1. Introduction
A total of 43,170 US women died from breast cancer (BC) in 2023, most from stage IV disease1-5. Despite some progress in prolonging survival, their overall median survival remains 2-3 years3,4. Treating patients with stage IV BC can prolong population averaged survival by only a few months6-8. Systemic treatment options vary in the wide range of scenarios generated by patient- and cancer-related variables, and include many possible options from combinations of chemo-, hormone-, bio-, or immune-therapy categories4, 9,10.
However, despite the grim overall numbers, there are some long-term survivors11,12. There are several variables linked to survival times. Key among these is the presentation of the disease. Patients with de novo presentation of stage IV BC have a median survival superior to that of patient with recurrent disease13-19. In one study, the median survival of patients with de novo metastatic BC was 41 months while the median survival of patients with recurrent BC was 25 months17. In patients with recurrent disease, shorter metastasis-free intervals of <24 months are associated with worse survival than longer metastasis-free intervals14,16,18,19. Neoadjuvant treatment for the original local disease had a significant positive association with survival20.
In patients with de novo diagnosis of stage IV disease, risk stratification models identified variables at the time of diagnosis positively and negativley associated with overall survival. These variables included age at diagnosis, particularly age over 80 years, premenopausal status, Karnofsky performance status, estrogen receptor (ER)-, progesterone receptor (PR)- and human epidermal growth factor receptor 2 (Her2)-status, tumor histology, grade, tumor size categories 0-3 vs. 4, lymph node status, number of metastatic organ systems from 1-4, number of metastases, number of metastatic sites, bone only metastases +/-, visceral metastases +/-, brain only metastases +/-, frontline therapy with Her2 antibody or hormone blockade, and combinations of these variables14,17,21-28.
In addition to these time-fixed variables, time-varying variables occurring after initial diagnosis of de-novo stage IV disease are predictive of survival. These include resection of the primary tumor,29-32, of locoregional metastases33, or of lung only metastases34. The impact of primary tumor resection on survival is affected by age, race, cohabitation status, income, tumor size, grade and histology, ER, PR or Her2 status, expression of Ki67 and CA15, alkaline phosphatase, lymphovascular invasion, lymph node status, metastasis to brain, liver, and lung, and chemotherapy27.29,30.
With respect to systemic therapy, although population- averaged survival extension is modest, there are multiple treatment- and circumstance-associated combinations that can result in prolonged survival in specific cases. Systemic treatment options consist of individual or combinations of categories of chemo-, hormone-, bio-, or immune-therapy4,9,10. Variables that impact survival include treatment timing, dosing intensity and density, duration, sequential or intermittent drug administration, alternating modalities, first or subsequent lines of treatment11,12, adverse events (AEs)9, particularly in association with other negative patient risk factors35-42. For exampler, long-term survival of Her2+ patients with Her2-targeted therapy, taxane-based, hormone maintenance therapy, nab-paclitaxel therapy, or multimodality therapy, with or without local treatment was dependent on hormone receptor expression, disease burden, soft tissue or bone vs visceral metastases, resection of the primary tumor or metastases and young age43-49, and on freedom from disease progression after a year of therapy50.
Overall, the highest responses in stage IV BC are observed with the first line of therapy, followed by rapidly diminishing returns11,12,42. Disease progression after the first few treatment regimens results in exceptionally heterogeneous disease, with cancer cells and tumor stroma selected for treatment resistance and an enhanced capacity to re-metastasize51. Decisions to treat to prolong survival also include considerations of treatment-induced AEs52-61. Therefore, it would be highly beneficial to patients and their caregivers to know the patient- and cancer-spcecific circumstances where a patient could expect long term survival benefits from specific treatmets. Population-based predictive normograms achieved concordance indices (C-index) below 0.700 using significant univariate risk factors17,62, which rose to 0.737 using Cox-regression modeling of five prognostic categories63. However, predictive models for individual patient survival are needed to navigate the exceptionally large number of potential circumstance specific scenarios.
Given the rich and variable population-based predictive data with significant variation and the general limited survival with stage IV BC, we have undertaken a Deep Learning (a subfield of artificial intelligence) approach to generate predictive modeling of individual patient survival from the SEER-Medicare data of time-fixed and time-varying covariates. Using this unique approach, we can identify patient and treatment scenarios that will result in long term survival. The models will help caregivers identify specific treatments from a wide variety of choices that will optimize survival in individual patients and unique circumstances.
2. Methods
2.1. Study Data: SEER-Medicare Linked Dataset
Unlike the SEER dataset, which does not provide data on cancer therapy, the SEER-Medicare (S-M) linked dataset created almost 30 years ago is considered a major source to assess cancer care and outcomes in the US64. The S-M linked dataset provides information about Medicare beneficiaries with cancer. This dataset includes cancer incidence data for about 26% of the US population in various regions. Medicare data have SEER cancer file, a person-level file that provides SEER demographic and clinical information for up to 10 primary cancer diagnoses, treatments, and mortality. Medicare files capture the fee-for-service claims from hospitals (MEDPAR), outpatient facilities (Outpatient), National Claims History (NCH), hospice care, home health agencies, and the Master Beneficiary Summary File (MBSF), which contains Medicare enrollment information. The CCflag file includes, for every patient, the date the patient was diagnosed with one of 22 chronic conditions. We use these data to compute a time-varying patient's comorbidity index. There were 883,053 BC patients during 1991-2016. The number of cancer patients and their corresponding records in each file were outlined previously65. The patient numbers in each file reflect the fact that some patients had recorded encounters in multiple files. Based on the Observational Medical Outcomes Partnership (OMOP) common data model, we fused the data in the various files. This enabled us to transform data contained within those disparate files into a common format (data model) as well as a common representation (terminologies, vocabularies, coding schemes).
2.2. Inclusion Criteria
In our study, we included women with the diagnosis of stage IV breast cancer, who have not had any other malignancy history except non-melanoma skin and eyelid cancer, as a standard in NCI clinical trials
66, included all comorbidities recorded at every visit from the date of diagnosis, averaged over the course of all the treatments assessed for each patient, all prior treatments, delineated age, race, marital status, breast cancer stage, grade, hormonal and Her2 status, and laterality. We included only patients who were enrolled in both Parts A and B of Medicare with no HMO enrollment 1 month prior to diagnosis with breast cancer and whose age at enrollment was 65 or older who qualified by age and not disability. The enrolled population was, therefore, skewed by age and only represented an elderly subset of the breast cancer patient population. We included patients who were enrolled in both Parts A and B of Medicare with no HMO enrollment from 1 month prior to diagnosis through 20 years following diagnosis, hospice or death to ensure that subjects were continuously enrolled in the proper parts of Medicare during the study period, as before
67. The number of entries, patients, mean age and comorbidity index are outlined in
Table 1.
2.3. Data Cleaning and Standardization
Data cleaning was implemented to remove duplicate records in the dataset. We included patients who have valid patient id, cancer type (breast, or breast plus skin, or eyelid), stage (I-III, IV), sex, date of diagnosis (month/year; in case of a missing day of the month, we assumed the first day of the month). A patient visit (inpatient, outpatient, and carrier claims) was included if it had a valid patient id and a valid date. We included only valid diagnoses, procedures, or HCPCS codes at a given visit. We applied the Melt transformation to convert a visit record (containing a mix of valid and missing codes) from wide format to stacked/long form, thus being able to delete invalid codes while keeping only valid ones for each visit. We performed data transformation and standardization, ensuring all features were numeric and standardized, and applied bucketing for continuous values, e.g., age and comorbidity index.
2.4. Treatments and Adverse Events: Encoding and Embeddings
We included all patients' ICD-9 and HCPCS codes. We annotated 141 HCPCS drug J codes: 82 chemotherapy drugs, 49 biotherapy drugs, and ten hormone therapy drugs, and consolidated related adverse events into 18 categories to enable a more comprehensive analysis of the variables associated with their occurrence, as before 67.
2.5. Predictive Modeling
2.5.1. Discrete Time-to-Event Data
In our dataset, as is in real life, patient follow-up visits occur on a given day with irregular gaps between two consecutive visits. A patient has two sets of covariates: time-fixed covariates (e.g., age at diagnosis) and time-varying covariates (e.g., current age, current comorbidity index, treatments administered at this visit and earlier visits, and adverse events). A patient's survival status is recorded at each visit while the patient is at risk, i.e., has not yet experienced the event (death in our case) of interest. We have 14,312 breast cancer stage IV patients and 1,880,153 entries over the period 1991-2015. Since we selected patients with ICD-9 diagnosis codes for this study, the study period was set to 1991-2015. Thus, our study is right-censored, i.e., some patients are not followed all the way to their event (death) time, resulting in censored time (2015-12-31) instead of event time. Thus, instead of observing the event (death in our case) at time, , we observe a possibly right-censored event time, is the censoring time (2015-12-31 in our case). We also observe the indicator labeling the observed event time as an event or a censored observation.
The general idea of prediction using this discrete-time framework is to build models predicting the likelihood of surviving each visit. In our case, the event of interest is death. Let be the time to death since diagnosis, and is the censoring time, and is a covariate vector. We are predicting the probability of a patient experiencing failure (death) by time . This probability, referred to as the cumulative incidence function, is given by An alternative to this probability is the survival function, and the hazard rate, The Cox proportional hazards (CPH)68 is the most used Model in survival analysis. It provides a semi-parametric hazard rate, that is the product of a baseline hazard, and a relative risk function, , i.e., given by: , where is a covariate vector, and is a parameter vector. The Model's proportionality hazard assumption, i.e., the effect of each patient covariate is the same at all values of the follow-up time, is unrealistic for most clinical situations69.
We applied the existing DL-based models DeepSurv70 to extend the Cox regression models with neural networks estimators71,72, and DeepHit71-74, Nnet-survival69, and Cox-Time74 to extend the Cox regression model with neural networks and overcome the proportionality assumption of the Cox model, as before75.
2.5.2. Time-Varying Covariates
The above methods, while dealing with discerete-time data they assume the covariates of a patient, , are time-fixed. In our case, a realistic prediction of the patient's survival needs to take into account not only the patient's marital status, race, hispanic, age, laterality, grade, hormonal and Her2 status, and laterality at the time of diagnosis, but also, the administered treatments, induced adverse events, and comorbidity index, and the age at each visit. Thus, being able to predict associations between specific treatments in specific scenarios and outcomes relating to quality of life, progression-free survival, and survival in the spectrum of stage IV cancer settings.
To achieve this objective, we propose to extend a patient covariate vector to include not only time-fixed covariates but also covariates that summarize the patient's history from previous visits. Specifically, for a given treatment, is a tally of the number of times was administered to this patient, , from the time of the diagnosis to the time of death/end of the study. Similarly, for a given adverse event inuced, is a tally of the number of times patient, , experienced , from the time of the diagnosis to the time of death/end of the study. We divide the age of a patient into 6 bins: <=65, 65< <=70, 70< <=75, 75< <=80, 80< <=85, >85. We have then, is a tally of the number of times the age of patient, , falls within the , from the time of the diagnosis to the time of death/end of the study. We handle a patient comorbidity index similar to a patient age. We divide the comorbidity index of a patient into 6 bins: <=2, 2< <=4, 4< <=6, 6< <=8, 8< <=10, >10. We have then, is a tally of the number of times the comorbidity index of patient, , falls within the 6 comorbidity bins: from the time of the diagnosis to the time of death/end of the study.
2.6. Experiments
2.6.1. Data Prepration
Our data consisted of patient-level records with a single row for each patient and a column for each patient covariate as described above. A patient had a column indicating the corresponding value of and a column indicating the patient event status at that time. The time horizon (1991-01-01 through 2015-012-31) was divided into months.
Our Model provides an estimate of a patient's complete survival curve, S(t); these estimated survival curves represent event probabilities as time functions.
2.6.2. Performance Metrics
In general, metrics for measuring the Model's predictive performance are often in terms of discrimination, i.e., the Model's ability to distinguish between patients with high and low risk of experiencing the event, and calibration, i.e., the agreement between the estimated and actual incidence of the event76.
At time 0, patients are event-free, and their survival status changes from one time period to another. Thus, performance metrics for assessing the predictive accuracy of time-to-event outcomes are time-dependent. In addition to accounting for time, performance metrics must also account for censoring. Two such metrics that are commonly used to measure the predictive accuracy of survival prediction models are: 1) time-dependent concordance index77, and time-dependent Brier score Brier score78-80. The concordance index, or C-index, referred to as the AUC (area under the receiver operating characteristics curve), is the most commonly used method to measure the Model's predictive accuracy. It estimates the probability that, for a randomly selected two patients, the predicted survival times of the two patients have the same ordering as their true survival times81. The C-index is a scale-free measure with 1 representing perfect discrimination and 0.5 representing discriminative ability similar to chance. Brier score, the predictive error, assesses the Model's calibration and discrimination ability, with lower values indicating better prediction. Similar to74, we use the time-dependent C-index by Antolini et al.77 and will use the integrated Brier score (IBS) by Graf et al.79 distribution.
2.6.3. Hyperparameter Tuning
Hyperparameters are neural network parameters fixed by design and which accounts for censored patients by weighting the score by the inverse of the estimated censoring not tuned by training but should be optimized. Hyperparameter optimization is an important part of deep learning, in general. We applied Amazon SageMaker Python SDK (software development kit), an open-source library, to fine-tune the model by identifying optimal values of the network's hyperparameters, as before
75.
Table 2 includes the list of the hyperparameters and their ranges. The experiments will be conducted using five-fold cross-validation using a Bayesian optimization search scheme
82. The Bayesian optimization scheme has been shown to outperform other state of the art global optimization algorithms on a number of challenging optimization benchmark functions
83.
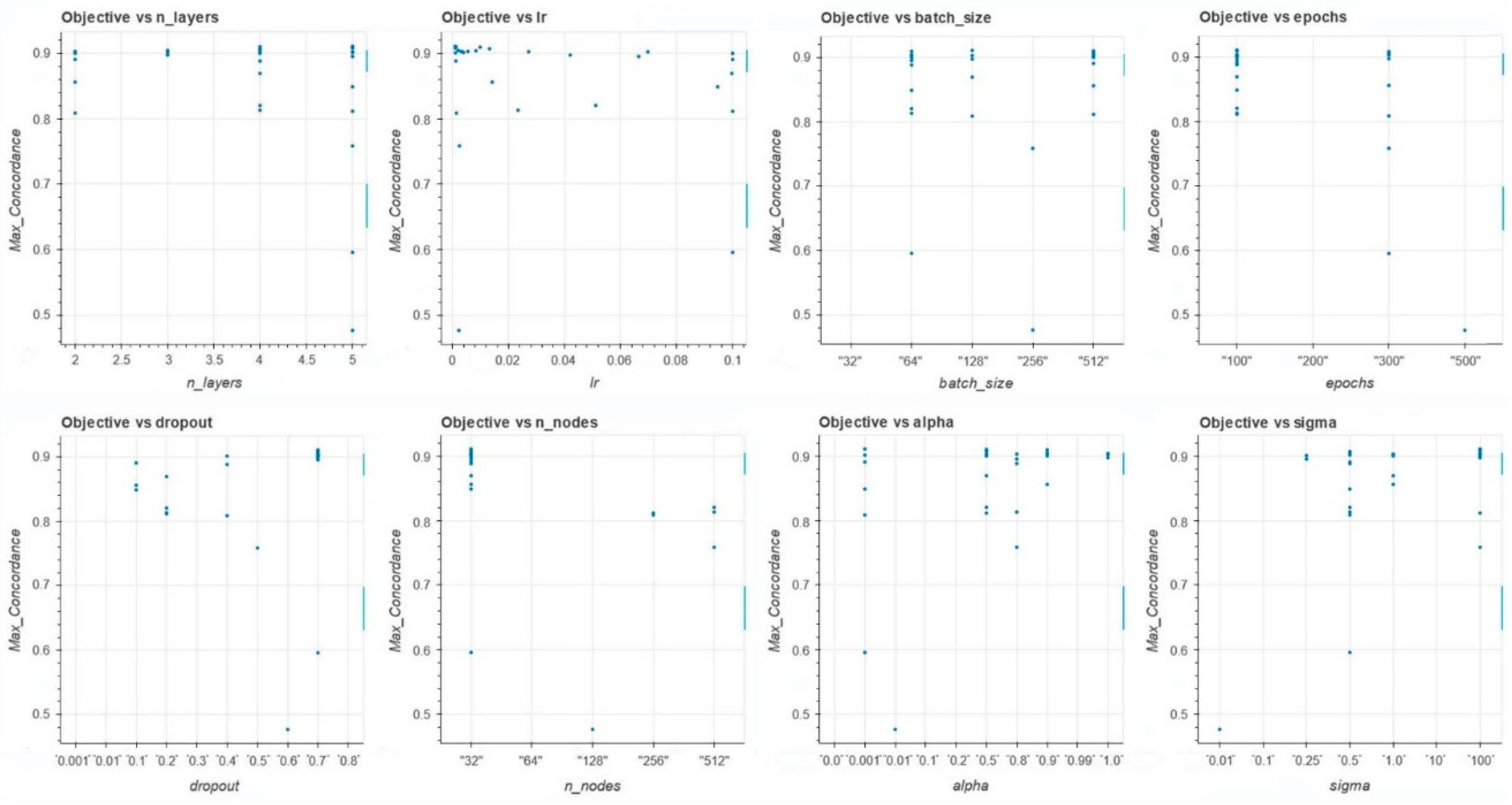
Figure 1 depicts the performance metric (time-dependent concordance) and each of the model's hyperparameters: n-layers, Ir, batch size, epochs, dropout, n_nodes, alpha, sigma.
The base model (vs. the extended Model)
We used the pycox package74. To validate our implementation, we applied the four models: DeepSurv, DeepHit, Nnet-survival, and Cox-Time to the real datasets METABRIC84 and SUPPORT85, which is publicly available on the Vanderbilt Biostatistics Web site. The experiments were conducted by five-fold cross-validation. As we demonstrated before75, the published results of each of these models using the Metabric and SUPPORT datasets are within our results' 95% confidence level, respectively (we used only the time-dependent C-index as the performance metric)86.
3. Results
3.1. Improved Predictive Capacity When Adding Time-Variable Covariates to Time Fixed Covariates
The results are presented in
Table 3. In terms of concordance, we observe the significant improvement of the model with the proposed extended patients' covariates compared with that of the patients' time-fixed covariates. For example, the DeepSurv model's prediction error is 4% when using the proposed extended patients' covariates versus over 32% when using the patients' time-fixed covariates.
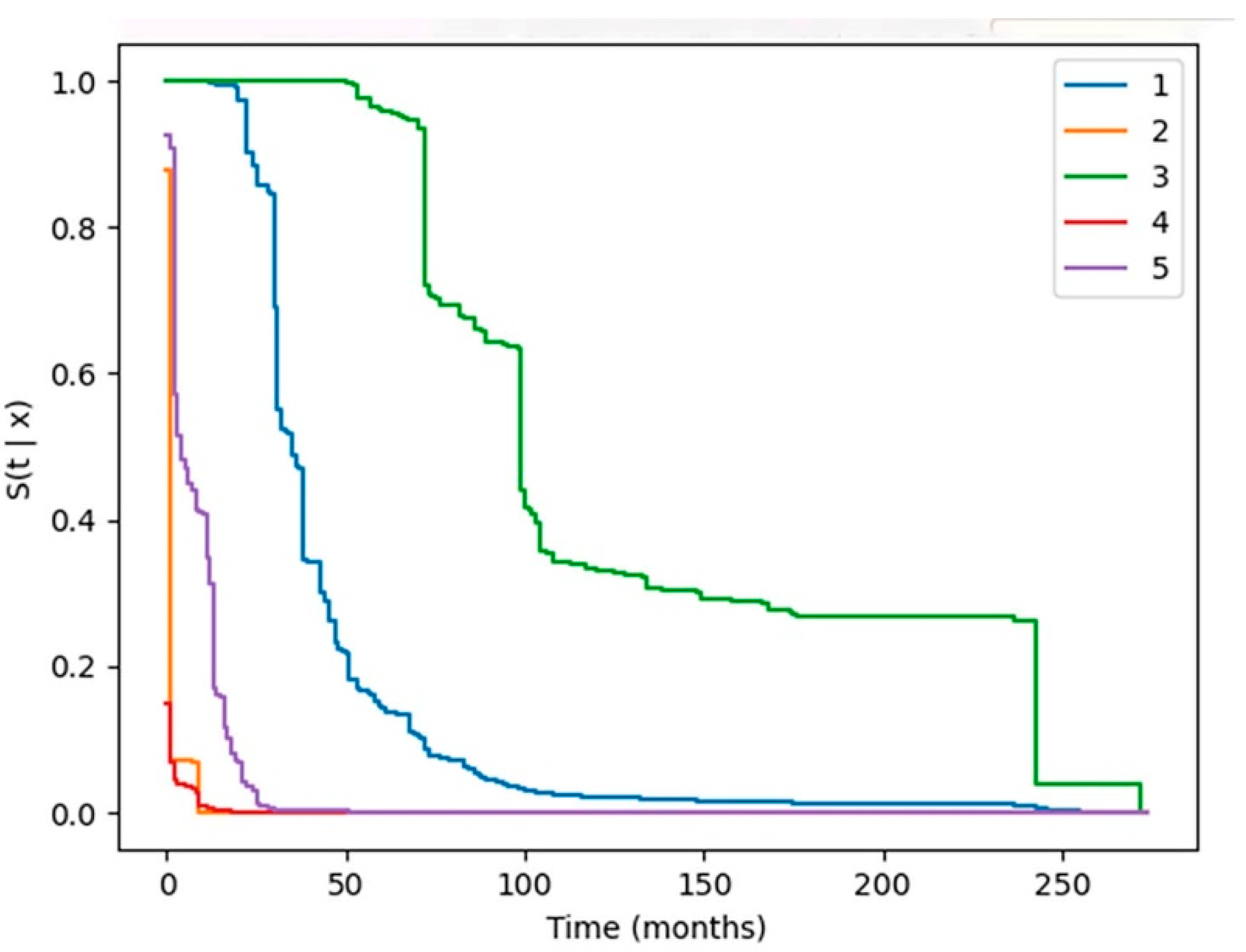
3.2. Predicited Survival Varies by Patient Covariates
The median survival of the five hypothetical patients ranges from less than one month to 100 months. The predictive models utilized the unique circumstances created by the interaction of the individual patient, cancer and treatment variables to generate the individual patient’s predicted survival curves. We cannot provide patient details because our DUA with the NCI SEER-Medicare does not permit including specific patient identifying information.
4. Discussion
Our results represent two significant achievements in predictive modeling of patient outcomes. The first achievement presents a unique and compelling opportunity to improve the prediction performance of the four deep learning models DeepSurv, DeepHit, Nnet-survival, and CoxTime, which outperform the Cox Proportional Hazard Model in survival analysis. Our models handle discrete-time distribution by extending the patients' covariates vectors to include both time-fixed covariates and covariates that summarize the patient's history from previous visits. This lowers the prediction error rate from 28-38% to 2-12% using the four deep learning models. The IBS can be viewed as the mean square error of prediction; lower values of the IBS indicate better predictive performance. The cause-specific time-dependent Concodrance-index accounts explicitly for censoring, and estimates the model's prediction error.
The second achievement is the application of the models to generate individual patients’ predictive survival curves, based on their unique patient, cancer and treatment features. This will permit predictive modeling in the myriad scenarios encountered with individual patients on likelihood of survival, AEs and impact of progressive therapy. The event pattern-based representation of associations relationships will enable us to perform reasoning and what-if analysis using temporal logic-based approaches; e.g., treating a patient who has progressed after multiple lines of systemic therapy and selecting the optimum treatment type most likely to prolong survival in the specific scenario generated from the conglomeration of the unique patient, cancer and treatment history variables. Outcomes from these models include the first demonstration that the occurrence of certain AE categories have a negative impact on survival65[Ada24]. In that light, the models will inform patients and treating physicians of the most efficacious next line of treatment to prolong survival, or of the likelihood of experiencing primarily adverse events with a minimal likelihood of prolonging survival. The concordance index and the Integrated Brier scores validates the accuracy of individual patient predicted survival curves. Prospective studies integrating clinical and genomic data may identify unique clinicogenomic features of MBC patients who can achieve durable disease control without prolonged chemotherapy47[Shi23].
There are several potential limitations to this study, however. Our patient population consisted of Medicare-enrolled patients and is restricted to patients over 65 years who qualified by age and not other medical conditions. Their age range, therefore, is not representative of the general population and their overall expected survival may be less than patients with stage IV BC of a younger age. We have consolidated AEs and treatment codes into general related categories to enable analysis. Thus, the impact of treatments and adverse events we have analyzed may vary among individual drugs or AEs collated into single categories. The AEs we considered as time-varying events are not graded in the SEER-Medicare data as minor or moderate vs. severe. Our future investigations based on data-driven hypotheses generated from this work will include a classification of non-severe and severe AEs, with the latter assigned to AEs associated with emergency department or hospital admissions or death.
5. Conclusions
These investigations demonstrate a significant improvement of the deep learning models DeepSurv, DeepHit, Nnet-survival, and CoxTime. These models outperform the Cox Proportional Hazard Model in survival analysis with the proposed extended patients' covariates compared with that of the patients' time-fixed covariates, reducing the error rate of concordance indices from 28-38% to 2-12%. The project also developed individual patients’ predictive survival curves, based on their unique patient, cancer and treatment and adverse events variables. The models provide a unique tool to medical care givers to generate realistic survival probabilities for individual patients with stage IV BC given their unique, specific circumstances. It will be a highly useful adjunct to the clinical decision-making in individual patients guiding potential next line of therapy.
Author Contributions
Conceptualization, N.A. and R.W.; Methodology, N.A. and R.W.; Software, 393 N.A. and R.W.; Validation, N.A. and R.W.; Formal analysis, N.A. and R.W.; Investigation, N.A. and 394 R.W.; Resources, N.A. and R.W.; Data curation, N.A. and R.W.; Writing—original draft, N.A. and 395 R.W.; Writing—review & editing, N.A. and R.W.; Visualization, N.A. and R.W.; Project administra- 396 tion, N.A. and R.W.
Funding
This research was funded by 1. Northeast Big Data Innovation Hub, USA, GG014586-02 (R.W. and N.A.); 2. 2020 Busch Biomedical Grant Program, USA (R.W. and N.A.); 3. AmazonWeb Services Health Equity Initiative (“HEI”) Program, USA, CC ADV 00011104 2023 TR. (R.W. and N.A.). This study used the linked SEER-Medicare database. The interpretation and reporting of these data are the sole responsibility of the authors. The authors acknowledge the efforts of the National Cancer Institute; Information Management Services (IMS), Inc.; and the Surveillance, Epidemiology, and End Results (SEER) Program tumor registries in the creation of the SEER-Medicare database and wish to thank them for their advice and review of the datasets designating the different treatment venues. The collection of cancer incidence data from the California Cancer Registry used in this study was supported by the California Department of Public Health pursuant to California Health and Safety Code Section 103885; Centers for Disease Control and Prevention’s (CDC) National Program of Cancer Registries, under cooperative agreement 1NU58DP007156; the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201800032I awarded to the University of California, San Francisco, contract HHSN261201800015I awarded to the University of Southern California, and contract HHSN261201800009I awarded to the Public Health Institute. The ideas and opinions expressed herein are those of the author(s) and do not necessarily reflect the opinions of the State of California, Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Rutgers Institutional Review Board under Exempt Review study number Pro20140000175.
Informed Consent Statement
Not applicable.
Data Availability Statement
Original data were obtained from SEER-Medicare under a two-tiered review process. SEER-Medicare data are available to investigators upon review.
Conflicts of Interest
The authors declare no conflict of interest.
References
- American Cancer Society Facts and Figures 2023. 2023. https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2021.html Downloaded 6-6-22.
- Gong, Y.; Liu, Y.-R.; Ji, P.; Hu, X.; Shao, Z.-M. Impact of molecular subtypes on metastatic breast cancer patients: a SEER population-based study. Sci. Rep. 2017, 7, 45411. [Google Scholar] [CrossRef] [PubMed]
- Wieder, R.; Shafiq, B.; Adam, N. Greater Survival Improvement In African American vs.Caucasian Women With Hormone Negative Breast Cancer. Journal of Cancer. 2020, 11, 2808–2820. [Google Scholar] [CrossRef] [PubMed]
- Cardoso F, Paluch-Shimon S, Senkus E, Curigliano G, Aapro MS, Andre F, Barrios CH, Bergh J, Bhattacharyya GS, Biganzoli L, et al. 5th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 5). Annals of Oncology. 2020, 31, 1623–1649. [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA: a Cancer Journal for Clinicians. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Xu, B.; Li, W.; Zhang, Q.; Shao, Z.; Li, Q.; Wang, X.; Li, H.; Sun, T.; Yin, Y.; Zheng, H.; et al. Pertuzumab, trastuzumab, and docetaxel for Chinese patients with previously untreated her2-positive locally recurrent or metastatic breast cancer (puffin): a phase iii, randomized, double-blind, placebo-controlled study. Breast Cancer Research & Treatment. 2020, 182, 689–697. [Google Scholar] [CrossRef]
- Swain, S.M.; Miles, D.; Kim, S.B.; Im, Y.H.; Im, S.A.; Semiglazov, V.; Ciruelos, E.; Schneeweiss, A.; Loi, S.; Monturus, E.; et al. CLEOPATRA study group Pertuzumab, trastuzumab, and docetaxel for her2-positive metastatic breast cancer (cleopatra): end-of-study results from a double-blind, randomised, placebo-controlled, phase 3 study. Lancet Oncology. 2020, 21, 519–530. [Google Scholar] [CrossRef]
- Schmid, P.; Rugo, H.S.; Adams, S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Dieras, V.; Henschel, V.; Molinero, L.; Chui, S.Y.; et al. IMpassion130 Investigators Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (impassion130): updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncology. 2020, 21, 44–59. [Google Scholar]
- Giuliano M, Schettini F, Rognoni C, Milani M, Jerusalem G, Bachelot T, De Laurentiis M, Thomas G, De Placido P, Arpino G, De Placido S, Cristofanilli M, Giordano A, Puglisi F, Pistilli B, Prat A, Del Mastro L, Venturini S, Generali D. (2019) Endocrine treatment versus chemotherapy in postmenopausal women with hormone receptor-positive, HER2-negative, metastatic breast cancer: a systematic review and network meta-analysis. Lancet Oncology. 20:1360-1369.
- Sambi, M.; Qorri, B.; Harless, W.; Szewczuk, M.R. Therapeutic Options for Metastatic Breast Cancer. Advances in Experimental Medicine & Biology. 2019, 1152, 131–172. [Google Scholar]
- Banerji, U.; Kuciejewska, A.; Ashley, S.; Walsh, G.; O'Brien, M.; Johnston, S.; Smith, I. Factors determining outcome after third line chemotherapy for metastatic breast cancer. Breast. 2007, 16, 359–366. [Google Scholar] [CrossRef]
- Tacca, O.; LeHeurteur, M.; Durando, X.; Mouret-Reynier, M.A.; Abrial, C.; Thivat, E.; Bayet-Robert, M.; Penault-Llorca, F.; Chollet, P. Metastatic breast cancer: overall survival related to successive chemotherapies. What do we gain after the third line? Cancer Investigation. 2009, 27, 81–85. [Google Scholar] [CrossRef]
- Yardley, D.A.; Kaufman, P.A.; Brufsky, A.; Yood, M.U.; Rugo, H.; Mayer, M.; Quah, C.; Yoo, B.; Tripathy, D. Treatment patterns and clinical outcomes for patients with de novo versus recurrent HER2-positive metastatic breast cancer. Breast Cancer Res. Treat. 2014, 145, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Lobbezoo, D.J.; van Kampen, R.J.; Voogd, A.C.; Dercksen, M.W.; van den Berkmortel, F.; Smilde, T.J.; van de Wouw, A.J.; Peters, F.P.; van Riel, J.M.; Peters, N.A.; et al. Prognosis of metastatic breast cancer: are there differences between patients with de novo and recurrent metastatic breast cancer? Br. J. Cancer. 2015, 112, 1445–1451. [Google Scholar] [CrossRef] [PubMed]
- Den Brok, W.D.; Speers, C.H.; Gondara, L.; Baxter, E.; Tyldesley, S.K.; Lohrisch, C.A. Survival with metastatic breast cancer based on initial presentation, de novo versus relapsed. Breast Cancer Res. Treat. 2017, 161, 549–556. [Google Scholar] [CrossRef]
- Yamamura, J.; Kamigaki, S.; Fujita, J.; Osato, H.; Komoike, Y. The Difference in Prognostic Outcomes Between De Novo Stage IV and Recurrent Metastatic Patients with Hormone Receptor-positive, HER2-negative Breast Cancer. In Vivo. 2018, 32, 353–358. [Google Scholar] [CrossRef]
- Barcenas, C.H.; Song, J.; Murthy, R.K.; Raghavendra, A.S.; Li, Y.; Hsu, L.; Carlson, R.W.; Tripathy, D.; Hortobagyi, G.N. Prognostic Model for De Novo and Recurrent Metastatic Breast Cancer. JCO Clin Cancer Inform. 2021, 5, 789–804. [Google Scholar] [CrossRef]
- File, D.M.; Pascual, T.; Deal, A.M.; Wheless, A.; Perou, C.M.; Claire Dees, E.; Carey, L.A. Clinical subtype, treatment response, and survival in De Novo and recurrent metastatic breast cancer. Breast Cancer Res. Treat. 2022, 196, 153–162. [Google Scholar] [CrossRef]
- de Maar, J.S.; Luyendijk, M.; Suelmann, B.B.M.; van der Kruijssen, D.E.W.; Elias, S.G.; Siesling, S.; van der Wall, E. Comparison between de novo and metachronous metastatic breast cancer: the presence of a primary tumour is not the only difference-a Dutch population-based study from 2008 to 2018. Breast Cancer Res. Treat. 2023, 198, 253–264. [Google Scholar] [CrossRef]
- Shen, T.; Gao, C.; Zhang, K.; Siegal, G.P.; Wei, S. Prognostic outcomes in advanced breast cancer: the metastasis-free interval is important. Human Pathology. 2017, 70, 70–76. [Google Scholar] [CrossRef]
- Dawood, S.; Broglio, K.; Ensor, J.; Hortobagyi, G.N.; Giordano, S.H. Survival differences among women with de novo stage IV and relapsed breast cancer. Ann. Oncol. 2010, 21, 2169–2174. [Google Scholar] [CrossRef]
- Eng, L.G.; Dawood, S.; Sopik, V.; Haaland, B.; Tan, P.S.; Bhoo-Pathy, N.; Warner, E.; Iqbal, J.; Narod, S.A.; Dent, R. Ten-year survival in women with primary stage IV breast cancer. Breast Cancer Res. Treat. 2016, 160, 145–152. [Google Scholar] [CrossRef]
- Klar, N.; Rosenzweig, M.; Diergaarde, B.; Brufsky, A. Features Associated With Long-Term Survival in Patients With Metastatic Breast Cancer. Clin Breast Cancer. 2019, 19, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Plichta, J.K.; Thomas, S.M.; Sergesketter, A.R.; Greenup, R.A.; Rosenberger, L.H.; Fayanju, O.M.; Kimmick, G.; Force, J.; Hyslop, T.; Hwang, E.S. A Novel Staging System for De Novo Metastatic Breast Cancer Refines Prognostic Estimates. Ann. Surg. 2022, 275, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Plichta, J.K.; Thomas, S.M.; Hayes, D.F.; Chavez-MacGregor, M.; Allison, K.; de Los Santos, J.; Hortobagyi, G.N. Novel prognostic staging system for patients with de novo metastatic breast cancer. Journal of Clinical Oncology. 2023, 41, 2546–2560. [Google Scholar] [CrossRef] [PubMed]
- Taskindoust, M.; Thomas, S.M.; Sammons, S.L.; Fayanju, O.M.; DiLalla, G.; Hwang, E.S.; Plichta, J.K. Survival Outcomes Among Patients with Metastatic Breast Cancer: Review of 47,000 Patients. Ann. Surg. Oncol. 2021, 28, 7441–7449. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, C.; Feng, Y.; Shen, C.; He, T.; Wang, Z.; Ma, L.; Du, Z. The prognostic role of surgery and a nomogram to predict the survival of stage IV breast cancer patients. Gland Surg. 2022, 11, 1224–1239. [Google Scholar] [CrossRef]
- Lv, Z.; Zhang, W.; Zhang, Y.; Zhong, G.; Zhang, X.; Yang, Q.; Li, Y. Metastasis patterns and prognosis of octogenarians with metastatic breast cancer: A large-cohort retrospective study. PLoS One. 2022, 17, e0263104. [Google Scholar] [CrossRef]
- Yoo, T.K.; Chae, B.J.; Kim, S.J.; Lee, J.; Yoon, T.I.; Lee, S.J.; Park, H.Y.; Park, H.K.; Eom, Y.H.; Kim, H.S.; et al. Identifying long-term survivors among metastatic breast cancer patients undergoing primary tumor surgery. Breast Cancer Res. Treat. 2017, 165, 109–118. [Google Scholar] [CrossRef]
- Yang, Y.S.; Chen, Y.L.; Di, G.H.; Jiang, Y.Z.; Shao, Z.M. Prognostic value of primary tumor surgery in de novo stage IV breast cancer patients with different metastatic burdens: a propensity score-matched and population-based study. Transl. Cancer Res. 2019, 8, 614–625. [Google Scholar] [CrossRef]
- Lin, Y.; Huang, K.; Zeng, Q.; Zhang, J.; Song, C. Impact of breast surgery on survival of patients with stage IV breast cancer: a SEER population-based propensity score matching analysis. PeerJ. 2020, 8, e8694. [Google Scholar] [CrossRef]
- Plichta, J.K.; Taskindoust, M.; Greenup, R.A. Surgery in the Setting of Metastatic Breast Cancer. Current Breast Cancer Reports. 2023, 15, 37–47. [Google Scholar] [CrossRef]
- Zhou, W.; Yue, Y.; Xiong, J.; Li, W.; Zeng, X. The role of locoregional surgery in de novo stage IV breast cancer: A meta-analysis of randomized controlled trials. Cancer Treat. Rev. 2024, 129, 102784. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Zhong, G.; Ye, W.; Jin, X. Assessing Surgical Benefits and Creating a Prognostic Model for Breast Cancer with Lung-only Metastasis: An Analysis of the National Cancer Database. Ann. Ital. Chir. 2024, 95, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Kwapisz, D. Oligometastatic breast cancer. Breast Cancer. 2019, 26, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Carrick, S.; Parker, S.; Thornton, C.E.; Ghersi, D.; Simes, J.; Wilcken, N. Single agent versus combination chemotherapy for metastatic breast cancer. Cochrane Database of Systematic Reviews. 2009, CD003372. [Google Scholar]
- Brufsky, A. M Delaying Chemotherapy in the Treatment of Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Advanced Breast Cancer. Clinical Medicine Insights. Oncology. 2015, 9, 137–147. [Google Scholar] [CrossRef]
- Rivera, E. Management of metastatic breast cancer: monotherapy options for patients resistant to anthracyclines and taxanes. American Journal of Clinical Oncology. 2010, 33, 176–185. [Google Scholar] [CrossRef]
- Sutherland, S.; Miles, D.; Makris, A. Use of maintenance endocrine therapy after chemotherapy in metastatic breast cancer. European Journal of Cancer. 2016, 69, 216–222. [Google Scholar] [CrossRef]
- Gennari, A.; D'amico, M.; Corradengo, D. Extending the duration of first-line chemotherapy in metastatic breast cancer: a perspective review. Therapeutic Advances in Medical Oncology. 2011, 3, 229–232. [Google Scholar] [CrossRef]
- Conforti, S.; Turano, S.; Minardi, S.; Locco, C.; Conforti, L.; Palazzo, S. Improvement of quality of life in third-line chemotherapy with lapatinib in a case of metastatic breast cancer. Tumori. 2013, 99, e136–e139. [Google Scholar] [CrossRef]
- Palumbo, R.; Sottotetti, F.; Riccardi, A.; Teragni, C.; Pozzi, E.; Quaquarini, E.; Tagliaferri, B.; Bernardo, A. Which patients with metastatic breast cancer benefit from subsequent lines of treatment? An update for clinicians. Therapeutic Advances in Medical Oncology. 2013, 5, 334–350. [Google Scholar] [CrossRef]
- Altundag, K.; Bondy, M.L.; Mirza, N.Q.; Kau, S.W.; Broglio, K.; Hortobagyi, G.N.; Rivera, E. Clinicopathologic characteristics and prognostic factors in 420 metastatic breast cancer patients with central nervous system metastasis. Cancer. 2007, 110, 2640–2647. [Google Scholar] [CrossRef] [PubMed]
- Yardley, D.A.; Tripathy, D.; Brufsky, A.M.; Rugo, H.S.; Kaufman, P.A.; Mayer, M.; Magidson, J.; Yoo, B.; Quah, C.; Ulcickas Yood, M. Long-term survivor characteristics in HER2-positive metastatic breast cancer from registHER. Br. J. Cancer. 2014, 110, 2756–2764. [Google Scholar] [CrossRef] [PubMed]
- Harano, K.; Lei, X.; Gonzalez-Angulo, A.M.; Murthy, R.K.; Valero, V.; Mittendorf, E.A.; Ueno, N.T.; Hortobagyi, G.N.; Chavez-MacGregor, M. Clinicopathological and surgical factors associated with long-term survival in patients with HER2-positive metastatic breast cancer. Breast Cancer Res. Treat. 2016, 159, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, E.; Saint-Martin, C.; Pierga, J.Y.; Brain, E.; Rouzier, R.; Savignoni, A.; Mouret-Fourme, E.; Dieras, V.; Piot, I.; Dubot, C.; et al. Long-term survival in HER2-positive metastatic breast cancer treated with first-line trastuzumab: results from the french real-life curie database. Breast Cancer Res. Treat. 2019, 178, 505–512. [Google Scholar] [CrossRef]
- Shin, J.; Kim, J.Y.; Oh, J.M.; Lee, J.E.; Kim, S.W.; Nam, S.J.; Park, W.; Park, Y.H.; Ahn, J.S.; Im, Y.H. Comprehensive Clinical Characterization of Decade-Long Survivors of Metastatic Breast Cancer. Cancers. 2023, 15, 4720. [Google Scholar] [CrossRef]
- Liu, B.; Liu, H.; Liu, M. Aggressive local therapy for de novo metastatic breast cancer: Challenges and updates. Oncol. Rep. 2023, 50, 163. [Google Scholar] [CrossRef]
- Kikuchi, M.; Fujii, T.; Honda, C.; Tanabe, K.; Nakazawa, Y.; Ogino, M.; Obayashi, S.; Shirabe, K. Characteristics of Patients With Metastatic Breast Cancer Who Survived more than 10 Years. Anticancer Res. 2023, 43, 217–221. [Google Scholar] [CrossRef]
- Battisti, N.M.L.; Tong, D.; Ring, A.; Smith, I. Long-term outcome with targeted therapy in advanced/metastatic HER2-positive breast cancer: The Royal Marsden experience. Breast Cancer Res. Treat. 2019, 178, 401–408. [Google Scholar] [CrossRef]
- Wieder, R. Fibroblasts as turned agents in cancer progression. Cancers. 2023, 15, 2014. [Google Scholar] [CrossRef]
- Rashid, N.; Koh, H.A.; Baca, H.C.; Li, Z.; Malecha, S.; Abidoye, O.; Masaquel, A. Clinical Impact of Chemotherapy-Related Adverse Events in Patients with Metastatic Breast Cancer in an Integrated Health Care System. Journal of Managed Care & Specialty Pharmacy. 2015, 21, 863–871. [Google Scholar]
- Tsai, H.T.; Isaacs, C.; Fu, A.Z.; Warren, J.L.; Freedman, A.N.; Barac, A.; Huang, C.Y.; Potosky, A.L. Risk of cardiovascular adverse events from trastuzumab (Herceptin(R)) in elderly persons with breast cancer: a population-based study. Breast Cancer Research & Treatment. 2014, 144, 163–170. [Google Scholar] [CrossRef]
- Du, X.L.; Osborne, C.; Goodwin, J.S. Population-based assessment of hospitalizations for toxicity from chemotherapy in older women with breast cancer. J. Clinical Oncology. 2002, 20, 4636–4642. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.; Calvo, V.; Ramirez-Merino, N.; O'Shaughnessy, J.; Brufsky, A.; Robert, N.; Vidal, M.; Munoz, E.; Perez, J.; Dawood, S.; et al. Adverse events risk associated with bevacizumab addition to breast cancer chemotherapy: a meta-analysis. Annals of Oncology. 2012, 23, 1130–1137. [Google Scholar] [CrossRef] [PubMed]
- Cashman, J.; Wright, J.; Ring, A. The treatment of comorbidities in older patients with metastatic cancer. Supportive Care in Cancer. 2010, 18, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Saif, M.W.; Lee, A.M.; Offer, S.M.; McConnell, K.; Relias, V.; Diasio, R.B. A DPYD variant (Y186C) specific to individuals of African descent in a patient with life-threatening 5-FU toxic effects: potential for an individualized medicine approach. Mayo Clinic Proceedings. 2014, 89, 131–136. [Google Scholar] [CrossRef]
- Modi, N.D.; Abuhelwa, A.Y.; Badaoui, S.; Shaw, E.; Shankaran, K.; McKinnon, R.A.; Rowland, A.; Sorich, M.J.; Hopkins, A.M. Prediction of severe neutropenia and diarrhoea in breast cancer patients treated with abemaciclib. Breast. 2021, 58, 57–62. [Google Scholar] [CrossRef]
- Sutton, A.L.; Felix, A.S.; Wahl, S.; Franco, R.L.; Leicht, Z.; Williams, K.P.; Hundley, W.G.; Sheppard, V.B. Racial disparities in treatment-related cardiovascular toxicities amongst women with breast cancer: a scoping review. Journal of Cancer Survivorship. 2022, 01210–02. [Google Scholar] [CrossRef]
- Foglietta, J.; Inno, A.; de Iuliis, F.; Sini, V.; Duranti, S.; Turazza, M.; Tarantini, L.; Gori, S. Cardiotoxicity of Aromatase Inhibitors in Breast Cancer Patients. Clinical Breast Cancer. 2017, 17, 11–17. [Google Scholar] [CrossRef]
- Swain, S.M.; Schneeweiss, A.; Gianni, L.; Gao, J.J.; Stein, A.; Waldron-Lynch, M.; Heeson, S.; Beattie, M.S.; Yoo, B.; Cortes, J.; et al. Incidence and management of diarrhea in patients with HER2-positive breast cancer treated with pertuzumab. Annals of Oncology. 2017, 28, 761–768. [Google Scholar] [CrossRef]
- Chen, M.S.; Liu, P.C.; Yi, J.Z.; Xu, L.; He, T.; Wu, H.; Yang, J.Q.; Lv, Q. Development and validation of nomograms for predicting survival in patients with de novo metastatic triple-negative breast cancer. Sci Rep. 2022, 12, 14659. [Google Scholar] [CrossRef]
- Tang, W.; Shao, M.; Fang, W.; Wang, J.; Fu, D. A Population-Based Research Utilized a Risk Stratification Model to Forecast the Overall Survival of Young Women With Diagnosed Stage IV Breast Cancer. Clin. Breast Cancer. 2023, 23, e523–e533. [Google Scholar] [CrossRef] [PubMed]
- Enewold, L.; Parsons, H.; Zhao, L.; Bott, D.; Rivera, D.R.; Barrett, M.J.; Virnig, B.A.; Warren, J.L. Updated overview of the SEER-Medicare data: enhanced content and applications. JNCI Monographs. 2020, 2020, 3–13. [Google Scholar]
- Adam, N, Wieder, R. Temporal Association Rule Mining: Race-Based Patterns of Treatment-Adverse Events in Breast Cancer Patients Using SEER–Medicare Dataset. Biomedicines 2024a, 12, 1213. [CrossRef] [PubMed]
- Perez, M.; Murphy, C.C.; Pruitt, S.L.; Rashdan, S.; Rahimi, A.; Gerber, D.E. Potential Impact of Revised NCI Eligibility Criteria Guidance: Prior Malignancy Exclusion in Breast Cancer Clinical Trials. J. Natl. Compr. Canc. Netw. 2022, 20, 792–799. [Google Scholar] [CrossRef]
- Wieder, R.; Adam, N. Racial Disparities in Breast Cancer Treatments and Adverse Events in the SEER-Medicare Data. Cancers. 2023, 15, 4333. [Google Scholar] [CrossRef]
- Cox, D.R. Regression models and life-tables. Journal of the Royal Statistical Society: Series B (Methodological). 1972, 34, 187–202. [Google Scholar] [CrossRef]
- Gensheimer, M.F.; Narasimhan, B. A scalable discrete-time survival model for neural networks. PeerJ. 2019, 7, e6257. [Google Scholar] [CrossRef]
- Katzman, J.L.; Shaham, U.; Cloninger, A.; Bates, J.; Jiang, T.T.; Kluger, Y. DeepSurv: hazards deep neural personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med. Res. Methodol. 2018, 18, 24. [Google Scholar] [CrossRef]
- Meir, T.; Gutman, R.; Gorfine, M. PyDTS: A Python Package for Discrete Time Survival Analysis with Competing Risks. arXiv 2022, arXiv:arXiv:2204.05731. [Google Scholar]
- Meir, T.; Gorfine, M. Discrete-time Competing-Risks Regression with or without Penalization. arXiv 2023, arXiv:arXiv:2303.01186. [Google Scholar]
- Lee, C.; Zame, W.; Yoon, J.; Van Der Schaar, M. Deephit: A deep learning approach to survival analysis with competing risks. In Proceedings of the AAAI conference on artificial intelligence 2018. 2018, 32, 2314–2321. [Google Scholar] [CrossRef]
- Kvamme, H.; Borgan, Ø.; Scheel, I. Time-to-event prediction with neural networks and Cox regression. Journal of Machine Learning Research. 2019, 20, 1–30. [Google Scholar]
- Adam, N.; Wieder, R. AI Survival Prediction Modeling: The Importance of Considering Treatments and Changes in Health Status Over Time. Cancers, 2024, 16, 3527. [Google Scholar] [CrossRef] [PubMed]
- Steyerberg, E.W.; Vickers, A.J.; Cook, N.R.; Gerds, T.; Gonen, M.; Obuchowski, N.; Pencina, M.J.; Kattan, M.W. Assessing the performance of prediction models: a framework for some traditional and novel measures. Epidemiol. 2010, 21, 128. [Google Scholar] [CrossRef]
- Antolini, L.; Boracchi, P.; Biganzoli, E. A time-dependent discrimination index for survival data. Statistics in Medicine. 2005, 24, 3927–3945. [Google Scholar] [CrossRef]
- Brier, G.W. Verification of forecasts expressed in terms of probability. Mon. Weather Rev. 1950, 78, 1–3. [Google Scholar] [CrossRef]
- Graf, E.; Schmoor, C.; Sauerbrei, W.; Schumacher, M. Assessment and comparison of prognostic classification schemes for survival data. Stat. Med. 1999, 18, 2529–2545. [Google Scholar] [CrossRef]
- Gerds, T.A.; Schumacher, M. Consistent estimation of the expected brier score in general survival models with right-censored event times. Biom. J. 2006, 48, 1029–1040. [Google Scholar] [CrossRef]
- Ishwaran, H.; Kogalur, U.B.; Blackstone, E.H.; Lauer, M.S. Random survival forests. Annals of Applied Statistics. 2008, 2, 841–860. [Google Scholar] [CrossRef]
- Snoek, J.; Larochelle, H.; Adams, R.P. Practical bayesian optimization of machine learning algorithms. Advances in neural information processing systems. 2012, 25. [Google Scholar]
- Jones, D.R. A taxonomy of global optimization methods based on response surfaces. Journal of Global Optimization. 2001, 21, 345–383. [Google Scholar] [CrossRef]
- Mucaki, E.J.; Baranova, K.; Pham, H.Q.; Rezaeian, I.; Angelov, D.; Ngom, A.; Rueda, L.; Rogan, P.K. Predicting Outcomes of Hormone and Chemotherapy. In the: Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) Study by Biochemically-inspired Machine Learning. F1000Res. 2016, 5, 2124.
- Connors, A.F.; Dawson, N.V.; Desbiens, N.A.; Fulkerson, W.J.; Goldman, L.; Knaus, W.A.; Lynn, J.; Oye, R.K.; Bergner, M.; Damiano, A.; et al. A controlled trial to improve care for seriously iII hospitalized patients: The study to understand prognoses and preferences for outcomes and risks of treatments (SUPPORT). JAMA. 1995, 274, 1591–1598. [Google Scholar] [CrossRef]
- Kvamme, H.; Borgan, Ø. Continuous and discrete-time survival prediction with neural networks. Lifetime data analysis. 2021, 27, 710–736. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).