1. Introduction
During an in vitro fertilization (IVF) cycle, fertilization check is performed 16-20 hours after insemination where zygotes are morphologically assessed for the presence of pronuclei [
1]. Normal fertilization requires proper segregation of the parental genomes for the developing embryo to acquire the correct biparental set of chromosomes [
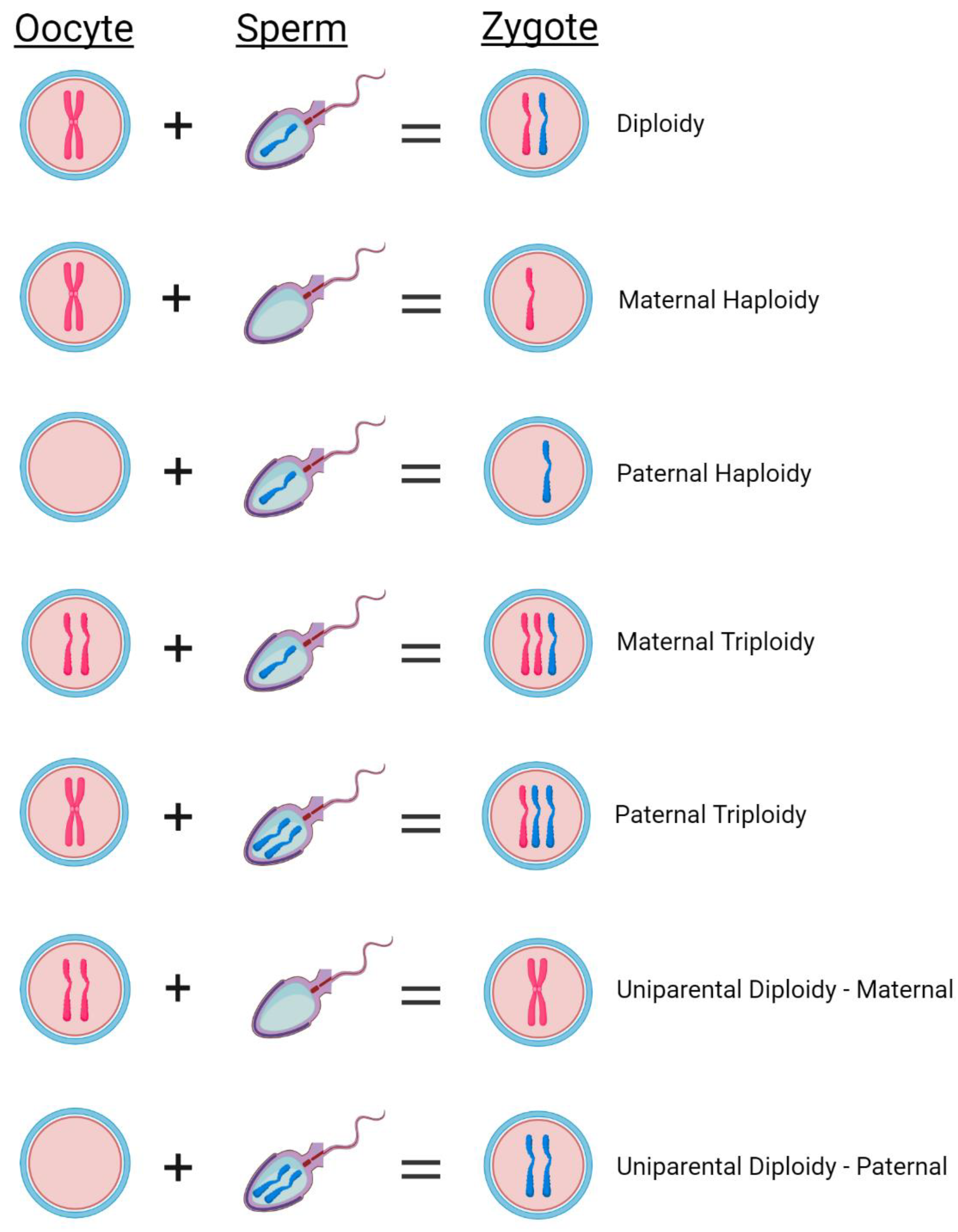
2]. Zygotes with 2 pronuclei (2PN) and two polar bodies are considered normally fertilized with the assumption that each pronucleus originated from each of the two parental gametes (
Figure 1) [
3,
4]. However, ploidy abnormalities are well documented in human conceptions. In fact it is estimated that ~10% of inseminated oocytes fail to fertilize normally and often when an alternate number of pronuclei are observed, these zygotes are discarded due to known chromosomal ploidy abnormalities that lead to implantation failure and/or pregnancy loss [
5,
6].
There are three different PN variations observed in IVF. First, the absence of any visible pronucleus (0PN) which could be the result of failed fertilization or that the pronuclei have disappeared by the time of fertilization check [
7]. Second, the presence of only 1 pronucleus (1PN) which is typically considered to be a haploid embryo [
8]. Haploid zygotes are not compatible with life due to the complete loss of half of the genome and have never been reported in prenatal testing [
2]. The presence of only a single pronucleus can occur due to errors during fertilization including failure of male or female chromatids to form a pronucleus, uncondensed sperm heads, ejection of the spermatozoon, fusion of the maternal and paternal pronucleus, or parthenogenesis (
Figure 1) [
9]. Lastly, 3 pronuclei (3PN), which are considered to be triploid embryos [
5], are the most common abnormality associated with first-trimester pregnancy loss [
10,
11]. There are two types of triploidy: maternally inherited (digynic) or paternally inherited (diandric) (
Figure 1). While paternally inherited triploidy commonly results in a molar pregnancy, maternally inherited triploidy results in intrauterine growth restriction and both types almost always result in pregnancy loss [
12]. The formation of 3PN zygotes during IVF after intracytoplasmic sperm injection (ICSI) is thought to occur as a result of a retention of the second polar body [
13] but 3PNs can also arise via polyspermic fertilization or oocyte-derived meiotic failure [
14]. Although rare, uniparental disomy (UPD) can also occur where both sets of chromosomes are inherited from only one parent (
Figure 1) [
15].
Oocytes with an alternate number of pronuclei can develop into blastocysts that are morphologically indistinguishable from 2PN embryos, though at significantly lower rates [
16]. 2PN zygotes are typically reported to have a blastocyst development rate of around 50% whereas 1PN zygotes are between 26-32% [
4,
17,
18]. 3PN zygotes also have reported lower blastulation rates compared to 2PN zygotes, at ~43% [
9]. Additionally, while 3PN zygotes only have a prevalence of approximately 1-3% in all pregnancies, they are much more predominant in ICSI and IVF and account for 15-18% of cytogenetically abnormal cases among spontaneous abortions [
13].
Studies have shown that some zygotes with an alternate number of pronuclei may actually have a normal chromosomal composition and could result in a euploid live birth [
18,
19,
20]. However, it is also well documented that true abnormally fertilized embryos will result in spontaneous pregnancy loss, including triploid conceptions like partial molar pregnancies so it’s imperative to confirm diploid fertilization prior to an embryo transfer [
6,
21,
22]. Current Next-generation sequencing (NGS) technologies utilized for Preimplantation genetic testing for aneuploidy (PGT-A) includes standardization where DNA is normalized to diploid and is therefore copy number neutral [
8,
15]. This renders the distinction between 23X and 46XX, or 46XX and 69XXX impossible. However, single nucleotide polymorphism (SNP) analysis, in conjunction with NGS-based PGT-A, can be used to measure chromosome ploidy by generating data that can be evaluated using SNP b-allele frequencies for each chromosome [
15,
23,
24,
25,
26]. To maximize the number of euploid embryos in an IVF cycle and minimize pregnancy failures/losses it’s important to have the ability to identify every normally fertilized zygote. This study aims to determine if molecular identification of ploidy status can identify diploid fertilization in zygotes.
2. Materials and Methods
2.1. Zygote Fertilization Check
Fertilization was assessed 16-18 hours post-ICSI where PN numeration was noted and zygotes were sequentially cultured (Sage, CooperSurgical Fertility Solutions) to the blastocyst stage. Upon identification of an inner cell mass (day 5, 6, or 7 of development), a trophectoderm biopsy was performed using laser dissection for PGT-A. Biopsied blastocysts were then vitrified using the Cryotop method as previously described [
27]. Alternate PN zygotes collected between January 2021 to December 2023 were included in this study: n=291 zygotes with no visible pronuclei (0PN), n=217 with 1 pronucleus (1PN) and n=172 with 3 pronuclei (3PN). Alternate PN zygotes were individually tracked to the blastocyst stage prior to performing a trophectoderm biopsy for subsequent PGT-A.
2.2. PGT-A
Biopsied TE cells were lysed and the isolated DNA was randomly fragmented and amplified (SurePlex DNA Amplification System, Vitrolife). NGS libraries were prepared from quantified dsDNA (High-Sensitivity Qubit Assay Kit, Life Technologies) using the VeriSeq™ PGS workflow (Vitrolife) followed by sequencing on a MiSeq instrument (Illumina) using single-end, dual index 36 base pair read sequencing. MiSeq Reporter software (Illumina) performed on-board secondary data analysis and sequencing data was analyzed using BlueFuse Multi software (Illumina).
2.3. SNP Analysis
Aliquots of amplified DNA were used for SNP analysis using the HumanKaryomap-12 Beadchip (Vitrolife) in accordance with manufacturer’s instructions. Briefly, samples were amplified and fragmented prior to an overnight hybridization to a beadchip containing ~300,000 informative markers. The next morning, beadchips were washed and nucleotides were labeled to extend the hybridized primers. Following staining of the primers, beadchips were scanned using the NextSeq 550 (Illumina). Data files were processed using Illumina’s open-source BeadArrayFiles and gtc2vcf.
2.4. Bioinformatic Data Analysis
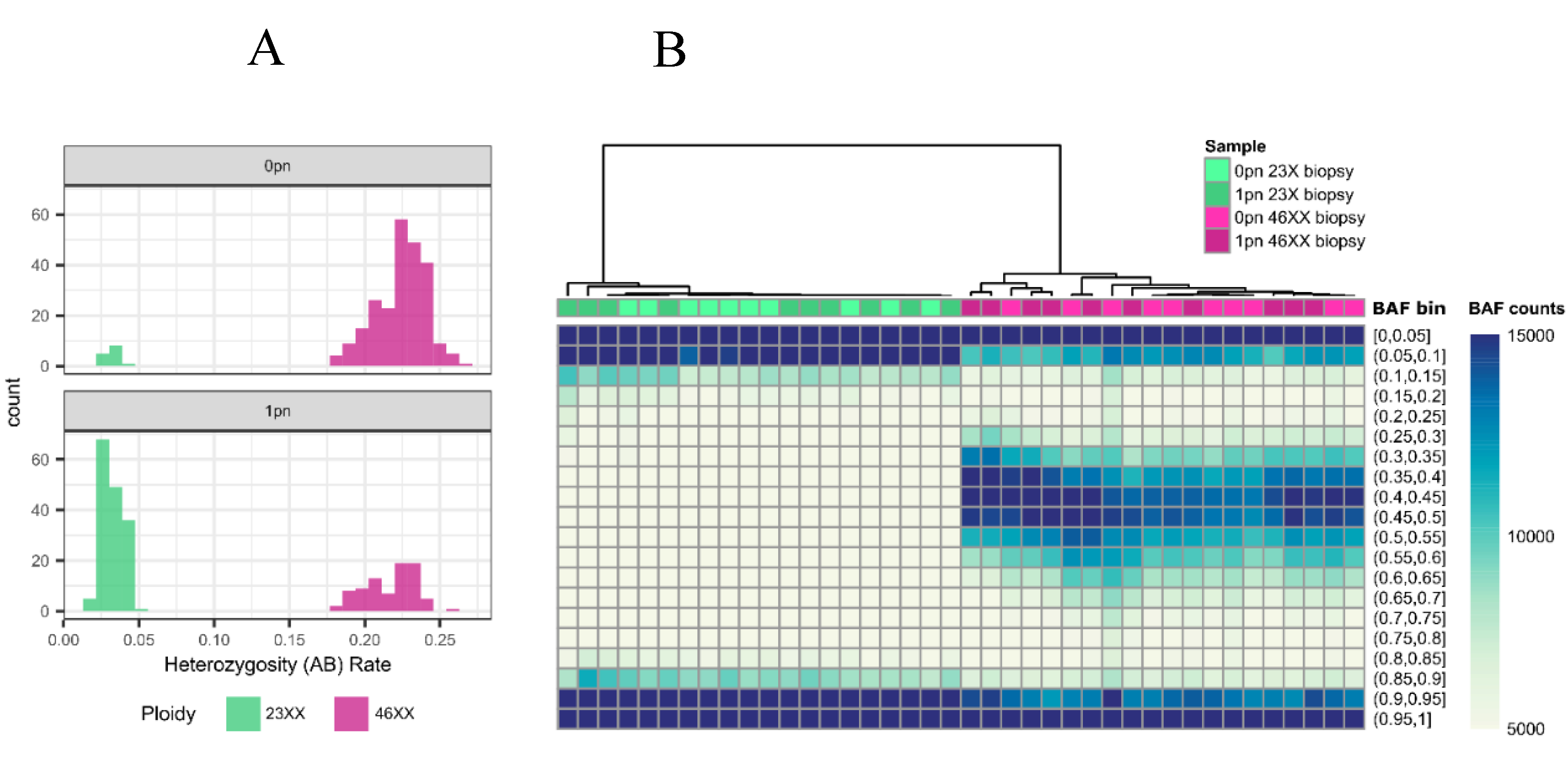
The proportion of heterozygous genotypes was calculated and directly compared to the range of an established library of haploid, trioploid and diploid genome results. Samples with an overall heterozygosity (AB) rate ≤ 5% were considered haploid while ≥ 18% were considered diploid (
Figure 2). For 0PN and 1PN blastocysts, GTC files were processed using Illumina’s open-source BeadArrayFiles v1.2.0 (
https://github.com/Illumina/BeadArrayFiles) and the v1 manifest file. For 3PN blastocysts, GTC files were processed using the open-source gtc2vcf (
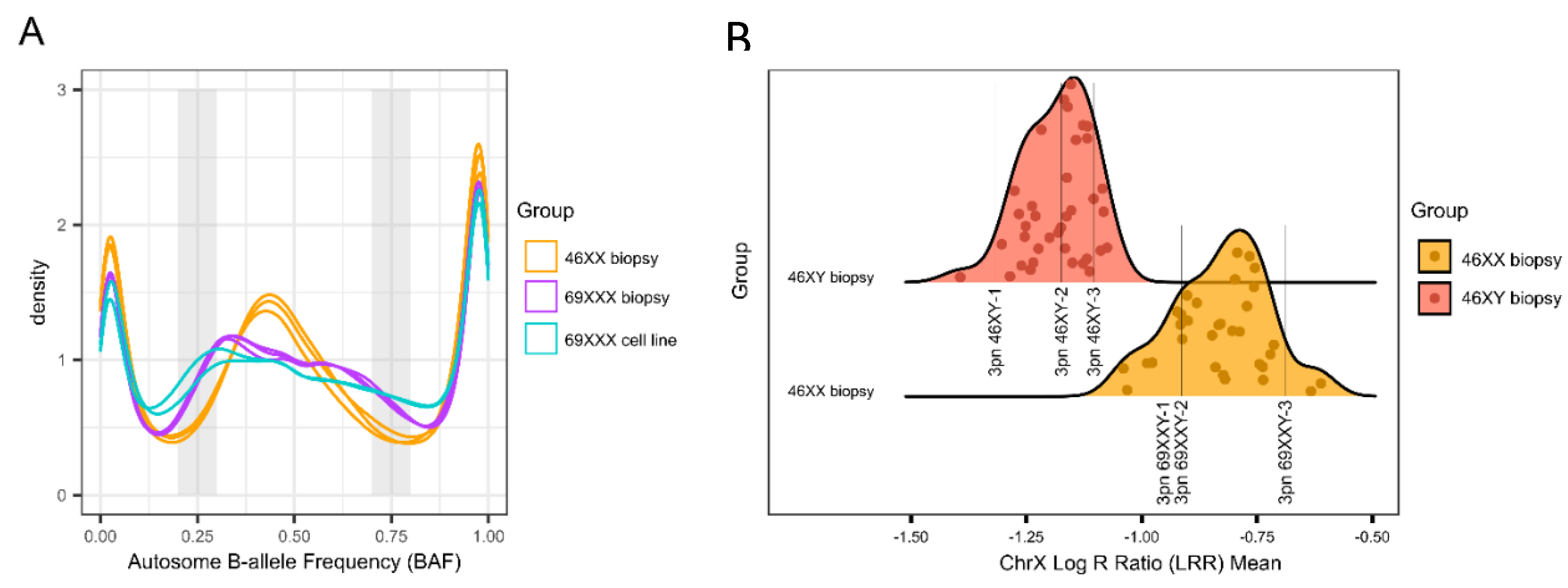
https://github.com/freeseek/gtc2vcf) and the v1 manifest and cluster files. All 3PN zygotes were compared to a library of known euploid samples (n = 37 46XY and n = 32 46XX) (
Figure 3). Samples exhibiting homozygosity on chromosome Y were tested for 69XXY while all others were tested for 69XXX. Potential 69XXY samples were tested by assessing the Log R Ratio (LRR) of chromosome X and comparing that to the euploid database to determine whether one or two X chromosomes were present (
Figure 3). A chromosome X mean LRR ≤ -1.0750221 (the maximum chrX mean LRR of a 46XY sample in the euploid database) is considered to be one copy while a mean LRR ≥ -1.0396700 (the minimum mean chrX LRR of a 46XX sample in the euploid database) is considered to represent two copies. Potential 69XXX samples were tested by assessing the B-allele Ratio (BAF) of autosomes. Two BAF regions (0.2-0.3 and 0.7-0.8) are compared to the euploid library. A BAF proportion < 5.89% within the 0.2-0.3 region and a BAF proportion < 4.31% within the 0.7-0.8 region were considered 46XX (
Figure 3). Statistical analysis was performed using an unpaired t-test or ANOVA where appropriate.
Figure 2.
0pn and 1pn Ploidy Analysis: (a) Histogram of heterozygosity (AB) rates for 0pn (n = 254) and 1pn (n = 242) biopsies. AB rates less than 0.05 are considered unfertilized (green), and rates greater than or equal to 0.18 are considered fertilized (pink). (b) Unsupervised hierarchical clustering heatmap of representative 0pn (n = 20) and 1pn (n = 20) biopsies that are fertilized (n = 20, pink) and unfertilized (n = 20, green). Biopsies determined by AB rate to be either 46XX or 23X cluster together regardless of pronuclear scoring.
Figure 2.
0pn and 1pn Ploidy Analysis: (a) Histogram of heterozygosity (AB) rates for 0pn (n = 254) and 1pn (n = 242) biopsies. AB rates less than 0.05 are considered unfertilized (green), and rates greater than or equal to 0.18 are considered fertilized (pink). (b) Unsupervised hierarchical clustering heatmap of representative 0pn (n = 20) and 1pn (n = 20) biopsies that are fertilized (n = 20, pink) and unfertilized (n = 20, green). Biopsies determined by AB rate to be either 46XX or 23X cluster together regardless of pronuclear scoring.
Figure 3.
3pn Ploidy Analysis: (a) Density plot of representative autosome B-allele frequency (BAF) for 46XX biopsies (n = 3, orange), 69XXX biopsies (n = 3, purple), and 69XXX cell lines (n = 2, blue). The shaded gray regions represent the two windows that are analyzed to determine ploidy status. (b) Density plot of ChrX log R ratio (LRR) means for representative euploid biopsies (n = 32 46XX, orange and n = 37 46XY, red). The two distributions are distinct and do not overlap. The LRR for representative examples of 3pn 46XY (n = 3) and 3pn 69XXY (n = 3) are shown.
Figure 3.
3pn Ploidy Analysis: (a) Density plot of representative autosome B-allele frequency (BAF) for 46XX biopsies (n = 3, orange), 69XXX biopsies (n = 3, purple), and 69XXX cell lines (n = 2, blue). The shaded gray regions represent the two windows that are analyzed to determine ploidy status. (b) Density plot of ChrX log R ratio (LRR) means for representative euploid biopsies (n = 32 46XX, orange and n = 37 46XY, red). The two distributions are distinct and do not overlap. The LRR for representative examples of 3pn 46XY (n = 3) and 3pn 69XXY (n = 3) are shown.
3. Results
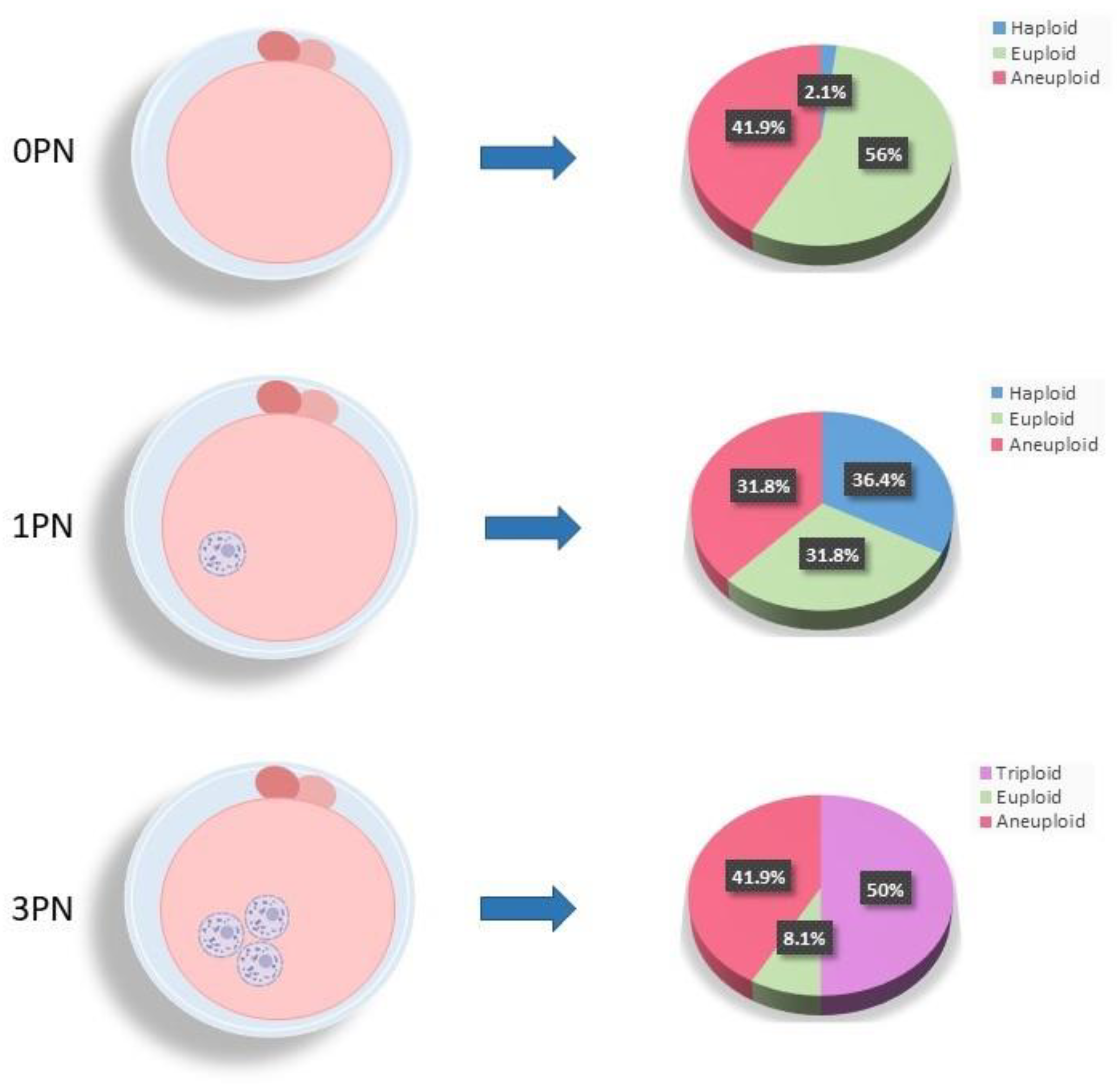
At the time of fertilization check on day 1 of embryo development when no visible pronuclei (0PN) were observed (n = 291; average maternal age = 36. 0 ± 4.2; average paternal age = 37.3 ± 5.5), the vast majority of blastocysts (98%) were found to be normally fertilized with 56% being euploid (n=163;
Figure 4) and 41.9% confirmed to be aneuploid (n=122;
Figure 4). Only a very small percentage (2.1%) of 0PN blastocysts were abnormally fertilized and haploid (n=6;
Figure 4). Upon analysis of the 1PN blastocysts (n = 217; average maternal age = 35.4 ± 4.0; average paternal age = 36.8 ± 5.8), just over a third (36.4%; n = 79;
Figure 4) contained haploid chromosomes (23XO) with another third (31.8%; n = 69;
Figure 4) identified as normally fertilized but aneuploid. The remaining third of the 1PN blastocysts (31.8%; n = 69;
Figure 4) were confirmed to be normally fertilized and diploid. Half (50%) of all 3PN blastocysts showed the presence of a third set of parental chromosomes confirming triploidy (n = 86; 69XXY = 59.3%; 69XXX = 40.7%;
Figure 4). Additionally, 41.9% (n = 72;
Figure 4) of the 3PN blastocysts were identified to be triploid and aneuploid, leaving only a very small percentage (n = 14; 8.1%;
Figure 4) found to be diploid with normal fertilization. Of note, only a very small proportion of biopsied blastocysts (1%) displaying the correct number of pronuclei for normal fertilization (2PN) were actually identified as triploid (69XXY) with a third set of parental chromosomes (data not shown). As expected, the average maternal age was significantly increased in chromosomally aneuploid embryos, regardless of PN numeration. Interestingly, a significant increase in mean maternal age was also observed for the triploid zygotes compared to 3pn zygotes with diploid fertilization (p < 0.05).
To date, there have been 74 euploid blastocyst transfers from zygotes originally identified with either 0PN (n=53), 1PN (n=17), or 3PN (n=4) resulting 16 ongoing clinical pregnancies and 32 healthy live births with no pregnancy or prenatal complications (
Table 1). This included 29 IVF cycles where no other euploid embryos were available (8% of 0PN zygotes and 3% of 1PN zygotes). Four transfers lead to a biochemical loss, two resulted in spontaneous miscarriage prior to the first ultrasound, one pregnancy was lost in a second trimester, and the remaining 19 had a negative hCG (
Table 1). Three of the four normally fertilized 3PN embryos transferred thus far have resulted in healthy live births (
Table 1).
4. Discussion
This study utilized SNP analysis to evaluate ploidy from a large cohort of embryos arising from 0PN, 1PN, and 3PN zygotes. A surprising number of blastocysts that were identified to have alternate pronuclear numeration at fertilization check on day 1 of embryonic development were actually found to be normally fertilized and containing two parental genomes.
Aside from aneuploidy, zygotes with no visible pronuclei were almost always diploid. It is evident that these embryos most likely had two pronuclei present but they had disappeared prior to fertilization check. Utilization of a time-lapse system would enable identification of pronuclei at any time during development. 1PN blastocysts on the other hand were diploid in roughly 2/3 of the cases. The other third, if transferred, would have resulted in pregnancy failure and/or loss. 3PN blastocysts were largely triploid with only a small percentage showing a diploid chromosome constitution. However, performing additional SNP analysis prevented this small percentage of 3PN embryos from being discarded, which allows for a greater number of embryos available for transfer, leading to a higher probability of conception.
There are many suggested mechanisms that might lead to monopronucleation including parthenogenesis, asynchronous pronuclei formation, early pronuclei fusion, and premature pronuclear breakdown among others [
3,
28,
29,
30]. Apart from parthenogenetic activation, all other mechanisms have the possibility of producing normal diploid blastocysts [
31]. Triploidy can be the result of a haploid oocyte fertilized by a haploid sperm, followed by a duplication of either the oocyte or sperm chromosomes. It can also occur when a haploid oocyte is fertilized by two haploid sperm or when an abnormal diploid oocyte is fertilized by a haploid sperm. A triploid conception will always result in spontaneous pregnancy loss including a partial molar pregnancy, whereas haploid embryos would typically fail to implant [
6].
Previous studies have shown the possibility of developmental perturbations, morphokinetic irregularities, and increased aneuploidy in abnormally fertilized zygotes [
5,
18,
32]. Kratka et al. were the first group to utilize a SNP array platform to characterize ploidy in human blastocysts [
15]. They found that triploidy was the most common abnormal fertilization outcome as a result of cell division errors largely occurring during meiosis. Tong et al. compared clinical outcomes from 0PN- and 1PN-derived blastocysts and found similar pregnancy, miscarriage, and live birth rates compared to that of their 2PN embryos [
4]. Another study found similar pregnancy rates between 0PN, 1PN, and 2PN blastocysts although they determined that 1PN zygotes resulted in higher miscarriage and lower live birth rates [
21]. Bradley et al. performed a retrospective analysis of 1PN embryos and found that they had lower developmental potential as determined by their reduced ability to blastulate by day 5. Additionally, they found that even though they were determined to have no genetic abnormalities via CGH analysis and biparental inheritance, they had lower than expected pregnancy outcomes [
31].
There were 29 cycles in this study where no other euploid embryo was identified offering these infertility patients the possibility of a FET and chance at conception. This is the largest cohort, to our knowledge, that includes 0PN, 1PN and 3PN embryos and confirms that a surprising number of zygotes with an alternate number of pronuclei at fertilization check are indeed diploid and can result in a live birth. Adding SNP analysis for identification of ploidy in cases of unknown pronuclei or alternate number of pronuclei, maximizes the number of diploid blastocysts available for utilization during an IVF cycle, thereby increasing cumulative live birth rates. This current study indicates promising clinical outcomes after the transfer of confirmed diploid blastocysts from an original zygote with an alternate number of pronuclei.
Author Contributions
Conceptualization: MGK, WBS, Methodology: BRM, LNH, RML, MGK, Analysis: BRM, MEH, MGK, Investigation: BRM, MEH, LNH, RML, MGK, Data Curation: BRM, MEH, LNH, RML, MGK, Writing – Original Draft Preparation: BRM, MGK, Writing – Review and Editing: BRM, MEH, LNH, RML, MGK, Supervision: MGK, WBS.
Funding
This research received no external funding.
Informed Consent
Informed consent was obtained from all subjects involved in the study.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Montag, M., H. van der Ven, and G. German Pronuclear Morphology Study, Evaluation of pronuclear morphology as the only selection criterion for further embryo culture and transfer: results of a prospective multicentre study. Hum Reprod, 2001. 16(11): p. 2384-9. [CrossRef]
- Capalbo, A., et al., An expert opinion on rescuing atypically pronucleated human zygotes by molecular genetic fertilization checks in IVF. Hum Reprod, 2024. 39(9): p. 1869-1878. [CrossRef]
- Nagy, Z.P., et al., Timing of oocyte activation, pronucleus formation and cleavage in humans after intracytoplasmic sperm injection (ICSI) with testicular spermatozoa and after ICSI or in-vitro fertilization on sibling oocytes with ejaculated spermatozoa. Hum Reprod, 1998. 13(6): p. 1606-12. [CrossRef]
- Tong, X., et al., Clinical outcomes of frozen-thawed blastocysts from zygotes with no or one pronucleus for in vitro fertilization and intracytoplasmic sperm injection cycles. Arch Gynecol Obstet, 2023. 308(3): p. 1015-1022. [CrossRef]
- Ezoe, K., et al., Human 1PN and 3PN zygotes recapitulate all morphokinetic events of normal fertilization but reveal novel developmental errors. Hum Reprod, 2022. 37(10): p. 2307-2319. [CrossRef]
- Capalbo, A., et al., Abnormally fertilized oocytes can result in healthy live births: improved genetic technologies for preimplantation genetic testing can be used to rescue viable embryos in in vitro fertilization cycles. Fertil Steril, 2017. 108(6): p. 1007-1015 e3.
- Paz, M.V., et al., Blastocysts Derived From 0PN Oocytes: Genetic And Clinical Results. JBRA Assist Reprod, 2020. 24(2): p. 143-146. [CrossRef]
- Destouni, A., et al., Genome-wide haplotyping embryos developing from 0PN and 1PN zygotes increases transferrable embryos in PGT-M. Hum Reprod, 2018. 33(12): p. 2302-2311. [CrossRef]
- Uzun, K.N., et al., Comparison of the rates for reaching the blastocyst stage between normal and abnormal pronucleus embryos monitored by a time-lapse system in IVF patients. J Turk Ger Gynecol Assoc, 2021. 22(2): p. 120-126. [CrossRef]
- Walsh, R. and A. Sharma, Extended survival of a premature infant with a postnatal diagnosis of complete triploidy. BMJ Case Rep, 2022. 15(2). [CrossRef]
- Kolarski, M., et al., Genetic Counseling and Prenatal Diagnosis of Triploidy During the Second Trimester of Pregnancy. Med Arch, 2017. 71(2): p. 144-147. [CrossRef]
- McFadden, D.E. and W.P. Robinson, Phenotype of triploid embryos. J Med Genet, 2006. 43(7): p. 609-12. [CrossRef]
- Mutia, K., et al., The Frequency of Chromosomal Euploidy Among 3PN Embryos. J Reprod Infertil, 2019. 20(3): p. 127-131.
- Li, M., et al., Three pro-nuclei (3PN) incidence factors and clinical outcomes: a retrospective study from the fresh embryo transfer of in vitro fertilization with donor sperm (IVF-D). Int J Clin Exp Med, 2015. 8(8): p. 13997-4003.
- Kratka, C., et al., Accurate detection and frequency of abnormal ploidy in the human blastocyst. F S Sci, 2023. 4(2S): p. 27-35. [CrossRef]
- Adelle Yun Xin Lim, C.S.S.L., Embryos Arising from Apronuclear (0PN) and Unipronuclear (1PN) Have Similar Euploidy Rates with Those from 2PN and Should be Considered for Transfer Fertiltiy and Reproduction, 2019. 1(2): p. 73-77.
- Kemper, J.M., et al., What happens to abnormally fertilized embryos? A scoping review. Reprod Biomed Online, 2023. 46(5): p. 802-807. [CrossRef]
- Chen, X., et al., Developmental Potential of Abnormally Fertilized Oocytes and the Associated Clinical Outcomes. Front Physiol, 2020. 11: p. 528424. [CrossRef]
- Mateo, S., et al., In vitro development and chromosome constitution of embryos derived from monopronucleated zygotes after intracytoplasmic sperm injection. Fertil Steril, 2013. 99(3): p. 897-902 e1. [CrossRef]
- Otsu, E., et al., Developmental potential and chromosomal constitution of embryos derived from larger single pronuclei of human zygotes used in in vitro fertilization. Fertil Steril, 2004. 81(3): p. 723-4. [CrossRef]
- Li, M., et al., Obstetric and neonatal outcomes after the transfer of vitrified-warmed blastocysts developing from nonpronuclear and monopronuclear zygotes: a retrospective cohort study. Fertil Steril, 2021. 115(1): p. 110-117. [CrossRef]
- Staessen, C. and A.C. Van Steirteghem, The chromosomal constitution of embryos developing from abnormally fertilized oocytes after intracytoplasmic sperm injection and conventional in-vitro fertilization. Hum Reprod, 1997. 12(2): p. 321-7. [CrossRef]
- Li, R., et al., Feasibility study of using unbalanced embryos as a reference to distinguish euploid carrier from noncarrier embryos by single nucleotide polymorphism array for reciprocal translocations. Prenat Diagn, 2021. 41(6): p. 681-689. [CrossRef]
- Treff, N.R., et al., SNP array-based analyses of unbalanced embryos as a reference to distinguish between balanced translocation carrier and normal blastocysts. J Assist Reprod Genet, 2016. 33(8): p. 1115-9. [CrossRef]
- Handyside, A.H., et al., Karyomapping: a universal method for genome wide analysis of genetic disease based on mapping crossovers between parental haplotypes. J Med Genet, 2010. 47(10): p. 651-8. [CrossRef]
- Griffin, D.K. and R.L. Gould, What is Karyomapping and where does it fit in the world of preimplantation genetic diagnosis (PGD)? Medical Research Archives, 2017. 5(6).
- Schoolcraft, W.B., et al., Live birth outcome with trophectoderm biopsy, blastocyst vitrification, and single-nucleotide polymorphism microarray-based comprehensive chromosome screening in infertile patients. Fertil Steril, 2011. 96(3): p. 638-40.
- Staessen, C., et al., Cytogenetic and morphological observations of single pronucleated human oocytes after in-vitro fertilization. Hum Reprod, 1993. 8(2): p. 221-3. [CrossRef]
- van der Heijden, G.W., et al., Parental origin of chromatin in human monopronuclear zygotes revealed by asymmetric histone methylation patterns, differs between IVF and ICSI. Mol Reprod Dev, 2009. 76(1): p. 101-8. [CrossRef]
- Azevedo, A.R., et al., Molecular cytogenetics of human single pronucleated zygotes. Reprod Sci, 2014. 21(12): p. 1472-82. [CrossRef]
- Bradley, C.K., et al., Clinical use of monopronucleated zygotes following blastocyst culture and preimplantation genetic screening, including verification of biparental chromosome inheritance. Reprod Biomed Online, 2017. 34(6): p. 567-574. [CrossRef]
- Zhao, H., et al., The aneuploidy testing of blastocysts developing from 0PN and 1PN zygotes in conventional IVF through TE-biopsy PGT-A and minimally invasive PGT-A. Front Reprod Health, 2022. 4: p. 966909. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).