Sci USA 119(26) (2022).
- S. Mitra, A. Kundagrami, Polyelectrolyte complexation of two oppositely charged symmetric polymers: A minimal theory, J Chem Phys 158(1) (2023).
- C.C. Pletka, R. Nepravishta, J. Iwahara. Detecting Counterion Dynamics in DNA-Protein Association. Angew Chem Int Edit 2020, 59, 1465–1468. [Google Scholar] [CrossRef]
- B.H. Yu, X. Wang, J. Iwahara, Measuring Local Electrostatic Potentials Around Nucleic Acids by Paramagnetic NMR Spectroscopy, J Phys Chem Lett 13(42) (2022).
- B.H. Yu, J. Iwahara. Experimental approaches for investigating ion atmospheres around nucleic acids and proteins. Comput Struct Biotec 2021, 19, 2279–2285. [Google Scholar] [CrossRef]
- M.T. Record, T.M. Lohman, P.L.D. Haseth. Ion Effects on Protein - Nucleic-Acid Interactions. Biophysical Journal 1976, 16, A14–A14. [Google Scholar]
- G.S., Manning. Limiting Laws and Counterion Condensation in Polyelectrolyte Solutions .I. Colligative Properties. J Chem Phys 1969, 51, 924. [Google Scholar]
- P.L. Privalov, A.I. Dragan, C. Crane-Robinson. Interpreting protein/DNA interactions: distinguishing specific from non-specific and electrostatic from non-electrostatic components. Nucleic Acids Research 2011, 39, 2483–2491. [Google Scholar] [CrossRef] [PubMed]
- A.I. Dragan, C.M. Read, C. Crane-Robinson. Enthalpy–entropy compensation: the role of solvation. European Biophysics Journal 2017, 46, 301–308. [Google Scholar] [CrossRef] [PubMed]
- J.J. Walkowiak, M. Ballauff, R. Zimmermann. Thermodynamic Analysis of the Interaction of Heparin with Lysozyme. Biomacromolecules 2020, 21, 4615–4625. [Google Scholar] [CrossRef]
- W. Malicka, R. Haag, M. Ballauff. Interaction of Heparin with Proteins: Hydration Effects. J Phys Chem B 2022, 126, 6250–6260. [Google Scholar] [CrossRef]
- C. Yigit, J. Heyda, J. Dzubiella, Charged patchy particle models in explicit salt: Ion distributions, electrostatic potentials, and effective interactions, J Chem Phys 143(6) (2015).
- C. Yigit, J. Heyda, M. Ballauff, J. Dzubiella, Like-charged protein-polyelectrolyte complexation driven by charge patches, J Chem Phys 143(6) (2015).
- Q. Ran, X. Xu, P. Dey. Interaction of human serum albumin with dendritic polyglycerol sulfate: Rationalizing the thermodynamics of binding. The Journal of Chemical Physics 2018, 149, 163324. [Google Scholar] [CrossRef]
- X. Xu, Q. Ran, P. Dey. Counterion-Release Entropy Governs the Inhibition of Serum Proteins by Polyelectrolyte Drugs. Biomacromolecules 2018, 19, 409–416. [Google Scholar] [CrossRef] [PubMed]
- C., Tanford. Extension of Theory of Linked Functions to Incorporate Effects of Protein Hydration. J Mol Biol 1969, 39, 539. [Google Scholar]
- D.P. Mascotti, T.M. Lohman. Thermodynamics of Single-Stranded Rna and DNA Interactions with Oligolysines Containing Tryptophan - Effects of Base Composition. Biochemistry 1993, 32, 10568–10579. [Google Scholar] [CrossRef]
- S. Bergqvist, R. O'Brien, J.E. Ladbury. Site-specific cation binding mediates TATA binding protein-DNA interaction from a hyperthermophilic archaeon. Biochemistry 2001, 40, 2419–2425. [Google Scholar] [CrossRef]
- S. Bergqvist, M.A. Williams, R. O'Brien. Reversal of halophilicity in a protein-DNA interaction by limited mutation strategy. Structure 2002, 10, 629–637. [Google Scholar] [CrossRef]
- K. Datta, V.J. LiCata. Thermodynamics of the binding of Thermus aquaticus DNA polymerase to primed-template DNA. Nucleic Acids Research 2003, 31, 5590–5597. [Google Scholar] [CrossRef] [PubMed]
- K. Datta, V.J. LiCata. Salt dependence of DNA binding by Thermus aquaticus and Escherichia coli DNA polymerases. Journal of Biological Chemistry 2003, 278, 5694–5701. [Google Scholar] [CrossRef]
- K. Datta, V.J. LiCata. Temperature dependence of DNA binding by Thermus aquaticus and Escherichia coli DNA polymerases. Biophysical Journal 2003, 84, 15a–15a. [Google Scholar]
- J.J. Walkowiak, M. Ballauff. Interaction of Polyelectrolytes with Proteins: Quantifying the Role of Water. Adv Sci 2021, 8, 2100661. [Google Scholar] [CrossRef] [PubMed]
- M.T. Record, E. Guinn, L. Pegram. Introductory Lecture: Interpreting and predicting Hofmeister salt ion and solute effects on biopolymer and model processes using the solute partitioning model. Faraday Discuss 2013, 160, 9–44. [Google Scholar] [CrossRef] [PubMed]
- K.A.V. Meulen, R.M. Saecker, M.T. Record. Formation of a wrapped DNA-protein interface: Experimental characterization and analysis of the large contributions of ions and water to the thermodynamics of binding IHF to H ' DNA. J Mol Biol 2008, 377, 9–27. [Google Scholar] [CrossRef]
- A. Chowdhury, D. Nettels, B. Schuler. Interaction Dynamics of Intrinsically Disordered Proteins from Single-Molecule Spectroscopy. Annu Rev Biophys 2023, 52, 433–462. [Google Scholar] [CrossRef]
- A. Chowdhury, A. Borgia, S. Ghosh. Driving forces of the complex formation between highly charged disordered proteins. Proc Natl Acad Sci U S A 2023, 120, e2304036120. [Google Scholar] [CrossRef]
- Z. Alexandrowicz, Osmotic and Donnan Equilibria in Polyacrylic Acid-Sodium Bromide Solutions, J Polym Sci 56(163) (1962) 115-&.
- Z., Alexandrowicz. On the Thermodynamical Interpretation of Osmotic and of Donnan Equilibria in Salt-Containing Polyelectrolyte Solutions .1. J Polym Sci 1960, 43, 325–336. [Google Scholar]
- Z., Alexandrowicz. Results of Osmotic and of Donnan Equilibria Measurements in Polymethacrylic Acid-Sodium Bromide Solutions .2. J Polym Sci 1960, 43, 337–349. [Google Scholar]
- J. Blaul, M. Wittemann, M. Ballauff, M. Rehahn, Osmotic coefficient of a synthetic rodlike polyelectrolyte in salt-free solution as a test of the Poisson-Boltzmann cell model, J Phys Chem B 104(30) (2000) 7077-7081.
- A. Guilleaume, J. Blaul, M. Ballauff. The distribution of counterions around synthetic rod-like polyelectrolytes in solution: a study by small-angle X-ray scattering and by anomalous small-angle X-ray scattering. Eur Phys J E Soft Matter 2002, 8, 299–309. [Google Scholar] [CrossRef] [PubMed]
- J.H. Ha, R.S. Spolar, M.T. Record, Role of the Hydrophobic Effect in Stability of Site-Specific Protein-DNA Complexes, J Mol Biol 209(4) (1989) 801-816.
- M. Ballauff, Denaturation of proteins: electrostatic effects vs. hydration, Rsc Adv 12(16) (2022) 10105-10113.
- X. Xu, M. Ballauff, Interaction of Lysozyme with a Dendritic Polyelectrolyte: Quantitative Analysis of the Free Energy of Binding and Comparison to Molecular Dynamics Simulations, The Journal of Physical Chemistry B 123(39) (2019) 8222-8231.
- Y.F. Liu, J.M. Sturtevant. Significant discrepancies between van't Hoff and calorimetric enthalpies .3. Biophys Chem 1997, 64, 121–126. [Google Scholar] [CrossRef] [PubMed]
- J. Bukala, P. Yavvari, J. Walkowiak, M. Ballauff, M. Weinhart, Interaction of Linear Polyelectrolytes with Proteins: Role of Specific Charge-Charge Interaction and Ionic Strength, Biomolecules 11(9) (2021).
- L.W. Chang, T.K. Lytle, M. Radhakrishna, J.J. Madinya, J. Velez, C.E. Sing, S.L. Perry, Sequence and entropy-based control of complex coacervates, Nat Commun 8 (2017).
- G.S. Manning, Approximate solutions to some problems in polyelectrolyte theory involving nonuniform charge distributions, Macromolecules 41(16) (2008) 6217-6227.
- B.B. Minsky, A. Atmuri, I.A. Kaltashov, P.L. Dubin, Counterion Condensation on Heparin Oligomers, Biomacromolecules 14(4) (2013) 1113-1121.
- A.D. Cardin, H.J.R. A.D. Cardin, H.J.R. Weintraub, Molecular Modeling of Protein-Glycosaminoglycan Interactions, Arteriosclerosis 9(1) (1989) 21-32.
- A.D. Cardin, D.A. Demeter, H.J.R. Weintraub. Molecular Design and Modeling of Protein Heparin Interactions. Methods in Enzymology 1991, 203, 556–583. [Google Scholar]
- T.R. Rudd, M.D. Preston, E.A. Yates. The nature of the conserved basic amino acid sequences found among 437 heparin binding proteins determined by network analysis. Mol Biosyst 2017, 13, 852–865. [Google Scholar] [CrossRef]
- K. Achazi, R. Haag, M. Ballauff, J. Dernedde, J.N. Kizhakkedathu, D. Maysinger, G. Multhaup, Understanding the Interaction of Polyelectrolyte Architectures with Proteins and Biosystems, Angew Chem Int Ed Engl 60(8) (2021) 3882-3904.
- K.A. Connors, D. Arndt, J.M. Rawlings, A.M.B. Hansen, M.W. Lam, H. Sanderson, S.E. Belanger, Environmental hazard of cationic polymers relevant in personal and consumer care products: A critical review, Integr Environ Asses 19(2) (2023) 312-325.
- W.S. Hall, R.J. Mirenda. Acute Toxicity of Waste-Water Treatment Polymers to Daphnia-Pulex and the Fathead Minnow (Pimephales-Promelas) and the Effects of Humic-Acid on Polymer Toxicity. Res J Water Pollut C 1991, 63, 895–899. [Google Scholar]
- J.L. Pereira, T. Vidal, F.J.M. Gonçalves, R.G. Gabriel, R. Costa, M.G. Rasteiro, Is the aquatic toxicity of cationic polyelectrolytes predictable from selected physical properties?, Chemosphere 202 (2018) 145-153.
- M.T. Record, P.L. Dehaseth, T.M. Lohman, Interpretation of Monovalent and Divalent-Cation Effects on Lac Repressor-Operator Interaction, Biochemistry 16(22) (1977) 4791-4796.
- B.M. Baker, K.P. Murphy, Evaluation of linked protonation effects in protein binding reactions using isothermal titration calorimetry, Biophysical Journal 71(4) (1996) 2049-2055.
- B.M. Baker, K.P. Murphy. Dissecting the energetics of a protein-protein interaction: The binding of ovomucoid third domain to elastase. J Mol Biol 1997, 268, 557–569. [Google Scholar] [CrossRef] [PubMed]
- R.E. Hileman, R.N. Jennings, R.J. Linhardt. Thermodynamic analysis of the heparin interaction with a basic cyclic peptide using isothermal titration calorimetry. Biochemistry 1998, 37, 15231–15237. [Google Scholar] [CrossRef]
- C.C. La, S.A. Smith, S. Vappala, R. Adili, C.E. Luke, S. Abbina, H.M.D. Luo, I. Chafeeva, M. Drayton, L.A. Creagh, M.D. Jaraquemada-Peláez, N. Rhoads, M.T. Kalathottukaren, P.K. Henke, S.K. Straus, C.G. Du, E.M. Conway, M. Holinstat, C.A. Haynes, J.H. Morrissey, J.N. Kizhakkedathu, Smart thrombosis inhibitors without bleeding side effects via charge tunable ligand design, Nat Commun 14(1) (2023).
- E. Schneck, F. Sedlmeier, R.R. Netz. Hydration repulsion between biomembranes results from an interplay of dehydration and depolarization. P Natl Acad Sci USA 2012, 109, 14405–14409. [Google Scholar] [CrossRef]
- A. Schlaich, J.O. Daldrop, B. Kowalik, M. Kanduc, E. Schneck, R.R. Netz, Water Structuring Induces Nonuniversal Hydration Repulsion between Polar Surfaces: Quantitative Comparison between Molecular Simulations, Theory, and Experiments, Langmuir (2024).
- C.G. Malmberg, A.A. Maryott. Dielectric Constant of Water from 0-Degrees-C to 100-Degrees-C. J Res Nat Bur Stand 1956, 56, 1–8. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




