1. Introduction
Bombyx mori nucleopolyhedrovirus (BmNPV) or
f65 (Bm65) is expressed at the early stages of BmNPV infection in BmN cells and is involved in the production of progeny virions. Previous studies have shown that Bm65 plays important roles in DNA damage repair, high efficiency of viral replication, and high production of progeny virions during the viral life cycle [
1,
2,
3].
Bm65 codes for a protein comprising 104 deduced amino acid residues with a molecular mass of approximately 12.2 kDa. Therefore, it is regarded as a relatively small protein (< 40 kDa) that can, in theory, passively diffuse between the cytoplasm and nucleus [
4,
5]. Surprisingly, Bm65 was reported to be located mainly in the nucleus, with a small proportion in the cytoplasm, and Bm65 was clustered into visible aggregates with some unknown components in the nucleus [
1]. Moreover, Li et al. reported the
76KRKCSK sequence to be an efficient nuclear localization signal of Bm65 that was directly involved in viral production [
6]. To date, the mechanism by which Bm65 aggregates in the nucleus and the dynamic shuttling of Bm65 between the nucleus and cytoplasm remain unclear. Therefore, it is necessary to further study the correlation of the Bm65-RPL13 interaction and the formation of Bm65 aggregates.
Previous studies have shown that Bm65 can form aggregates and accumulate in the nucleus of BmNPV-infected BmN cells [
1,
3]. Similar studies have reported that some proteins aggregate in the nucleus or cytoplasm of virus-infected cells [
7,
8,
9,
10]. Various cellular factors can affect protein aggregate accumulation and regulate viral propagation and cellular function, even in the occurrence of diseases in humans [
11,
12]. However, the mechanism by which Bm65 accumulates in the nucleus of BmNPV-infected cells is unclear. It is speculated that host proteins are indispensable for the formation of Bm65 aggregates in the nucleus. Therefore, it is necessary to identify scientific issues concerning the formation mechanism of Bm65 aggregates and the correlation between Bm65 aggregates and viral propagation.
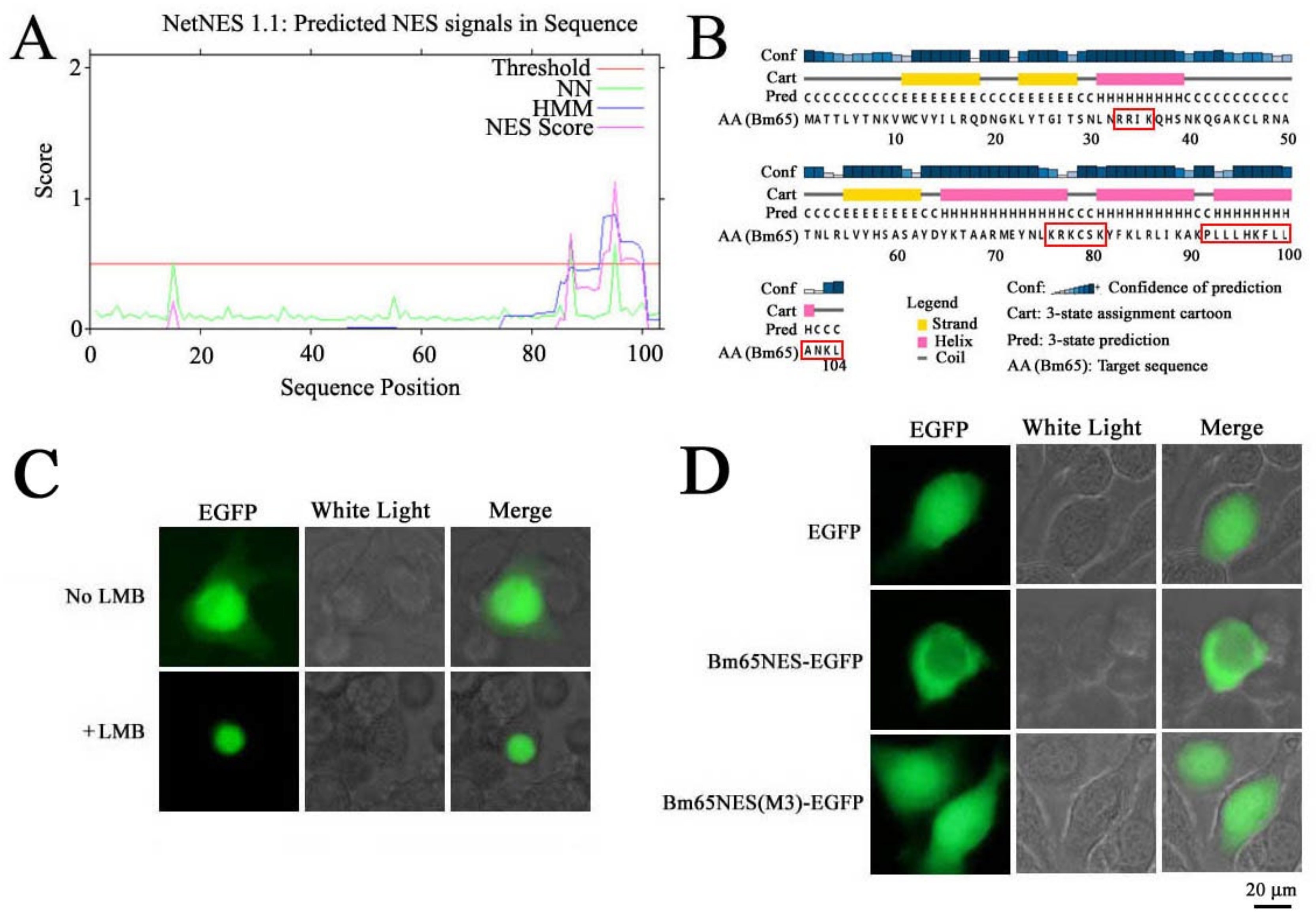
A leucine-rich region was clustered at the C terminus of Bm65, which was predicted to be a potential nuclear export signal (NES) using NetNES1.1 software. To date, the function of the leucine-rich region of Bm65 is unclear. NESs play an important role in maintaining the dynamic shutting balance of target proteins between the cytoplasm and nucleus and are also involved in the functional regulation of target proteins. Leucine-rich NESs are among the most widely studied signals that can be recognized by chromosome region maintenance 1. Human immunodeficiency virus (HIV) Rev was the first identified NES-containing protein [
13,
14]. To date, all identified NESs are leucine-rich, and are often involved in intracellular biochemical processes, such as gene expression, signal transduction, autophagy, viral replication, and infection [
15,
16,
17,
18]. However, incorrect protein localization can cause cellular abnormalities involving protein aggregation, biosynthesis, and metabolic disorders, resulting in the occurrence of some diseases in humans [
19,
20].
The putative NES was amplified from the Bm65 sequence and fused with enhanced green fluorescent protein (EGFP), which was used to study the intracellular distribution of Bm65. Further research was conducted to study the effect of Bm65 NES on viral propagation. Additionally, the ribosomal protein L13 (RPL13), in conjunction with rRNA, was reported to interact with Bm65 via Co-Immunoprecipitation and Liquid Chromatograph-Mass Spectrometer (LC-MS/MS) analysis [
21]. RPL13 is a major component of ribosomes involved in protein biosynthesis. Thus, Bm65 NES mutations were created to demonstrate the binding site of Bm65 with RPL13. Meanwhile, the mutations were also used to further study the role of Bm65 NES in the viral life cycle. In a word, some mutations in the Bm65 NES were made to study the dynamic shuttling between nucleus and cytoplasm of Bm65, as well as the effect on the accumulation of Bm65 aggregates and viral propagation.
2. Materials and Methods
Recombinant Viruses, Plasmids, and Cells
BmNPV bacmid (Bm-bacmid) with a deletion of Bm65 (Bm
Bm65KO) was constructed using homologous recombination and maintained in our laboratory [
3]. Two Bm-Bacmids of the wild type and
Bm65-deleted type harboring the pMON7124 helper plasmid propagate in the
Escherichia coli strain DH10B, which could be used to produce recombinant viruses through transposition. Three recombinant plasmids, namely pMD18T-P
Bm65-Bm65-flag, HTB-P
ie1-EGFP, and HTB-P
ie1-EGFP-sv40-P
Bm65-Bm65-flag, were maintained in our laboratory [
3]. Some viruses, including wild type BmNPV, vBm
Bm65(NES-M1), vBm
Bm65(NES-M2), vBm
Bm65(NES-M3), vBm
Bm65M10, and vBm
Bm65M11 were generated for further research in the study. Plasmids of HTB-P
ie1-Bm65NES-EGFP and HTB-P
ie1-Bm65(NESM3)-EGFP were constructed to study the intracellular distribution of fluorescence signal. BmN cells were used for transfection or infection in the study.
Computer-Assisted Sequence Analysis
Transfection and Fluorescence Microscopy
BmN cells (106 cells/dish) were seeded into 35-mm dishes and incubated at 27°C for 16–24 h before transfection. Recombinant DNA molecules (2 µg/dish) and 5 µl Cellfectin (Invitrogen Life Technology, USA) were mixed in a total volume of 200 µl TC-100 serum-free medium, followed by an additional 45 min of incubation at 27°C. Then, 800 µl serum-free medium was added to the DNA-Cellfectin solution, before finally overlaying onto BmN cells, followed by 5 h of additional incubation at 27°C. After the incubated cells were washed with serum-free TC-100 medium, 2 ml of TC-100 medium containing 10% fetal bovine serum was added to each dish for further culture. Fluorescence microscopy (Olympus-IX73-DP80, Japan) was used to observe fluorescence signals in BmN cells expressing target proteins at selected time points.
Recombinant Viruses for Expression of Bm65 and Bm65 Mutants
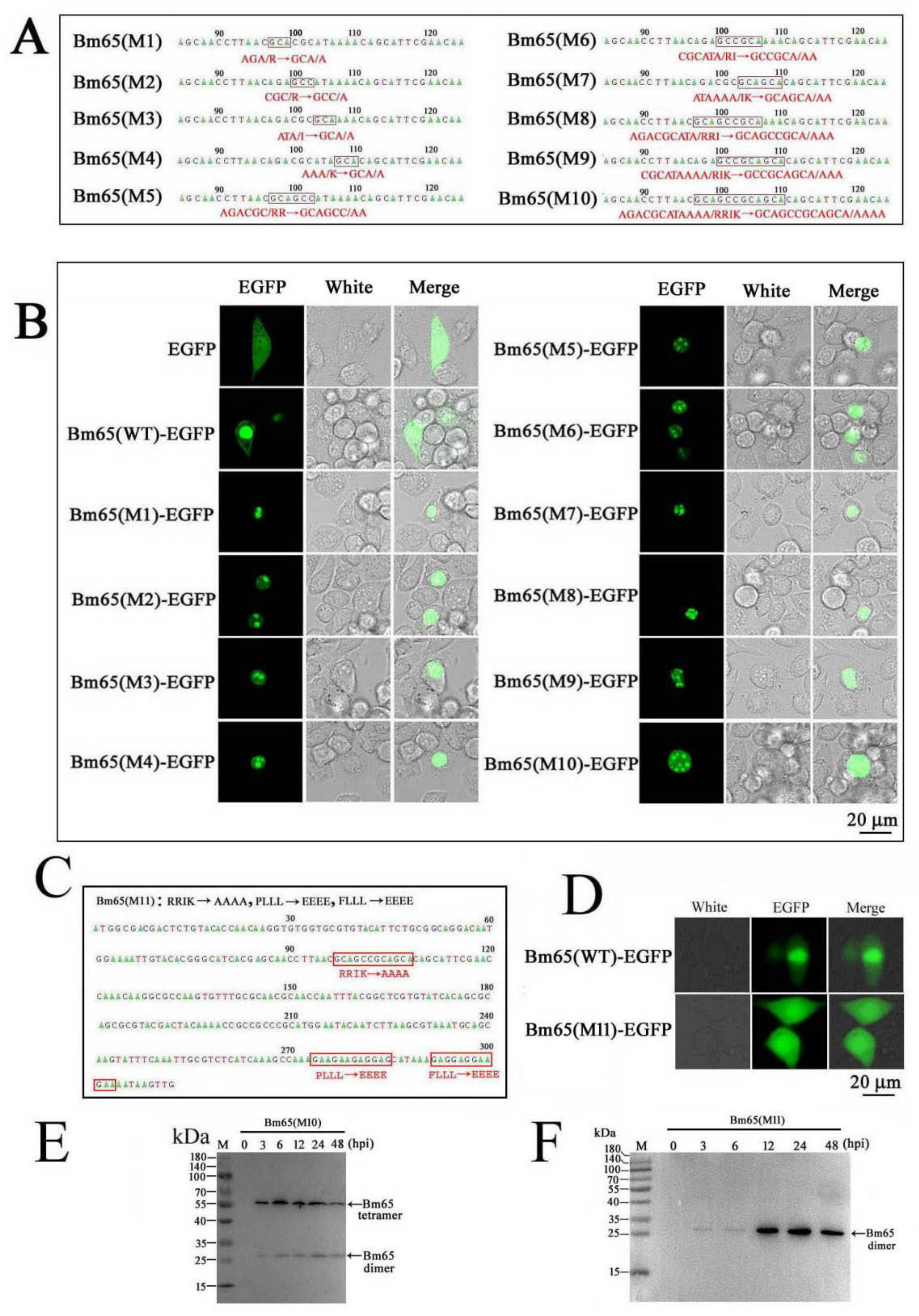
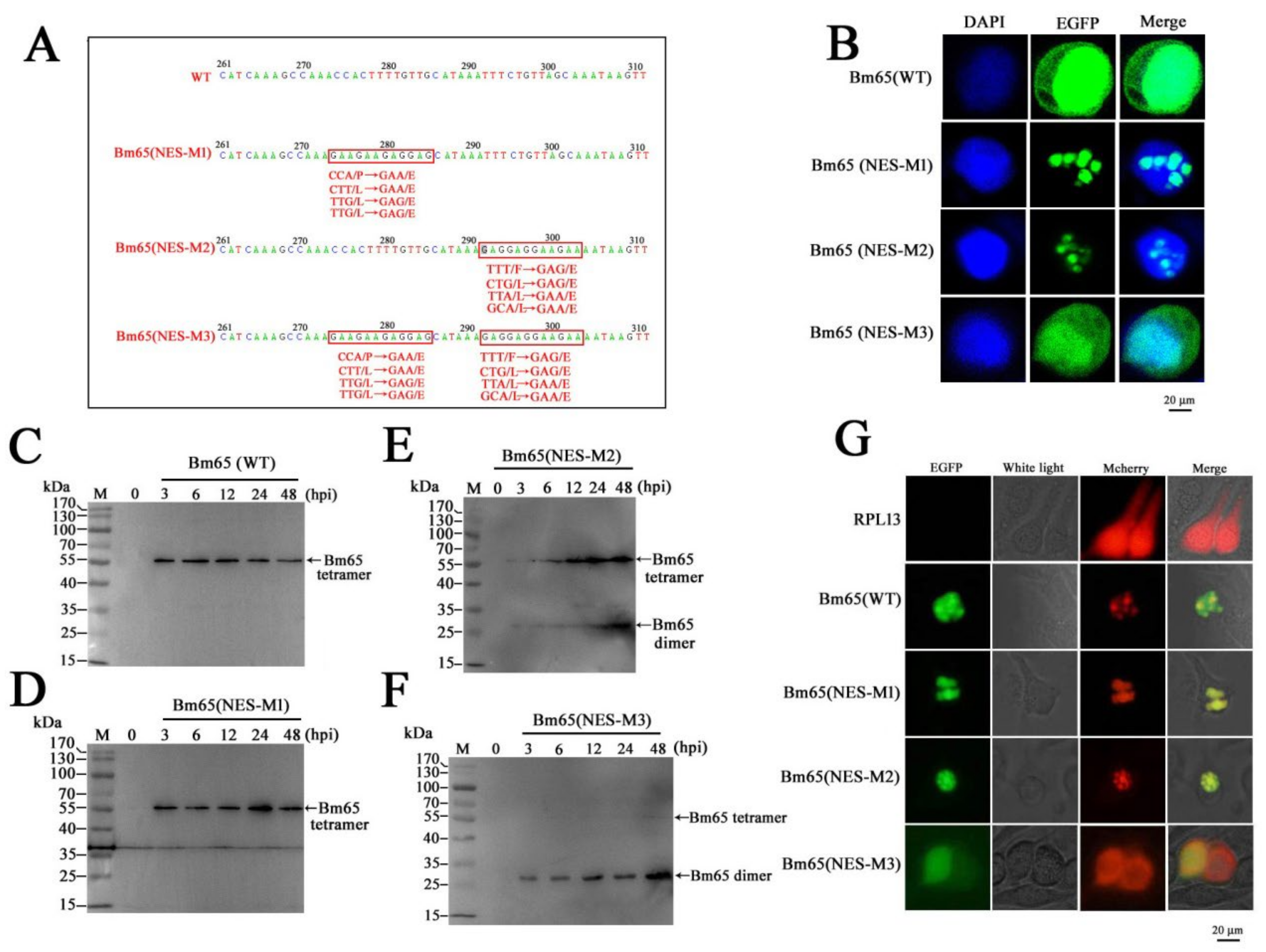
Several mutations, including 92P(E)L(E)L(E)L(E), 98F(E)L(E)L(E)L(E), and 92P(E)L(E)L(E)L(E)98F(E)L(E)L(E)L(E), were introduced in the Bm65 sequence according to the instructions of the MutanBEST Kit (TaKaRa). Briefly, pMD18T-PBm65-Bm65-flag was used as a template to introduce 92P(E)L(E)L(E)L(E) and 98F(E)L(E)L(E)L(E) in the Bm65 sequence using primer pairs of Bm65-flag-F/NES-M1-R, and Bm65-flag-F/NES-M2-R, respectively. Additionally, pMD18T-PBm65-Bm65 (NES-M1)-Flag, containing the mutation of 92P(E)L(E)L(E)L(E), was used as a template to introduce the mutation of 92P(E)L(E)L(E)L(E)98F(E)L(E)L(E)L(E) in the Bm65 sequence using Bm65-flag-F and NES-M2-R by PCR. The three recombinant plasmids were digested with SpeI and XhoI, and the resulting purified target DNA fragment was ligated with HTB-Pie1-egfp-sv40-PBm65-Bm65-flag digested with the same enzymes to generate HTB-Pie1-egfp-sv40-PBm65-Bm65(NES-M1/M2/M3)-Flag. Transposition between HTB-Pie1-egfp-sv40-PBm65-Bm65(NES-M1/M2/M3)-Flag and BmBm65KO was introduced to generate BmBm65(NES-M1/M2/M3)-Flag−GFP, which was selected by blue–white screening and further confirmed by PCR using M13 primers. pMD18T-PBm65-Bm65(NES-M3)-Flag was used as a template to introduce 33R(A)R(A)I(A)K(A) into the Bm65(NES-M3) sequence using Bm65(M11)-F and Bm65(M11)-R. Subsequently, HTB-Pie1-egfp-sv40-PBm65-Bm65(M10/M11)-Flag was constructed via enzyme digestion and ligation. Additionally, BmBm65M10 and BmBm65M11 were generated by transposition between BmBm65KO and HTB-Pie1-egfp-sv40-PBm65-Bm65(M10/M11)-Flag using blue–white screening and PCR confirmation.
Construction of Recombinant Plasmids
Bm65NES-F and Bm65NES-R were used to amplify truncated Bm65 fused with EGFP from HTB-Pie1-Bm65-EGFP to produce Bm65NES-EGFP. The DNA fragment was ligated with HTB-Pie1-Bm65-EGFP digested with EcoRI and XhoI to produce HTB-Pie1-Bm65NES-EGFP. pMD18T-PBm65-Bm65(NES-M3)-Flag was used as a template to amplify a 79-bp Bm65NES(M3) fragment using Bm65NES-F and Bm65(NESM3)-R, which was ligated with HTB-Pie1-Bm65-EGFP and digested with EcoRI and PstI to generate HTB-Pie1-Bm65NES(M3)-EGFP.
To generate a series of Bm65 mutants in the 33RRIK region, pMD18T-Bm65 was used as a template to amplify Bm65 mutants with different primer pairs according to the instructions of the MutanBEST Kit (TaKaRa). Briefly, primer pairs of Bm65(M1)-F/Bm65M-R, Bm65(M2)-F/Bm65M-R, Bm65(M3)-F/Bm65M-R, and Bm65(M4)-F/Bm65M-R were used to make single point mutations in 33RRIK, resulting in the generation of 33R(A)RIK, 33RR(A)IK, 33RRI(A)K, and 33RRIK(A), respectively. Bm65(M5)-F/Bm65M-R, Bm65(M6)-F/Bm65M-R and Bm65(M7)-F/Bm65M-R were used to make two point mutations in 33RRIK, resulting in the generation of 33R(A)R(A)IK, 33RR(A)I(A)K, and 33RRI(A)K(A). Bm65(M8)-F/Bm65M-R and Bm65(M9)-F/Bm65M-R were used to produce three point mutations in 33RRIK, resulting in the generation of 33R(A)R(A)I(A)K and 33RR(A)I(A)K(A). Bm65(M9)-F and Bm65M-R were used to introduce 33R(A)R(A)I(A)K(A) in the Bm65 sequence. Subsequently, these Bm65 mutants were used as a template to amplify target DNA using Bm65-F and Bm65-R, which were ligated into HTB-Pie1-Bm65-EGFP digested with EcoRI and PstI to generate the corresponding recombinant plasmids.
Confocal Microscopy Analysis
BmN cells (1 × 10
5) were seeded into a 35-mm glass-bottom cell culture dish (NSET), and 2 µg of each plasmid was used for transfection. Bm65 and Bm65 mutants fused with EGFP were transiently expressed in BmN cells under control of the
ie1 promoter. The cell culture supernatants were removed 48 h after transfection (hpt). Subsequently, some treatment was performed as following. Briefly, the cells were washed three times with phosphate buffered saline (PBS; 2.6 mM KCl, 0.136M NaCl, 8 mM Na
2HPO4, 2 mM KH
2PO4, pH 7.4), fixed with 4% paraformaldehyde for 15 min, washed three times with PBS for 10 min, and permeabilized in 0.1% Triton X100 for 15 min. Finally, the cells were stained with DAPI (60 µg/ml, Sigma) for 10 min and washed three times with PBS. Confocal microscopy was used to observe fluorescence signals as previously described [
1,
6].
Viral Growth Curve Analysis
Virus growth curve analysis was performed as previously described to determine the effect of Bm65 mutants on viral propagation [
3]. Briefly, BmN cells (10
6 cells/well) were seeded into six-well plates and cultured overnight. The next day, 2.0 µg of recombinant acid DNA was used for transfection of BmN cells to generate recombinant viruses. The supernatants of transfected BmN cells containing recombinant viruses were harvested at selected time points. The Budded Virus (BV) titer was determined using 50% tissue culture infective dose (TCID50) end-point dilution, as described previously [
3]. The presence of green fluorescence in BmN cells indicated successful viral infection. The experiments were repeated three times. Statistical analysis was performed using a single-factor analysis of variance.
Quantitative Analysis of Viral DNA Synthesis
To further study the effect of mutations in
92PLLLHKFLLA of Bm65 on viral replication, quantitative real-time PCR (qPCR) analysis was performed to examine the copies of the viral genome in recombinant virus-infected BmN cells, as described previously [
22]. Briefly, some recombinant viruses, including wild type Bm65NES-M1, Bm65NES-M2, and Bm65NES-M3 BmNPV, were used to infect BmN cells for comparative analysis of viral genome copies. Total DNA was extracted from each sample using a Universal Genomic DNA Extraction Kit (TaKaRa) according to the manufacturer’s instructions. The total DNA was resuspended in 150 μl of sterile water. qPCR analysis was performed with 10 μl of the extracted DNA and Hot Start PCR Master Mix III (Chaoshi-Bio) according to the manufacturer’s instructions using qPCR-F/qPCR-R targeting the 170-bp region of
GP64. All primers used in the study are listed in
Table 1.
3. Results
Inhibition of Viral Propagation by Mutations in the 92PLLLHKFLLA of Bm65
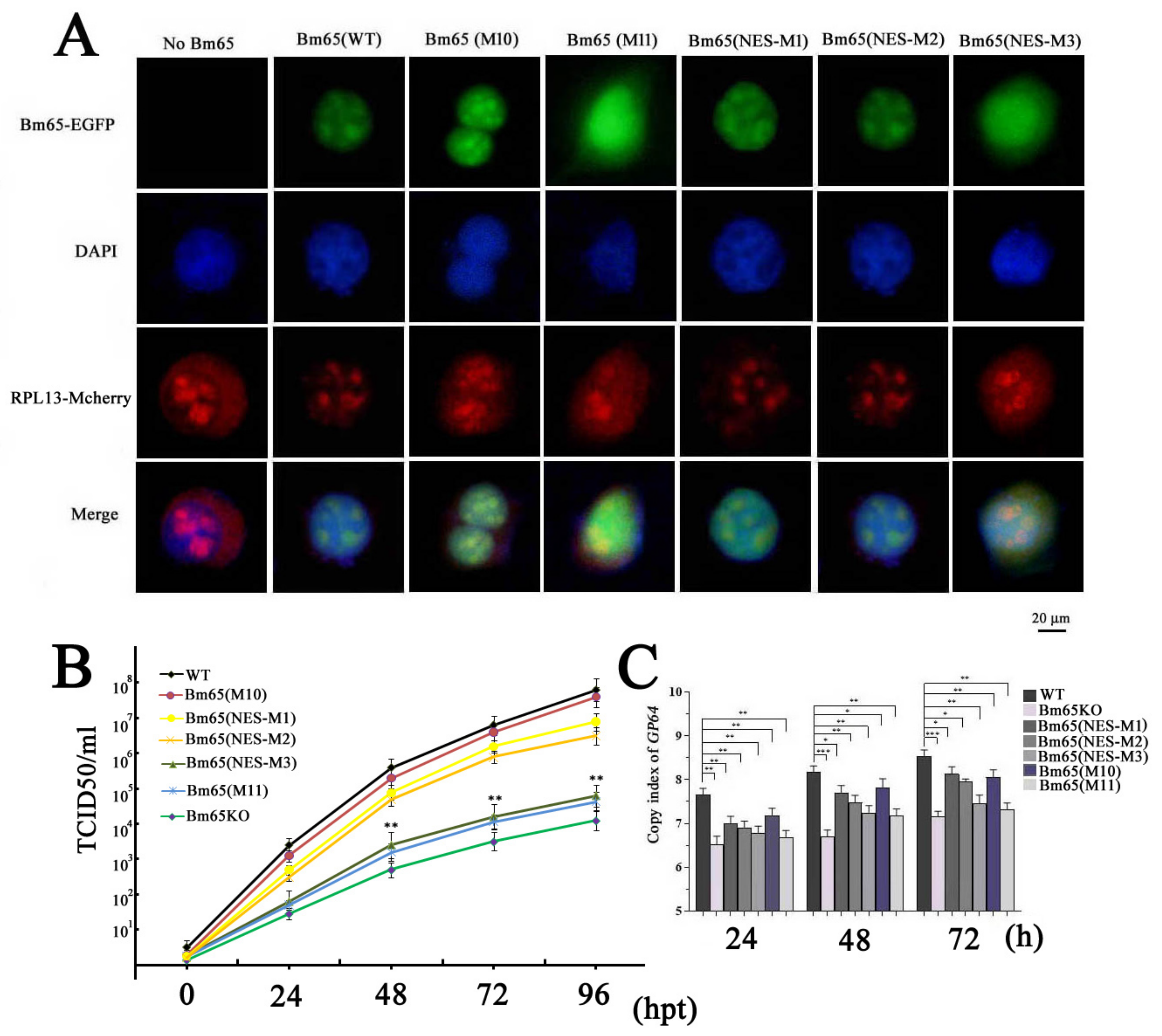
To demonstrate the correlation between Bm65 aggregates and the interaction between Bm65 and RPL13, co-transfection of BmN cells with two recombinant plasmids was made to express RPL13 and Bm65 or Bm65 mutants, respectively. RPL13 and Bm65 were fused with mCherry and EGFP, respectively. Thus, the intracellular distribution of green and red fluorescence using confocal microscopy was used to examine the co-localization of Bm65 and RPL13 in BmN cells. The results showed fluorescence co-localization between RPL13 and wild-type Bm65 or the three Bm65 mutants, including Bm65(M10), in BmN cells, Bm65(NES-M1), and Bm65(NES-M2) (
Figure 4A). However, Bm65(M11) and Bm65(NES-M3) of Bm65 mutants did not co-localize with RPL13 in BmN cells (
Figure 4A). The mutation of
92PLLLHKFLLA into
92EEEEHKEEEE completely blocked the interaction between RPL13 and Bm65(M11) or Bm65(NES-M3), resulting in the failure of fluorescence co-localization and Bm65 aggregation in BmN cells.
The TCID50 endpoint dilution in BmN cells was measured to determine the titers of the recombinant viruses. The results indicated that recombinant viruses with the mutations of
92EEEEHKEEEE in the Bm65 sequence all showed a decreased tendency in the slope of the growth curves compared to the WT, Bm65(M10), Bm65(NES-M1), and Bm65(NES-M2) viruses (
Figure 4B). The statistical analysis revealed a significant difference in the viral titers between Bm65 (NES-M3) or Bm65(M11) and the Bm65 wild type virus from 48 to 96 hpt. To further study the effect of mutations in the
92PLLLHKFLLA region of Bm65 on viral propagation, several recombinant viruses were used to infect BmN cells to compared the viral genome copies. Mutations in the
92PLLLHKFLLA gene were introduced into the genomes of the Bm65(M11), Bm65(NES-M3), and Bm65KO viruses. For this purpose, qPCR analysis was performed to examine the copy number of
gp64 to compare the efficiency of viral propagation, and the copies of
gp64 were also used to analyze the effect of the mutations on viral propagation. Compared to the Bm65, Bm65(M10), Bm65(NES-M1), and Bm65(NES-M2), the mutations in the
92PLLLHKFLLA of Bm65(M11), Bm65(NES-M3), and Bm65KO viruses obviously inhibited viral propagation, resulting in a decrease in
gp64 copy number (
Figure 4C). There was a significant difference (p < 0.001) in
gp64 copy numbers between the Bm65 and Bm65(M11) or Bm65(NES-M3). Additionally, western blotting analysis confirmed that the expression of GP64 and VP39 in BmN cells infected with wild-type BmNPV was significantly higher than that in BmN cells infected with the Bm65 mutant with
92PLLLHKFLLA BmNPV (data not shown). This result indicated that mutations in the
92PLLLHKFLLA region of Bm65 could sharply inhibit viral propagation and downregulate the expression of GP64 and VP39.
4. Discussion
BmNPV, a typical baculovirus with a circular double-stranded DNA genome, can quickly spread viral infection among silkworms on a large scale, causing great economic loss. Silkworms have evolved some effective strategies to combat an invaded microorganism in long-term evolutionary processes [
23,
24], and some silkworm proteins have been reported to participate in the resistant response to viral invasion and propagation [
25]. BmNPV replication and propagation partly depend on the complicated interaction between the virus and silkworm. Bm65 is a BmNPV-encoded protein with a small weight of 12.2 kDa. In theory, Bm65 should exhibit a uniform distribution in BmN cells according to cell size. However, Bm65 exists mainly in the nuclei of BmNPV-infected BmN cells, and can spontaneously form Bm65 aggregates accumulated in the nucleus at late stages of viral infection. In addition, hydrophobic amino acids have been shown to promote the accumulation of Bm65 aggregates in the nucleus. We aimed to reveal the role of
33RRIK in the nuclear entry of Bm65 via point mutation. The subcellular localization results showed that the mutations in
33RRIK did not block nuclear entry of the Bm65 mutant, but clearly accelerated the accumulation of Bm65 in the nucleus of BmN cells (
Figure 2B). However, the components of Bm65 aggregates remain unclear. Therefore, it is essential to specify the specific components of Bm65 aggregates and define their roles in viral propagation. Meanwhile, it will be helpful for us to further demonstrate the interactions between Bm65 and host proteins, as well as the mechanism of Bm65 aggregate formation.
Some ribosomal proteins are not only the requisite components of ribosomes but also directly participate in the regulation of host immunity and viral propagation. To date, diverse ribosomal proteins have been reported to interact with viral proteins to regulate viral biosynthesis, viral replication, and host immune responses. For example, RPL4 interacts with VP3 to regulate the replication of infectious bursal disease virus (IBDV) [
26]. RPL18 is a well-known critical factor due to its interaction with many viral proteins from Arabidopsis thaliana, Ebola viruses, and dengue viruses [
6]. RPL13 promotes IRES-driven translation of foot and mouth disease in a DDX3-dependent helicase [
27]. Guan et al. reported that RPL13 participates in the innate immune response induced by foot-and-mouth disease virus [
28]. Therefore, ribosomal proteins are a class of multifunctional proteins that play diverse roles during viral replication, virion assembly, and the antiviral immune response. In general, the viral strategy exploits host cellular resources for the production and spread of progeny virions. Some host cell proteins directly participate in viral replication, assembly, and maturation processes. In addition, some host proteins can be hijacked to inhibit antiviral immune responses, thereby facilitating viral propagation. We considered that BmNPV hijacking RPL13 could facilitate viral replication to improve viral production, which was confirmed by qPCR and TCID50 analyses (
Figure 4).
Zhang et al. previously identified RPL13 as a stress-inducible gene that plays a regulatory role in NF-κB signaling [
29]. Moreover, RPL13 participates in viral translation and host innate immune response [
27,
28]. Recently, RPL13 was reported to interact with Bm65 to repair UV-induced DNA damage [
21]. Therefore, RPL13 could function as an important binding factor with viral protein to regulate the interactions between BmNPV and silkworm. This also demonstrates the scientific correlation between the interactions and Bm65 aggregates.
In the present study, the hydrophobic region of
92PLLLHKFLLA was identified as an efficient NES of Bm65 and was involved in the dynamic transport of Bm65 from the nucleus to cytoplasm (
Figure 1). Furthermore, a complete mutation in
92PLLLHKFLLA region could block the interaction between Bm65 and RPL13, resulting in the failure of Bm65 aggregate production in the nucleus. Additionally, western blot analysis confirmed that the mutations inhibited the formation of the Bm65 tetramer and viral propagation. The
76KRKCSK is an efficient nuclear localization signal (NLS) for nuclear import of Bm65, but Bm65 mutants that lose the ability to form stable protein complexes (> 40 kDa) could not stably exist in the nuclei of BmN cells. Therefore, it is reasonable to speculate that the uniform distribution of the Bm65 mutant in BmN cells occurs through passive diffusion. However, the specific components of Bm65 aggregates and their roles in viral propagation remain unclear, and further research is required to clarify these issues.
Author Contributions
Conceptualization, G.L. and Q.T.; Methodology, G.L., Y.L., H.C., F.Z., Z.H., Z.G., and Q.T.; Software, Z.G.; Validation, Q.T.; Formal analysis, W.L., H.C., and Z.H.; Investigation, G.L., W.L., and Y.L.; Resources, K.C.; Data curation, W.L.; Writing—original draft, G.L., W.L., and Y.L.; Writing—review & editing, H.C. and Feifei Zhu; Supervision, Q.T.; Project administration, Q.T.; Funding acquisition, Q.T.
Funding
This research was supported by the National Natural Science Foundation of China (No. 32072794).
Institutional Review Board Statement
This study is not applicable.
Informed Consent Statement
This statement is not applicable.
Data Availability Statement
All required data are available in the manuscript. Additional data can be provided upon request.
Acknowledgments
We are grateful to Manli Wang and Zhihong Hu from the Wuhan Institute of Virology for providing VP39 antibodies.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tang, Q; Hu, Z.Y; Yang, Y.H; Wu, H.L; Qiu, L.P; Chen, K.P; Li, G.H. Overexpression of Bm65 correlates with reduced susceptibility to inactivation by UV light. J Invertebr Pathol, 2015, 127:87-92. [CrossRef]
- Tang, Q; Wu, P; Hu, Z.Y; Yang, Y.H; Qiu, L.P; Liu, H.Q; Zhu, S.Y; Guo, Z.J; Xia, H.C; Chen, K.P, Li, G.H. Evidence for the role of BmNPV Bm65 protein in the repair of ultraviolet-induced DNA damage. J Invertebr Pathol, 2017, 149:82-86. [CrossRef]
- Li, G.H; Qi, X.Y; Chen, H.Q; Hu, Z.Y; Chen, F.Y; Deng, L; Guo, Z.J; Chen, K.P; Tang, Q. The Motif of 76KRKCSK in Bm65 Is an Efficient Nuclear Localization Signal Involved in Production of Infectious Virions. Front Microbiol, 2019, 10:2739. [CrossRef]
- Li, G.H.; Qi, X.Y.; Hu, Z.Y.; Tang, Q. Mechanisms Mediating Nuclear Trafficking Involved in Viral Propagation by DNA Viruses. Viruses, 2019, 11(11):1035. [CrossRef]
- Fu, X; Liang, C; Li, F; Wang, L; Wu, X; Lu, A; Xiao, G; Zhang, G. The Rules and Functions of Nucleocytoplasmic Shuttling Proteins. Int J Mol Sci, 2018, 19(5):1445. [CrossRef]
- Li, S. Regulation of Ribosomal Proteins on Viral Infection. Cells, 2019, 8(5):508. [CrossRef]
- Guo, Z.J; Tao, L.X; Dong, X.Y; Yu, M.H; Tian, T; Tang, X.D. Characterization of aggregate/aggresome structures formed by polyhedrin of Bombyx mori nucleopolyhedrovirus. Sci Rep. 2015, 5:14601. [CrossRef]
- Xu, X; Zhou, X; Nan, H; Zhao, Y; Bai, Y; Ou, Y; Chen, H. Aggregation of AcMNPV LEF-10 and Its Impact on Viral Late Gene Expression. PLoS One, 2016, 11(5):e0154835. [CrossRef]
- Nan, H; Chen, H; Tuite, M.F; Xu, X. A viral expression factor behaves as a prion. Nat Commun, 2019, 10(1):359. [CrossRef]
- Yoo, Y.S; Park, Y.J; Lee, H.S; Oanh, N.T.K; Cho, M.Y; Heo, J; Lee, E.S; Cho, H; Park, Y.Y; Cho, H. Mitochondria ubiquitin ligase, MARCH5 resolves hepatitis B virus X protein aggregates in the liver pathogenesis. Cell Death Dis, 2019, 10(12):938. [CrossRef]
- Marreiros, R; Müller-Schiffmann, A; Bader, V; Selvarajah, S; Dey, D; Lingappa, V.R; Korth, C. Viral capsid assembly as a model for protein aggregation diseases: Active processes catalyzed by cellular assembly machines comprising novel drug targets. Virus Res, 2015, 207:155-64. [CrossRef]
- Jouanne, M; Rault, S; Voisin-Chiret, A.S. Tau protein aggregation in Alzheimer's disease: An attractive target for the development of novel therapeutic agents. Eur J Med Chem, 2017, 139:153-167.
- Fornerod, M; Ohno, M; Yoshida, M; Mattaj, I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell, 1997, 90(6):1051-60. [CrossRef]
- Fischer, U; Huber, J; Boelens, W.C; Mattaj, I.W; Lührmann, R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell, 1995, 82(3):475-83. [CrossRef]
- Behrens, R.T; Aligeti, M; Pocock, G.M; Higgins, C.A; Sherer, N.M. Nuclear Export Signal Masking Regulates HIV-1 Rev Trafficking and Viral RNA Nuclear Export. J Virol, 2017, 91(3):e02107-16. [CrossRef]
- Cheng, J.H.; Lai, G.H.; Lien, Y.Y.; Sun, F.C.; Hsu, S.L.; Chuang, P.C.; Lee, M.S. Identification of nuclear localization signal and nuclear export signal of VP1 from the chicken anemia virus and effects on VP2 shuttling in cells. Virol J, 2019, 16(1):45. [CrossRef]
- Meng, W; Wang, X.J; Wang, H.R. Targeting nuclear proteins for control of viral replication. Crit Rev Microbiol, 2019, 45(5-6):495-513. [CrossRef]
- Rodríguez-Vargas, J.M; Rodríguez, M.I; Majuelos-Melguizo, J; García-Diaz, Á; González-Flores, A; López-Rivas, A; Virág, L; Illuzzi, G; Schreiber, V; Dantzer, F; Oliver, F.J. Autophagy requires poly(adp-ribosyl)ation-dependent AMPK nuclear export. Cell Death Differ, 2016, 23(12):2007-2018. [CrossRef]
- Juarez-Navarro, K; Ayala-Garcia, V.M; Ruiz-Baca, E; Meneses-Morales, I; Rios-Banuelos, J.L; Lopez-Rodriguez, A. Assistance for Folding of Disease-Causing Plasma Membrane Proteins. Biomolecules, 2020, 10(5):728. [CrossRef]
- Yuan, S; Chu, H; Huang, J; Zhao, X; Ye, Z.W; Lai, P.M; Wen, L; Cai, J.P; Mo, Y; Cao, J; Liang, R; Poon, V.K; Sze, K.H; Zhou, J; To, K.K; Chen, Z; Chen, H; Jin, D.Y; Chan, J.F; Yuen, K.Y. Viruses harness YxxØ motif to interact with host AP2M1 for replication: A vulnerable broad-spectrum antiviral target. Sci Adv, 2020, 6(35):eaba7910. [CrossRef]
- Tang, Q; Tang, J; Chen, C; Zhu, F.F; Yu, Q; Chen, H.Q; Chen, L; Ma, S.S; Chen, K.P; Li, G.H. Bombyx mori RPL13 participates in UV-induced DNA damage repair of B. mori nucleopolyhedrovirus through interaction with Bm65. Insect Mol Biol, 2024.
- Shi, A; Hu, Z; Zuo, Y; Wang, Y; Wu, W; Yuan, M; Yang, K. Autographa californica Multiple Nucleopolyhedrovirus ac75 Is Required for the Nuclear Egress of Nucleocapsids and Intranuclear Microvesicle Formation. J Virol, 2018, 92(4):e01509-17. [CrossRef]
- Jiang, L; Goldsmith, M.R; Xia, Q. Advances in the Arms Race Between Silkworm and Baculovirus. Front Immunol, 2021, 12:628151. [CrossRef]
- Hu, Z; Zhu, F; Chen, K. The Mechanisms of Silkworm Resistance to the Baculovirus and Antiviral Breeding. Annu Rev Entomol, 2023, 68:381-399. [CrossRef]
- Kong, M; Zuo, H; Zhu, F; Hu, Z; Chen, L; Yang, Y; Lv, P; Yao, Q; Chen, K. The interaction between baculoviruses and their insect hosts. Dev Comp Immunol, 2018, 83:114-123. [CrossRef]
- Chen, Y; Lu, Z; Zhang, L; Gao, L; Wang, N; Gao, X; Wang, Y; Li, K; Gao, Y; Cui, H; Gao, H; Liu, C; Zhang, Y; Qi, X; Wang, X. Ribosomal protein L4 interacts with viral protein VP3 and regulates the replication of infectious bursal disease virus. Virus Res. 2016, 211:73-8. [CrossRef]
- Han, S; Sun, S; Li, P; Liu, Q; Zhang, Z; Dong, H; Sun, M; Wu, W; Wang, X; Guo, H. Ribosomal Protein L13 Promotes IRES-Driven Translation of Foot-and-Mouth Disease Virus in a Helicase DDX3-Dependent Manner. J Virol, 2020, 94(2):e01679-19. [CrossRef]
- Guan, J; Han, S; Wu, J; Zhang, Y; Bai, M; Abdullah, S.W; Sun, S; Guo, H. Ribosomal Protein L13 Participates in Innate Immune Response Induced by Foot-and-Mouth Disease Virus. Front Immunol, 2021, 12:616402. [CrossRef]
- Zhang, H.X; Liu, Z.X; Sun, Y.P; Zhu, J; Lu, S.Y; Liu, X.S; Huang, Q.H; Xie, Y.Y; Zhu, H.B; Dang, S.Y; Chen, H.F; Zheng, G.Y; Li, Y.X; Kuang, Y; Fei, J; Chen, S.J; Chen, Z; Wang, Z.G. Rig-I regulates NF-κB activity through binding to Nf-κb1 3'-UTR mRNA. Proc Natl Acad Sci U S A, 2013, 110(16):6459-64.).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).