Submitted:
31 October 2024
Posted:
05 November 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
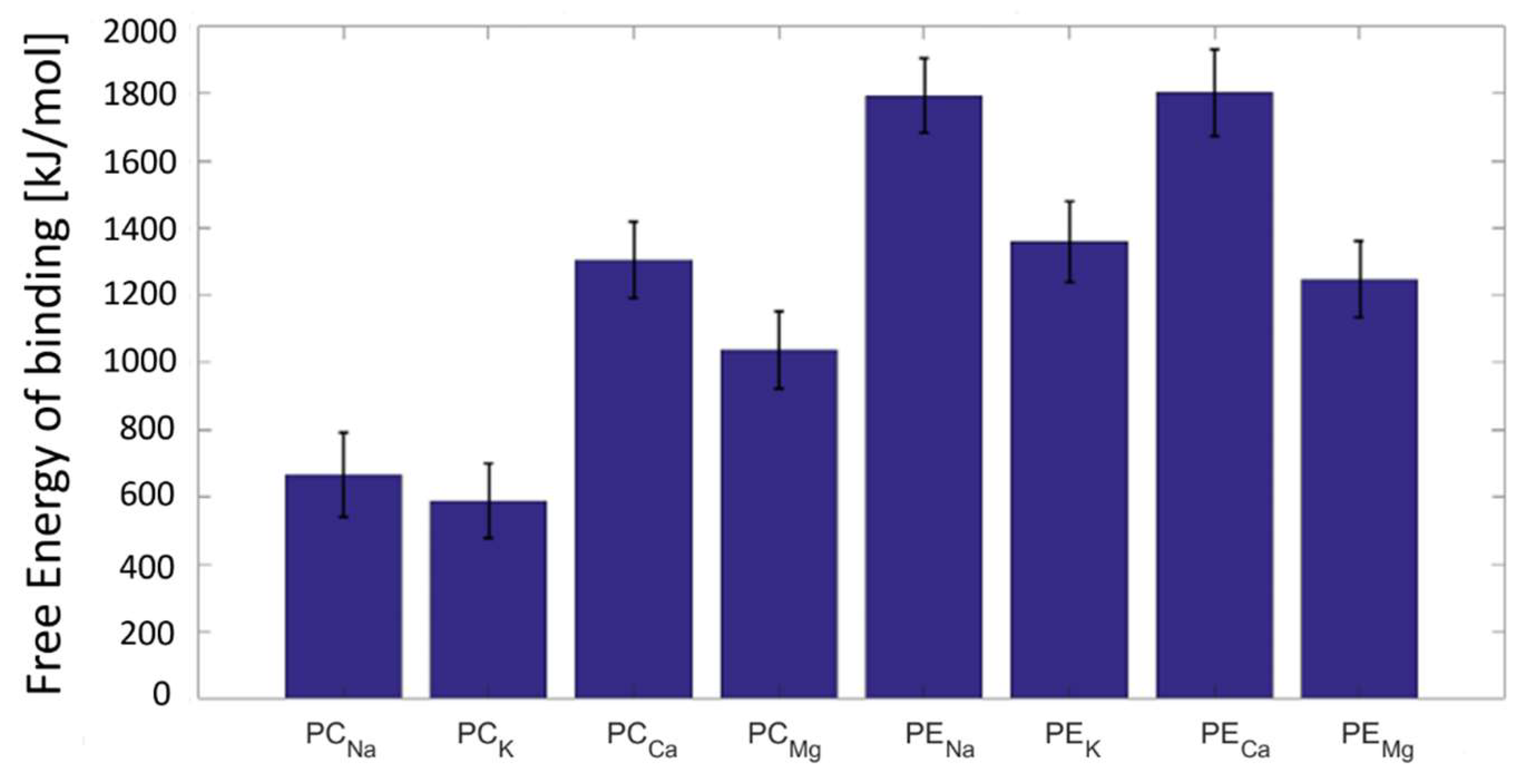
2.1. Free energy of Binding
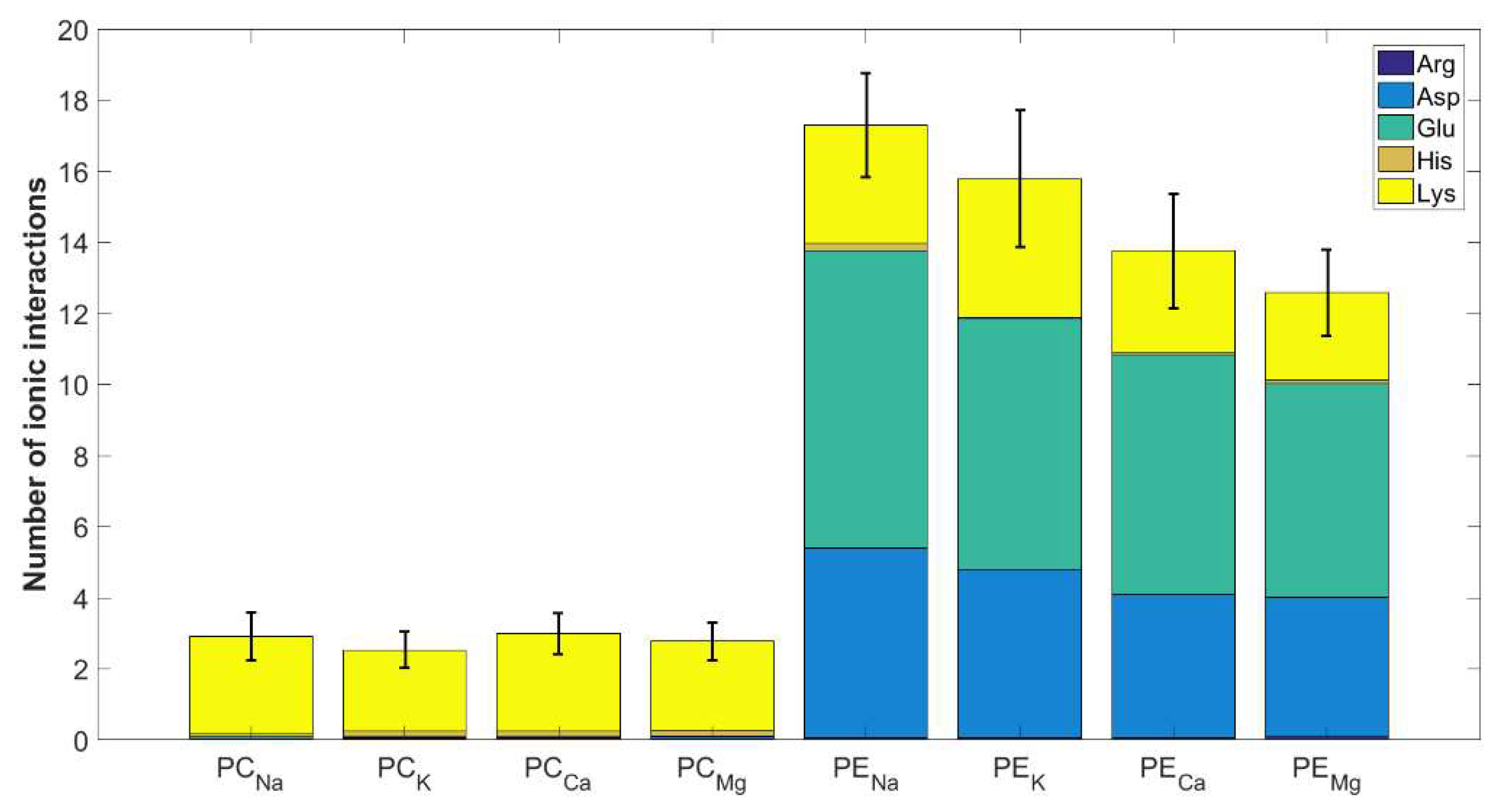
2.2. Intermolecular Contacts Identification
3. Results and Discussion
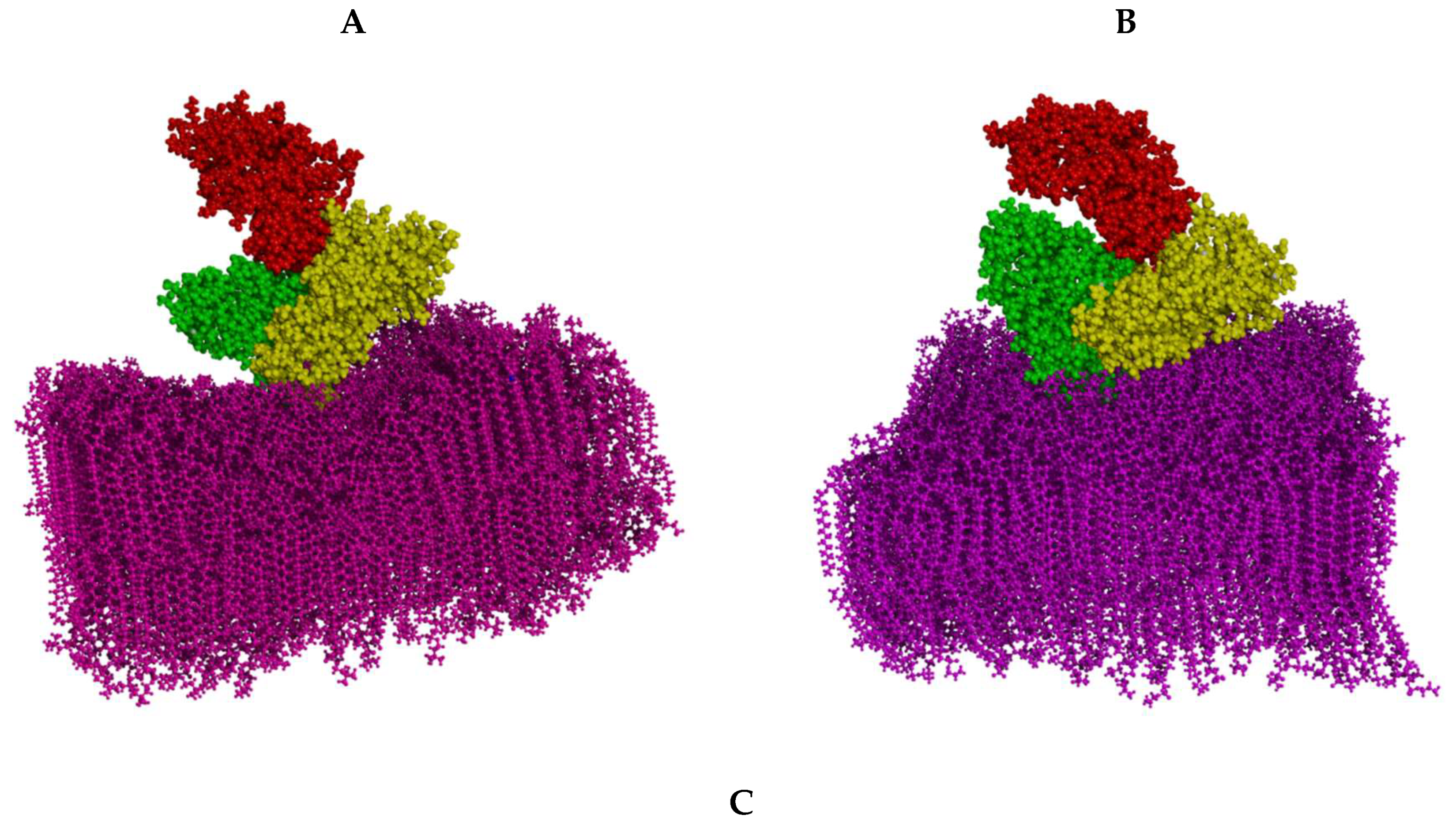
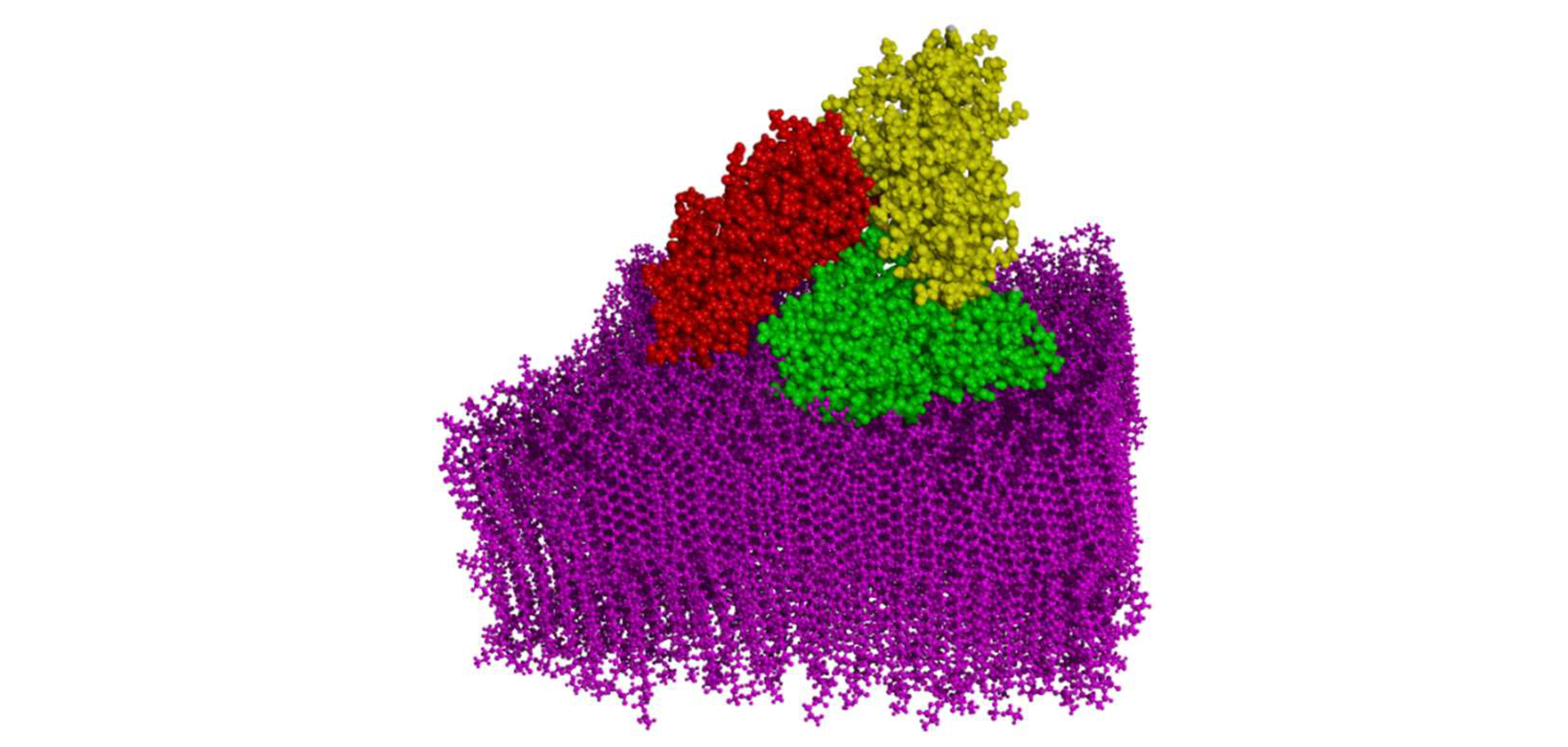
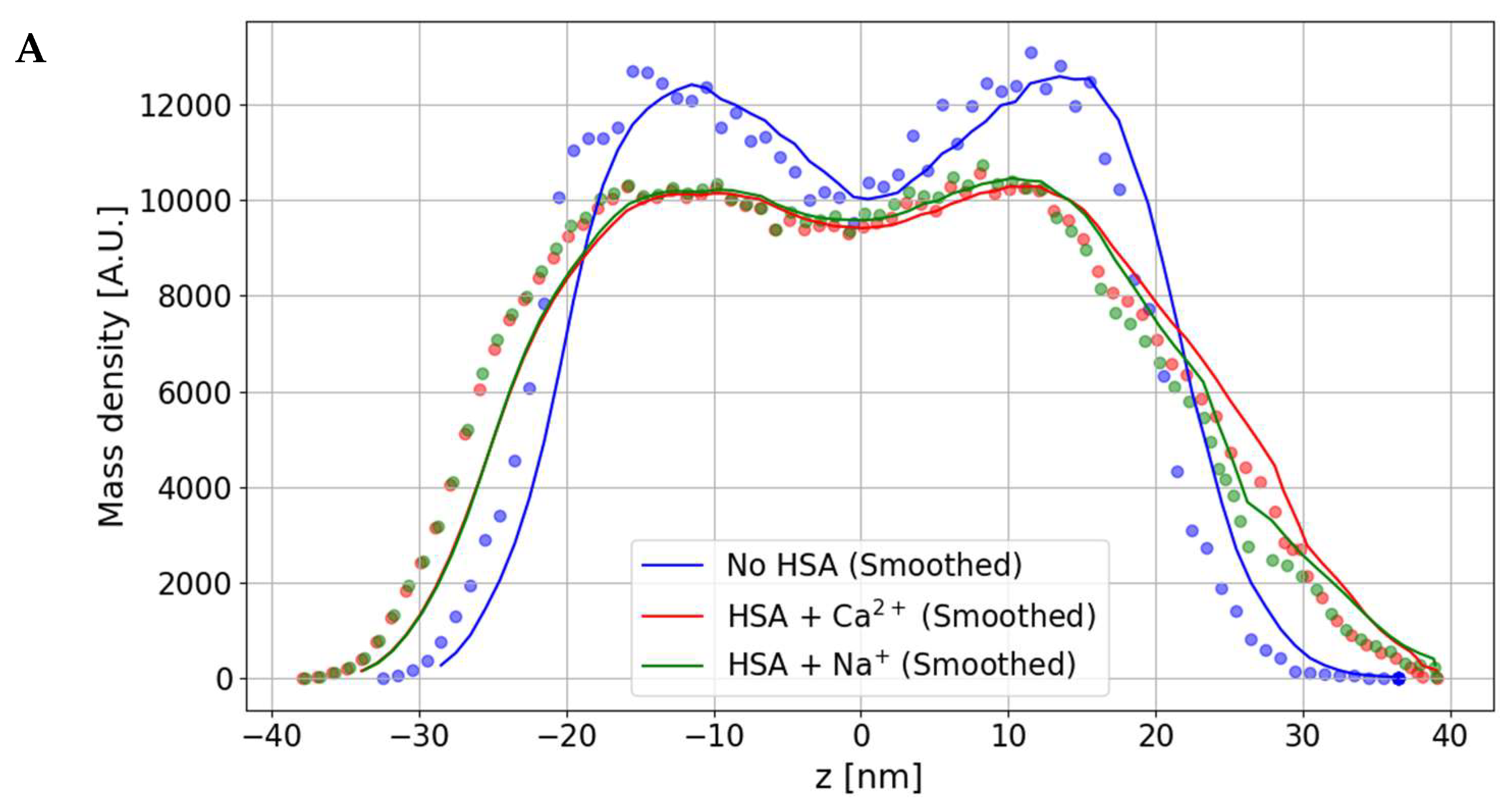
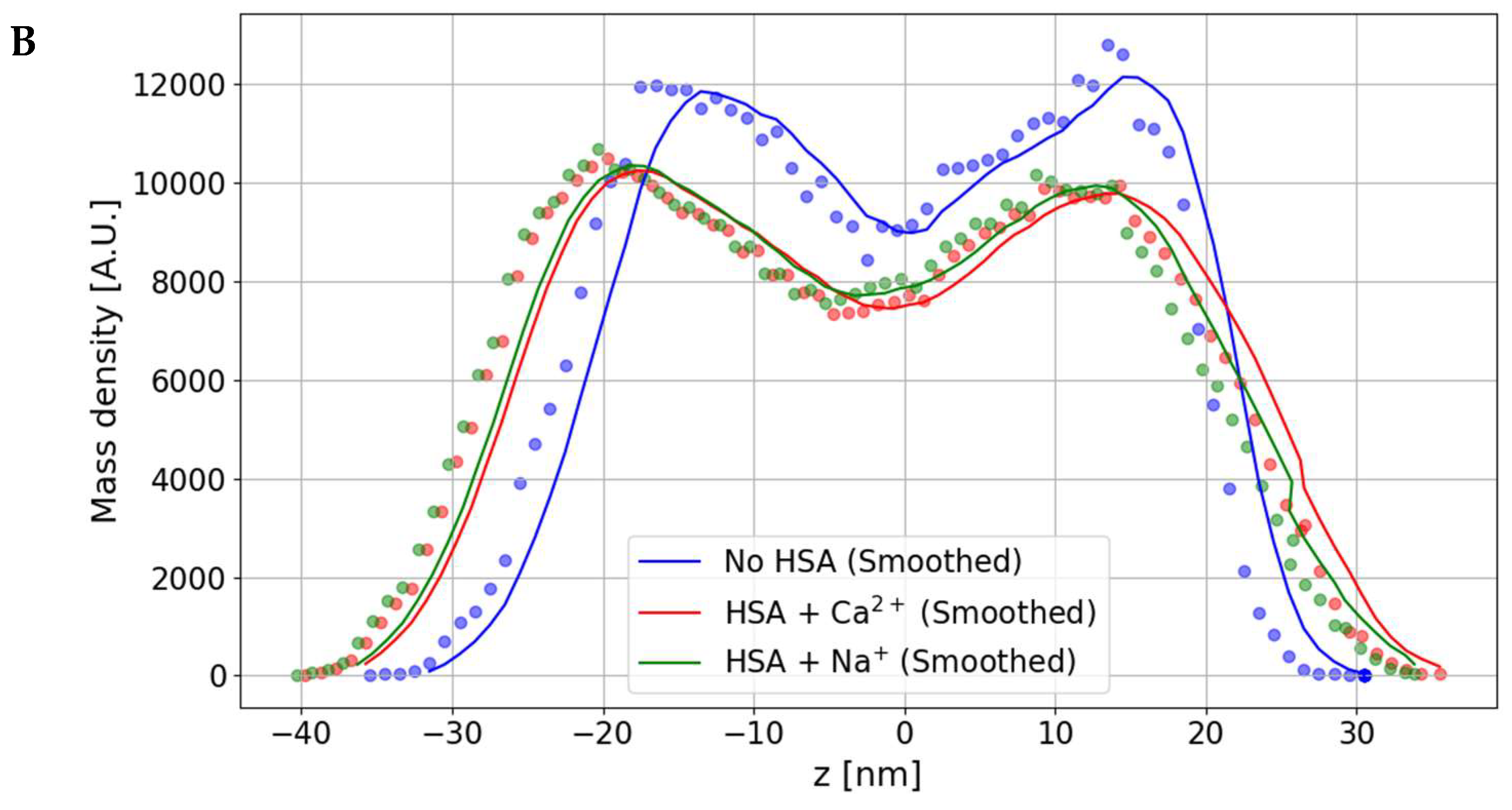
3.1. The Effect of HSA Binding on Bilayer Shape and Density
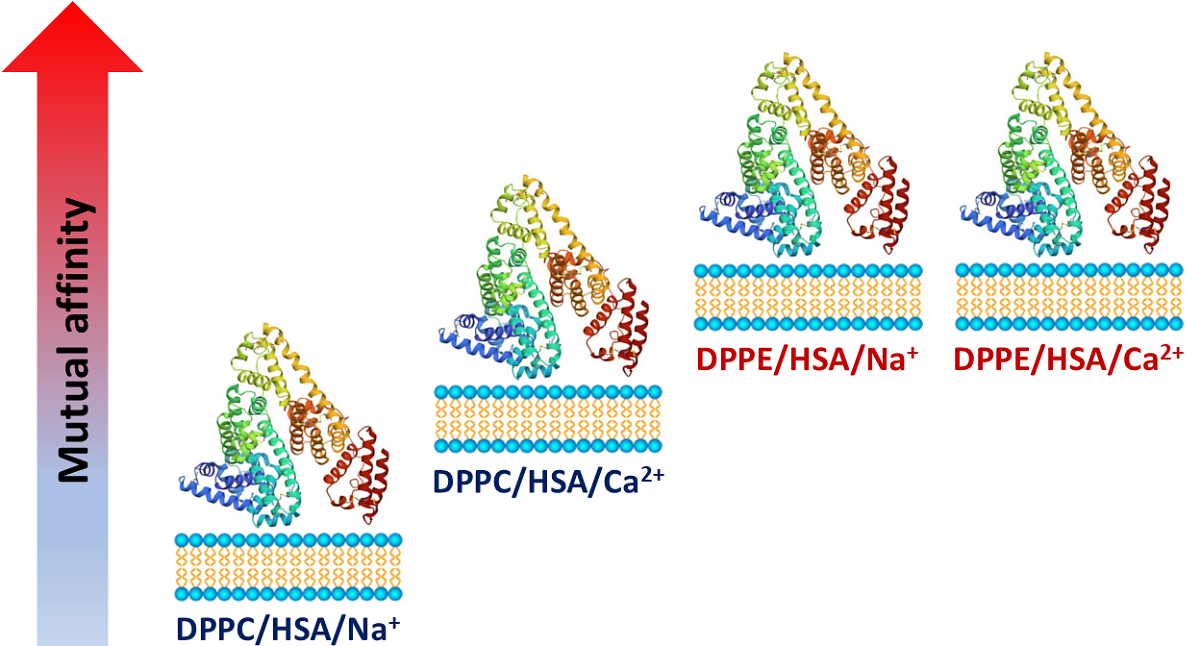
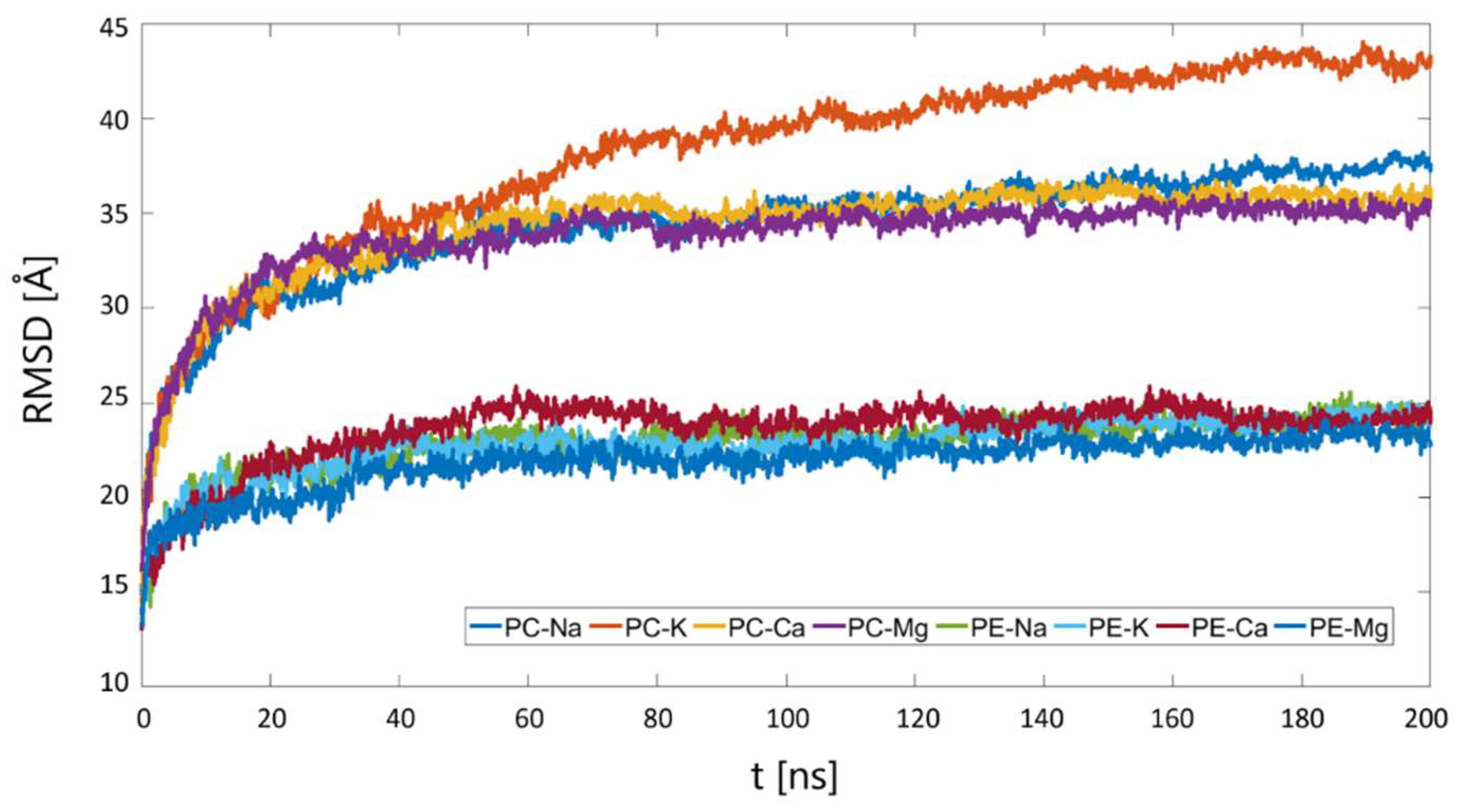
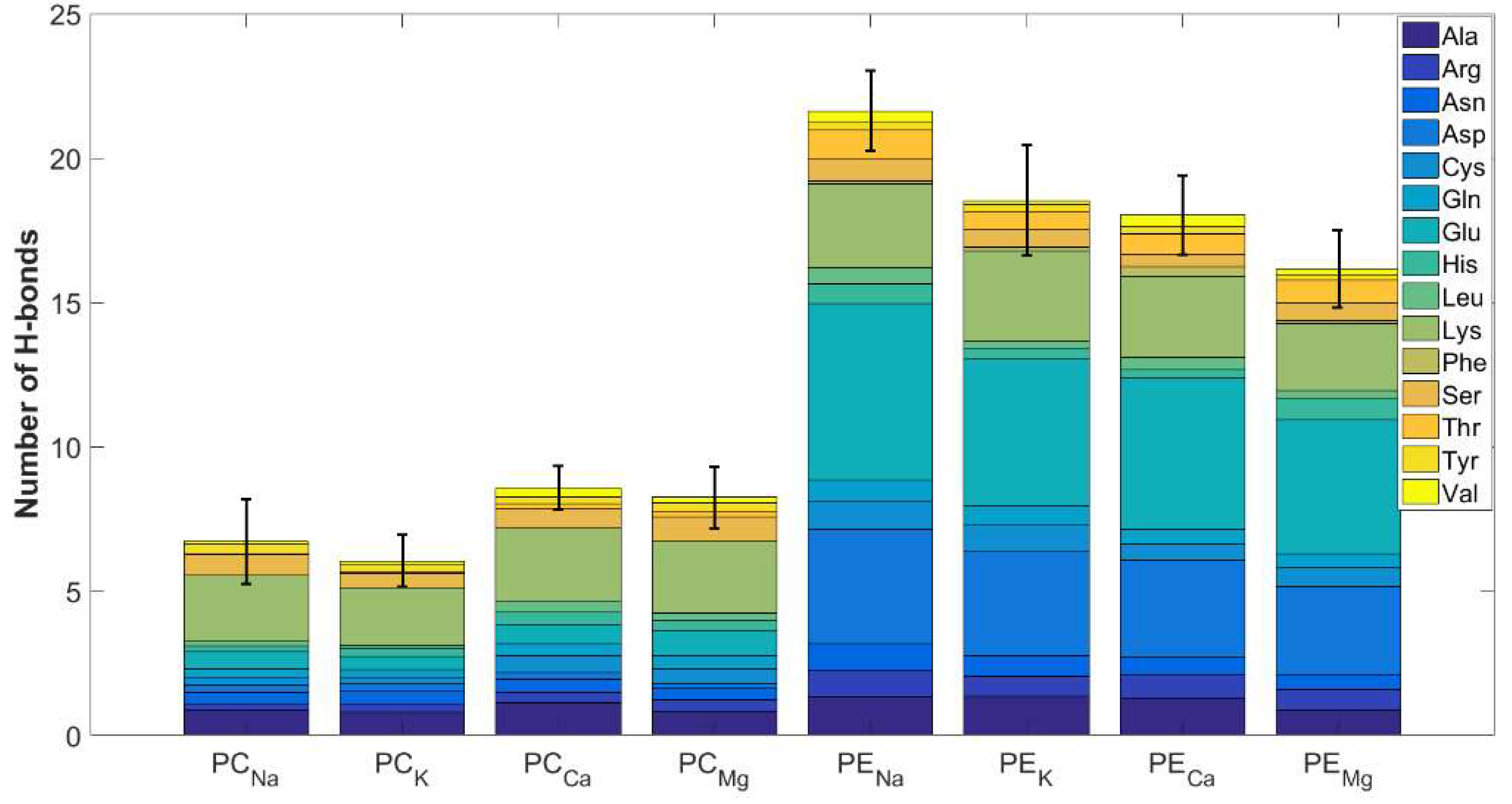
3.2. The Affinity of HSA for DPPC and DPPE in the Presence of Mono- and Divalent Cations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dai, Y.; Tang, H.; Pang, S. The Crucial Roles of Phospholipids in Aging and Lifespan Regulation. Front. Physiol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Zachowski, A. Phospholipids in animal eukaryotic membranes: transverse asymmetry and movement. Biochem. J. 1993, 294, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Scott, H.L.; Bolmatov, D.; Podar, P.T.; Liu, Z.; Kinnun, J.J.; Doughty, B.; Lydic, R.; Sacci, R.L.; Collier, C.P.; Katsaras, J. Evidence for long-term potentiation in phospholipid membranes. Proc. Natl. Acad. Sci. 2022, 119. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.; Sarangi, N.K.; Keyes, T.E. Role of phosphatidylserine in amyloid-beta oligomerization at asymmetric phospholipid bilayers. Phys. Chem. Chem. Phys. 2023, 25, 7648–7661. [Google Scholar] [CrossRef]
- Chauhan, A.; Ray, I.; Chauhan, V.P.S. Interaction of amyloid beta-protein with anionic phospholipids: Possible involvement of Lys28 and C-terminus aliphatic amino acids. Neurochem. Res. 2000, 25, 423–429. [Google Scholar] [CrossRef]
- Lee, G.; Pollard, H.B.; Arispe, N. Annexin 5 and apolipoprotein E2 protect against Alzheimer’s amyloid-β-peptide cytotoxicity by competitive inhibition at a common phosphatidylserine interaction site. Peptides 2002, 23, 1249–1263. [Google Scholar] [CrossRef]
- Toimil, P.; Prieto, G.; Miñones, J.; Trillo, J.M.; Sarmiento, F. Monolayer and Brewster angle microscopy study of human serum albumin—Dipalmitoyl phosphatidyl choline mixtures at the air–water interface. Colloids Surfaces B Biointerfaces 2012, 92, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Nsairat, H.; Khater, D.; Sayed, U.; Odeh, F.; Al Bawab, A.; Alshaer, W. Liposomes: structure, composition, types, and clinical applications. Heliyon 2022, 8. [Google Scholar] [CrossRef]
- Makouie, S.; Bryś, J.; Małajowicz, J.; Koczoń, P.; Siol, M.; Palani, B.K.; Bryś, A.; Obranović, M.; Mikolčević, S.; Gruczyńska-Sękowska, E. A Comprehensive Review of Silymarin Extraction and Liposomal Encapsulation Techniques for Potential Applications in Food. Appl. Sci. 2024, 14, 8477. [Google Scholar] [CrossRef]
- De Leo, V.; Maurelli, A.M.; Giotta, L.; Catucci, L. Liposomes containing nanoparticles: preparation and applications. Colloids Surfaces B Biointerfaces 2022, 218, 112737. [Google Scholar] [CrossRef]
- Youness, R.A.; Mohamed, A.H.; Efthimiadou, E.K.; Mekky, R.Y.; Braoudaki, M.; Fahmy, S.A. A Snapshot of Photoresponsive Liposomes in Cancer Chemotherapy and Immunotherapy: Opportunities and Challenges. ACS Omega 2023, 8, 44424–44436. [Google Scholar] [CrossRef] [PubMed]
- Jebastin, K.; Narayanasamy, D. Rationale utilization of phospholipid excipients: a distinctive tool for progressing state of the art in research of emerging drug carriers. J. Liposome Res. 2023, 33, 1–33. [Google Scholar] [CrossRef]
- Ferraz, M.P. Advanced Nanotechnological Approaches for Biofilm Prevention and Control. Appl. Sci. 2024, 14, 8137. [Google Scholar] [CrossRef]
- Veldhuizen, R.; Nag, K.; Orgeig, S.; Possmayer, F. The role of lipids in pulmonary surfactant. Biochim. Biophys. Acta - Mol. Basis Dis. 1998, 1408, 90–108. [Google Scholar] [CrossRef] [PubMed]
- Wüstneck, R.; Perez-Gil, J.; Wüstneck, N.; Cruz, A.; Fainerman, V.B.; Pison, U. Interfacial properties of pulmonary surfactant layers. Adv. Colloid Interface Sci. 2005, 117, 33–58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, Y.E.; Fan, Q.; Zuo, Y.Y. On the Low Surface Tension of Lung Surfactant. Langmuir 2011, 27, 8351–8358. [Google Scholar] [CrossRef]
- Qin, S.-S.; Yu, Z.-W.; Yu, Y.-X. Structural and Kinetic Properties of α-Tocopherol in Phospholipid Bilayers, a Molecular Dynamics Simulation Study. J. Phys. Chem. B 2009, 113, 16537–16546. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Shi, R.; Hao, C.; Chen, H.; Zhang, L.; Li, J.; Xu, G.; Sun, R. Behavior of lysozyme adsorbed onto biological liquid crystal lipid monolayer at the air/water interface. Chinese Phys. B 2016, 25, 090506. [Google Scholar] [CrossRef]
- Nobre, T.M.; Pavinatto, F.J.; Caseli, L.; Barros-Timmons, A.; Dynarowicz-Łątka, P.; Oliveira, O.N. Interactions of bioactive molecules & nanomaterials with Langmuir monolayers as cell membrane models. Thin Solid Films 2015, 593, 158–188. [Google Scholar] [CrossRef]
- Endo, S.; Goss, K.U. Serum albumin binding of structurally diverse neutral organic compounds: Data and models. Chem. Res. Toxicol. 2011, 24, 2293–2301. [Google Scholar] [CrossRef]
- Fasano, M.; Curry, S.; Terreno, E.; Galliano, M.; Fanali, G.; Narciso, P.; Notari, S.; Ascenzi, P. The extraordinary ligand binding properties of human serum albumin. IUBMB Life 2005, 57, 787–796. [Google Scholar] [CrossRef] [PubMed]
- De Simone, G.; Di Masi, A.; Ascenzi, P. Serum albumin: A multifaced enzyme. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Opriș, O.; Mormile, C.; Lung, I.; Stegarescu, A.; Soran, M.-L.; Soran, A. An Overview of Biopolymers for Drug Delivery Applications. Appl. Sci. 2024, 14, 1383. [Google Scholar] [CrossRef]
- Yang, W.J.; Wu, H.B.; Zhang, C.; Zhong, Q.; Hu, M.J.; He, J.L.; Li, G.A.; Zhu, Z.Y.; Zhu, J.L.; Zhao, H.H.; et al. Exposure to 2,4-dichlorophenol, 2,4,6-trichlorophenol, pentachlorophenol and risk of thyroid cancer: a case-control study in China. Environ. Sci. Pollut. Res. 2021, 28, 61329–61343. [Google Scholar] [CrossRef]
- Neligan, P.J. Fluid and electrolyte balance. Anaesth. Intensive Care Med. 2021, 22, 169–173. [Google Scholar] [CrossRef]
- Maurya, P.; Singh, S.; Mishra, N.; Pal, R.; Singh, N.; Parashar, P.; Saraf, S.A. Albumin-based nanomaterials in drug delivery and biomedical applications. In Biopolymer-Based Nanomaterials in Drug Delivery and Biomedical Applications; Elsevier, 2021; pp. 465–496.
- Peters, T. All About Albumin; Elsevier, 1995; ISBN 9780125521109.
- Belinskaia, D.A.; Voronina, P.A.; Shmurak, V.I.; Jenkins, R.O.; Goncharov, N. V. Serum Albumin in Health and Disease: Esterase, Antioxidant, Transporting and Signaling Properties. Int. J. Mol. Sci. 2021, 22, 10318. [Google Scholar] [CrossRef]
- Kragh-Hansen, U.; Chuang, V.T.G.; Otagiri, M. Practical Aspects of the Ligand-Binding and Enzymatic Properties of Human Serum Albumin. Biol. Pharm. Bull. 2002, 25, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, G.; Ahn, S.N. Structure, enzymatic activities, glycation and therapeutic potential of human serum albumin: A natural cargo. Int. J. Biol. Macromol. 2019, 123, 979–990. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, F.A.; Wynalda, M.A. Albumin-catalyzed metabolism of prostaglandin D2. Identification of products formed in vitro. J. Biol. Chem. 1983, 258, 11713–11718. [Google Scholar] [CrossRef]
- Fitzpatrick, F.A.; Liggett, W.F.; Wynalda, M.A. Albumin-eicosanoid interactions. A model system to determine their attributes and inhibition. J. Biol. Chem. 1984, 259, 2722–2727. [Google Scholar] [CrossRef]
- WATANABE, H.; TANASE, S.; NAKAJOU, K.; MARUYAMA, T.; KRAGH-HANSEN, U.; OTAGIRI, M. Role of Arg-410 and Tyr-411 in human serum albumin for ligand binding and esterase-like activity. Biochem. J. 2000, 349, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Curry, S. Plasma albumin as a fatty acid carrier. Adv. Mol. Cell Biol. 2003, 33, 29–46. [Google Scholar]
- Shojai, S.; Haeri Rohani, S.-A.; Moosavi-Movahedi, A.A.; Habibi-Rezaei, M. Human serum albumin in neurodegeneration. Rev. Neurosci. 2022, 33, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Kőszegi, T.; Poór, M. Ochratoxin A: Molecular Interactions, Mechanisms of Toxicity and Prevention at the Molecular Level. Toxins (Basel). 2016, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- Belinskaia, D.A.; Voronina, P.A.; Goncharov, N. V. Integrative Role of Albumin: Evolutionary, Biochemical and Pathophysiological Aspects. J. Evol. Biochem. Physiol. 2021, 57, 1419–1448. [Google Scholar] [CrossRef]
- Galántai, R.; Bárdos-Nagy, I. The interaction of human serum albumin and model membranes. Int. J. Pharm. 2000, 195, 207–218. [Google Scholar] [CrossRef]
- Dimitrova, M.N.; Matsumura, H.; Dimitrova, A.; Neitchev, V.Z. Interaction of albumins from different species with phospholipid liposomes. Multiple binding sites system. Int. J. Biol. Macromol. 2000, 27, 187–194. [Google Scholar] [CrossRef]
- de Souza, N.C.; Caetano, W.; Itri, R.; Rodrigues, C.A.; Oliveira, O.N.; Giacometti, J.A.; Ferreira, M. Interaction of small amounts of bovine serum albumin with phospholipid monolayers investigated by surface pressure and atomic force microscopy. J. Colloid Interface Sci. 2006, 297, 546–553. [Google Scholar] [CrossRef]
- Peng, Q.; Wei, X.-Q.; Shao, X.-R.; Zhang, T.; Zhang, S.; Fu, N.; Cai, X.-X.; Zhang, Z.-R.; Lin, Y.-F. Nanocomplex Based on Biocompatible Phospholipids and Albumin for Long-Circulation Applications. ACS Appl. Mater. Interfaces 2014, 6, 13730–13737. [Google Scholar] [CrossRef]
- Mariam, J.; Sivakami, S.; Dongre, P.M. Albumin corona on nanoparticles – a strategic approach in drug delivery. Drug Deliv. 2016, 23, 2668–2676. [Google Scholar] [CrossRef]
- Murakami, T.; Yarimitsu, S.; Nakashima, K.; Sawae, Y.; Sakai, N. Influence of synovia constituents on tribological behaviors of articular cartilage. Friction 2013, 1, 150–162. [Google Scholar] [CrossRef]
- Seror, J.; Kampf, N.; Maroudas, A.; Klein, J. Nanotribological Investigation of the Role of Proteoglycans in Biolubrication. In Proceedings of the Volume 3: Design; Tribology; Education; ASMEDC; 2008; pp. 513–519. [Google Scholar]
- Lin, W.; Klein, J. Recent Progress in Cartilage Lubrication. Adv. Mater. 2021, 33. [Google Scholar] [CrossRef]
- Stevenson, H.; Cann, P.M. Protein Content of Model Synovial Fluid and CoCrMo Wear. Biotribology 2021, 26, 100172. [Google Scholar] [CrossRef]
- Mirea, D.A.; Trunfio-Sfarghiu, A.-M.; Matei, C.I.; Munteanu, B.; Piednoir, A.; Rieu, J.P.; Blanchin, M.G.; Berthier, Y. Role of the biomolecular interactions in the structure and tribological properties of synovial fluid. Tribol. Int. 2013, 59, 302–311. [Google Scholar] [CrossRef]
- Li, Y.; Yuan, Z.; Yang, H.; Zhong, H.; Peng, W.; Xie, R. Recent Advances in Understanding the Role of Cartilage Lubrication in Osteoarthritis. Molecules 2021, 26, 6122. [Google Scholar] [CrossRef]
- Dėdinaitė, A.; Wieland, D.C.F.; Bełdowski, P.; Claesson, P.M. Biolubrication synergy: Hyaluronan – Phospholipid interactions at interfaces. Adv. Colloid Interface Sci. 2019, 274, 102050. [Google Scholar] [CrossRef] [PubMed]
- Siódmiak, J.; Bełdowski, P.; Augé, W.; Ledziński, D.; Śmigiel, S.; Gadomski, A. Molecular Dynamic Analysis of Hyaluronic Acid and Phospholipid Interaction in Tribological Surgical Adjuvant Design for Osteoarthritis. Molecules 2017, 22, 1436. [Google Scholar] [CrossRef] [PubMed]
- Raczyński, P.; Górny, K.; Bełdowski, P.; Yuvan, S.; Dendzik, Z. Application of Graphene as a Nanoindenter Interacting with Phospholipid Membranes—Computer Simulation Study. J. Phys. Chem. B 2020, 124, 6592–6602. [Google Scholar] [CrossRef]
- Bełdowski, P.; Kruszewska, N.; Yuvan, S.; Dendzik, Z.; Goudoulas, T.; Gadomski, A. Capstan-like mechanism in hyaluronan–phospholipid systems. Chem. Phys. Lipids 2018, 216, 17–24. [Google Scholar] [CrossRef]
- Bełdowski, P.; Yuvan, S.; Dėdinaitė, A.; Claesson, P.M.; Pöschel, T. Interactions of a short hyaluronan chain with a phospholipid membrane. Colloids Surfaces B Biointerfaces 2019, 184, 110539. [Google Scholar] [CrossRef]
- Raczyński, P.; Górny, K.; Bełdowski, P.; Marciniak, B.; Pöschel, T.; Dendzik, Z. Influence of silicon nanocone on cell membrane self-sealing capabilities for targeted drug delivery—Computer simulation study. Arch. Biochem. Biophys. 2023, 749, 109802. [Google Scholar] [CrossRef] [PubMed]
- Bełdowski, P.; Przybyłek, M.; Raczyński, P.; Dedinaite, A.; Górny, K.; Wieland, F.; Dendzik, Z.; Sionkowska, A.; Claesson, P.M. Albumin–Hyaluronan Interactions: Influence of Ionic Composition Probed by Molecular Dynamics. Int. J. Mol. Sci. 2021, 22, 12360. [Google Scholar] [CrossRef] [PubMed]
- Przybyłek, M.; Tuwalska, A.; Ledziński, D.; Śmigiel, S.; Sionkowska, A.; Białas, I.; Bełdowski, P. Effect of Nanohydroxyapatite on Silk Fibroin–Chitosan Interactions—Molecular Dynamics Study. Appl. Sci. 2024, 14, 4131. [Google Scholar] [CrossRef]
- Bonté, F.; Juliano, R.L. Interactions of liposomes with serum proteins. Chem. Phys. Lipids 1986, 40, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Petrache, H.I.; Tristram-Nagle, S.; Harries, D.; Kučerka, N.; Nagle, J.F.; Parsegian, V.A. Swelling of phospholipids by monovalent salt. J. Lipid Res. 2006, 47, 302–309. [Google Scholar] [CrossRef]
- Cordomí, A.; Edholm, O.; Perez, J.J. Effect of Ions on a Dipalmitoyl Phosphatidylcholine Bilayer. A Molecular Dynamics Simulation Study. J. Phys. Chem. B 2008, 112, 1397–1408. [Google Scholar] [CrossRef]
- Gurtovenko, A.A.; Vattulainen, I. Ion Leakage through Transient Water Pores in Protein-Free Lipid Membranes Driven by Transmembrane Ionic Charge Imbalance. Biophys. J. 2007, 92, 1878–1890. [Google Scholar] [CrossRef]
- Tantipolphan, R.; Rades, T.; McQuillan, A.J.; Medlicott, N.J. Adsorption of bovine serum albumin (BSA) onto lecithin studied by attenuated total reflectance Fourier transform infrared (ATR-FTIR) spectroscopy. Int. J. Pharm. 2007, 337, 40–47. [Google Scholar] [CrossRef]
- Kitaoka, H.; Yokoyama, Y.; Sakka, T.; Nishi, N. Salting-out and Competitive Adsorption of Ethanol into Lipid Bilayer Membranes: Conflicting Effects of Salts on Ethanol–Membrane Interactions Studied by Molecular Dynamics Simulations. J. Phys. Chem. B 2024, 128, 7596–7604. [Google Scholar] [CrossRef]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A Point-Charge Force Field for Molecular Mechanics Simulations of Proteins Based on Condensed-Phase Quantum Mechanical Calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Costanzo, L. Di; Christie, C.; Duarte, J.M.; Dutta, S.; Feng, Z.; et al. Protein Data Bank: the single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019, 47, D520–D528. [Google Scholar]
- Krieger, E.; Vriend, G. YASARA View—molecular graphics for all devices—from smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Im, W. Automated Builder and Database of Protein/Membrane Complexes for Molecular Dynamics Simulations. PLoS One 2007, 2, e880. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Vargyas, M.; Vasko-Szedlar, J.; Roux, B.; Im, W. PBEQ-Solver for online visualization of electrostatic potential of biomolecules. Nucleic Acids Res. 2008, 36, W270–W275. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L. Quantum and statistical mechanical studies of liquids. 10. Transferable intermolecular potential functions for water, alcohols, and ethers. Application to liquid water. J. Am. Chem. Soc. 1981, 103, 335–340. [Google Scholar] [CrossRef]
- Mark, P.; Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Schuler, L.D.; Daura, X.; Van Gunsteren, W.F.; Rapold, R.F.; Suter, U.W.; Darden, T.T.A.; York, D.; Pedersen, L.G.; Fuchs, P.F.J.; Hansen, H.S.; et al. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 2001, 81, 3586–3616. [Google Scholar]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Holst, M.; Baker, N.; Wang, F. Adaptive multilevel finite element solution of the Poisson-Boltzmann equation I. Algorithms and examples. J. Comput. Chem. 2000, 21, 1319–1342. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef] [PubMed]
- M. F. Brown Curvature forces in membrane lipid–protein interactions, Biochemistry 2012, 51, 9782–9795. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Isidoro, R.; Díaz-Salazar, A.J.; Costas, M. Biophysical study of the effect of ovalbumin and lysozyme in DMPC/sphingomyelin/cholesterol bilayers. J. Therm. Anal. Calorim. 2024, 149, 1219–1229. [Google Scholar] [CrossRef]
- Khodam Hazrati, M.; Vácha, R. Membrane Adsorption Enhances Translocation of Antimicrobial Peptide Buforin 2. J. Phys. Chem. B 2024, 128, 8469–8476. [Google Scholar] [CrossRef]
- Nylund, M.; Fortelius, C.; Palonen, E.K.; Molotkovsky, J.G.; Mattjus, P. Membrane curvature effects on glycolipid transfer protein activity. Langmuir 2007, 23, 11726–11733. [Google Scholar] [CrossRef]
- Jones, M.N. Surfactants in membrane solubilisation. Int. J. Pharm. 1999, 177, 137–159. [Google Scholar] [CrossRef]
- Richardson, J.D.; Van Lehn, R.C. Free Energy Analysis of Peptide-Induced Pore Formation in Lipid Membranes by Bridging Atomistic and Coarse-Grained Simulations. J. Phys. Chem. B 2024, 128, 8737–8752. [Google Scholar] [CrossRef]
- Meyuhas, D.; Lichtenberg, D. The effect of albumin on the state of aggregation and phase transformations in phosphatidylcholine-sodium cholate mixtures. Biochim. Biophys. Acta - Biomembr. 1995, 1234, 203–213. [Google Scholar] [CrossRef]
- Al-Ayed, M.S. Biophysical studies on the liposome-albumin system. Indian J. Biochem. Biophys. 2006, 43, 186–189. [Google Scholar]
- Rivel, T.; Ramseyer, C.; Yesylevskyy, S. The asymmetry of plasma membranes and their cholesterol content influence the uptake of cisplatin. Sci. Rep. 2019, 9, 5627. [Google Scholar] [CrossRef] [PubMed]
- Melcrová, A.; Pokorna, S.; Vošahlíková, M.; Sýkora, J.; Svoboda, P.; Hof, M.; Cwiklik, L.; Jurkiewicz, P. Concurrent Compression of Phospholipid Membranes by Calcium and Cholesterol. Langmuir 2019, 35, 11358–11368. [Google Scholar] [CrossRef]
- Wiig, H.; Kolmannskog, O.; Tenstad, O.; Bert, J.L. Effect of charge on interstitial distribution of albumin in rat dermis in vitro. J. Physiol. 2003, 550, 505–514. [Google Scholar] [CrossRef]
- Jachimska, B.; Pajor, A. Physico-chemical characterization of bovine serum albumin in solution and as deposited on surfaces. Bioelectrochemistry 2012, 87, 138–146. [Google Scholar] [CrossRef]
- Huang, P.; Loew, G.H. Interaction of an Amphiphilic Peptide with a Phospholipid Bilayer Surface by Molecular Dynamics Simulation Study. J. Biomol. Struct. Dyn. 1995, 12, 937–956. [Google Scholar] [CrossRef] [PubMed]
- Balali-Mood, K.; Bond, P.J.; Sansom, M.S.P. Interaction of monotopic membrane enzymes with a lipid bilayer: A coarse-grained MD simulation study. Biochemistry 2009, 48, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Mansourian, M.; Mahnam, K.; Madadkar-Sobhani, A.; Fassihi, A.; Saghaie, L. Insights into the human A1 adenosine receptor from molecular dynamics simulation: structural study in the presence of lipid membrane. Med. Chem. Res. 2015, 24, 3645–3659. [Google Scholar] [CrossRef]
- Gedeon, P.C.; Indarte, M.; Surratt, C.K.; Madura, J.D. Molecular dynamics of leucine and dopamine transporter proteins in a model cell membrane lipid bilayer. Proteins Struct. Funct. Bioinforma. 2010, 78, 797–811. [Google Scholar] [CrossRef]
- Yi, X.; Zhang, Y.; Gong, M.; Yu, X.; Darabedian, N.; Zheng, J.; Zhou, F. Ca 2+ Interacts with Glu-22 of Aβ(1–42) and Phospholipid Bilayers to Accelerate the Aβ(1–42) Aggregation Below the Critical Micelle Concentration. Biochemistry 2015, 54, 6323–6332. [Google Scholar] [CrossRef]
- Sahu, S.K.; Aradhyam, G.K.; Gummadi, S.N. Calcium binding studies of peptides of human phospholipid scramblases 1 to 4 suggest that scramblases are new class of calcium binding proteins in the cell. Biochim. Biophys. Acta - Gen. Subj. 2009, 1790, 1274–1281. [Google Scholar] [CrossRef]
- Filoteo, A.G.; Enyedi, A.; Penniston, J.T. The lipid-binding peptide from the plasma membrane Ca2+ pump binds calmodulin, and the primary calmodulin-binding domain interacts with lipid. J. Biol. Chem. 1992, 267, 11800–11805. [Google Scholar] [CrossRef] [PubMed]
- Mohan, M.S.; Bates, R.G.; Hiller, J.M.; Brand, M.J. Measurement of sodium in albumin solutions with ion-selective electrodes. Clin. Chem. 1978, 24, 580–584. [Google Scholar] [CrossRef] [PubMed]
- van Os, G.A.J.; Koopman-van Eupen, J.H.M. The interaction of sodium, potassium, calcium, and magnesium with human serum albumin, studied by means of conductivity measurements. Recl. des Trav. Chim. des Pays-Bas 1957, 76, 390–400. [Google Scholar] [CrossRef]
- Liu, C.; Min, F.; Liu, L.; Chen, J.; Ren, B.; Lv, K.; Tan, Y. Experimental study on the effect of ions on the surface hydration characteristics of fine quartz. Physicochem. Probl. Miner. Process. 2022, 58. [Google Scholar] [CrossRef]
- Hills, B.A. The Biology of Surfactant; Cambridge University Press: Cambridge, New York, New Rochelle, Melbourne, Sidney, 1988. [Google Scholar]
- Cho, D.; Narsimhan, G.; Franses, E.I. Interactions of Spread Lecithin Monolayers with Bovine Serum Albumin in Aqueous Solution. Langmuir 1997, 13, 4710–4715. [Google Scholar] [CrossRef]
- Phang, T.-L.; Franses, E.I. Expulsion of bovine serum albumin from the air/water interface by a sparingly soluble lecithin lipid. J. Colloid Interface Sci. 2004, 275, 477–487. [Google Scholar] [CrossRef]
- Conti Nibali, V.; Branca, C.; Wanderlingh, U.; D’Angelo, G. Intermolecular Hydrogen-Bond Interactions in DPPE and DMPC Phospholipid Membranes Revealed by Far-Infrared Spectroscopy. Appl. Sci. 2021, 11, 10038. [Google Scholar] [CrossRef]
- Leekumjorn, S.; Sum, A.K. Molecular investigation of the interactions of trehalose with lipid bilayers of DPPC, DPPE and their mixture. Mol. Simul. 2006, 32, 219–230. [Google Scholar] [CrossRef]
- Pantusa, M.; Sportelli, L.; Bartucci, R. Spectroscopic and calorimetric studies on the interaction of human serum albumin with DPPC/PEG:2000-DPPE membranes. Eur. Biophys. J. 2008, 37, 961–973. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, Z.; Wang, J.; Zong, W.; Liu, R. The interaction mechanism between anionic or cationic surfactant with HSA by using spectroscopy, calorimetry and molecular docking methods. J. Mol. Liq. 2016, 224, 1008–1015. [Google Scholar] [CrossRef]
- Adamczyk, O.; Szota, M.; Rakowski, K.; Prochownik, M.; Doveiko, D.; Chen, Y.; Jachimska, B. Bovine Serum Albumin as a Platform for Designing Biologically Active Nanocarriers—Experimental and Computational Studies. Int. J. Mol. Sci. 2023, 25, 37. [Google Scholar] [CrossRef] [PubMed]
- Michnik, A. Thermal stability of bovine serum albumin DSC study. J. Therm. Anal. Calorim. 2003, 71, 509–519. [Google Scholar] [CrossRef]
- Hamilton, J.A.; Era, S.; Bhamidipati, S.P.; Reed, R.G. Locations of the three primary binding sites for long-chain fatty acids on bovine serum albumin. Proc. Natl. Acad. Sci. U. S. A. 1991, 88, 2051–2054. [Google Scholar] [CrossRef] [PubMed]
- Yarimitsu, S.; Nakashima, K.; Sawae, Y.; Murakami, T. Influence of phospholipid and protein constituents on tribological properties of artificial hydrogel cartilage material. J. Biomech. Sci. Eng. 2013, 8, 257–267. [Google Scholar] [CrossRef]
- Krishna, D.V.; Sankar, M.R. Bioinspired artificial synovial fluid for in vitro frictional behavior of bovine articular cartilage and auxiliary biomaterials. J. Mol. Liq. 2023, 388, 122836. [Google Scholar] [CrossRef]
- Janicka, K.; Beldowski, P.; Majewski, T.; Urbaniak, W.; Petelska, A.D. The Amphoteric and Hydrophilic Properties of Cartilage Surface in Mammalian Joints: Interfacial Tension and Molecular Dynamics Simulation Studies. Molecules 2019, 24, 2248. [Google Scholar] [CrossRef]
- Chatterjee, A.; Dubey, D.K.; Sinha, S.K. Nanoscale friction and adhesion mechanisms in articular cartilage top layer hydrated interfaces: Insights from atomistic simulations. Appl. Surf. Sci. 2021, 550, 149216. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




