1. Introduction
Infectious diseases pose a significant burden on health systems worldwide due to high morbidity and mortality rates. According to the World Health Organization (WHO), millions of people die from infectious diseases each year, often because early and accurate diagnosis is not achieved [
1]. This reality highlights the critical importance of rapid and accurate diagnosis of infectious diseases.
For many years, traditional diagnostic methods used in clinical microbiology laboratories were generally based on culture-based techniques. However, these techniques are not only time-consuming but also inadequate for pathogens that are difficult or impossible to cultivate under laboratory conditions [
2]. This limitation is particularly evident in the case of slow-growing microorganisms or pathogens requiring highly specific culture conditions. Additionally, the sensitivity of immunological tests may significantly decrease during the early stages of infection, when specific antibody formation has not yet occurred [
3].
In recent years, advancements in molecular biology have led to significant transformations in the diagnosis of infectious diseases, ushering in a new era in diagnostic processes. Among these advancements, the Polymerase Chain Reaction (PCR), developed by Kary Mullis in the late 1980s, stands out [
4]. The primary advantage of PCR is its ability to rapidly and sensitively amplify the genetic material of the infectious agent, enabling specific detection. This capability has quickly made PCR an indispensable tool in clinical microbiology laboratories and established it as the gold standard for diagnosing many infectious diseases. During the COVID-19 pandemic, PCR technology became the most widely used diagnostic method worldwide [
5,
6]. The rapid and reliable results provided by PCR-based tests during the pandemic have once again highlighted the critical importance of molecular diagnostic methods for public health and clinical practice.
One of the most remarkable innovations following PCR is Next-Generation Sequencing (NGS) technology. NGS allows for the sequencing of entire genomes of pathogens in a single run, enabling the detection of new strains and genetic variations [
7]. This technology offers significant advantages, particularly in tracking antibiotic-resistant strains, identifying virulence factors, and understanding the molecular epidemiology of outbreaks. With the growing availability of bioinformatics tools and data analysis capabilities, NGS is increasingly being used in clinical diagnostics. The rapid identification of resistance genes and virulence factors has greatly accelerated the management of infections [
8].
Molecular diagnostic methods have expanded beyond PCR and NGS to include innovative approaches such as CRISPR-based diagnostic systems. In addition to its gene-editing potential, CRISPR (short for “clustered regularly interspaced short palindromic repeats”) technology stands out as a precise and rapid method for detecting specific DNA or RNA sequences in the diagnosis of infectious diseases. This technology marks the beginning of a new era in the diagnosis of infectious diseases [
9,
10].
However, the implementation of molecular diagnostic methods in clinical practice presents certain challenges, especially in low- and middle-income countries. The lack of advanced infrastructure, trained personnel, and sufficient funding poses significant barriers to the widespread adoption of these technologies [
11]. Nonetheless, ongoing efforts to develop portable and more affordable PCR and NGS systems may help overcome these challenges [
12,
13].
With the rapid advancement of molecular diagnostic methods, the advantages and limitations of each technology have become highly significant for clinical applications. Techniques such as PCR, NGS, Loop-Mediated Isothermal Amplification (LAMP), and CRISPR offer speed, sensitivity, and specificity in the diagnosis of infectious diseases, yet each has its own distinct strengths and weaknesses.
Table 1 provides a comparative overview of the advantages, disadvantages, and applications of these technologies, offering guidance on which methods may be preferred under different conditions in molecular diagnostic processes.
This review will provide a detailed examination of the role of molecular diagnostic methods in the diagnosis of infectious diseases, how they are used in clinical applications, their advantages, and the challenges encountered. Particular emphasis will be placed on advanced diagnostic methods such as PCR and NGS, highlighting the advantages they offer over traditional methods and discussing potential future developments.
2. Molecular Diagnostic Methods
Molecular diagnostic methods are advanced biotechnological tools that enable the rapid, sensitive, and specific detection of infectious diseases. The slow nature and limited sensitivity of traditional diagnostic methods pose a significant disadvantage, especially for infections that require prompt intervention. In contrast, molecular methods target the genetic material (DNA and RNA) of pathogens, providing faster and more accurate results. The ability of these techniques to be used in the early stages of infection and to detect even low amounts of pathogenic material makes them indispensable in clinical applications [
14,
15].
Today, the most commonly used techniques in molecular diagnostics include PCR, Real-Time PCR (qPCR), Multiplex PCR, NGS, LAMP, and CRISPR-based diagnostic methods. These techniques not only accelerate diagnostic processes but also provide critical information about the genetic structure of infectious agents, contributing to treatment planning.
2.1. Molecular Diagnostic Methods
PCR is considered one of the cornerstones of molecular diagnostics and has been a revolutionary development in the diagnosis of infectious diseases. Developed by Kary Mullis in 1983, this technique allows for the rapid amplification of a specific segment of target DNA millions of times, enabling the detection of even extremely small amounts of genetic material [
4]. This high sensitivity has made PCR an indispensable tool in the diagnosis of infectious diseases. Today, PCR is used to detect a wide range of microorganisms, including viruses, bacteria, fungi, and parasites, and is regarded as the "gold standard" in clinical microbiology laboratories [
16].
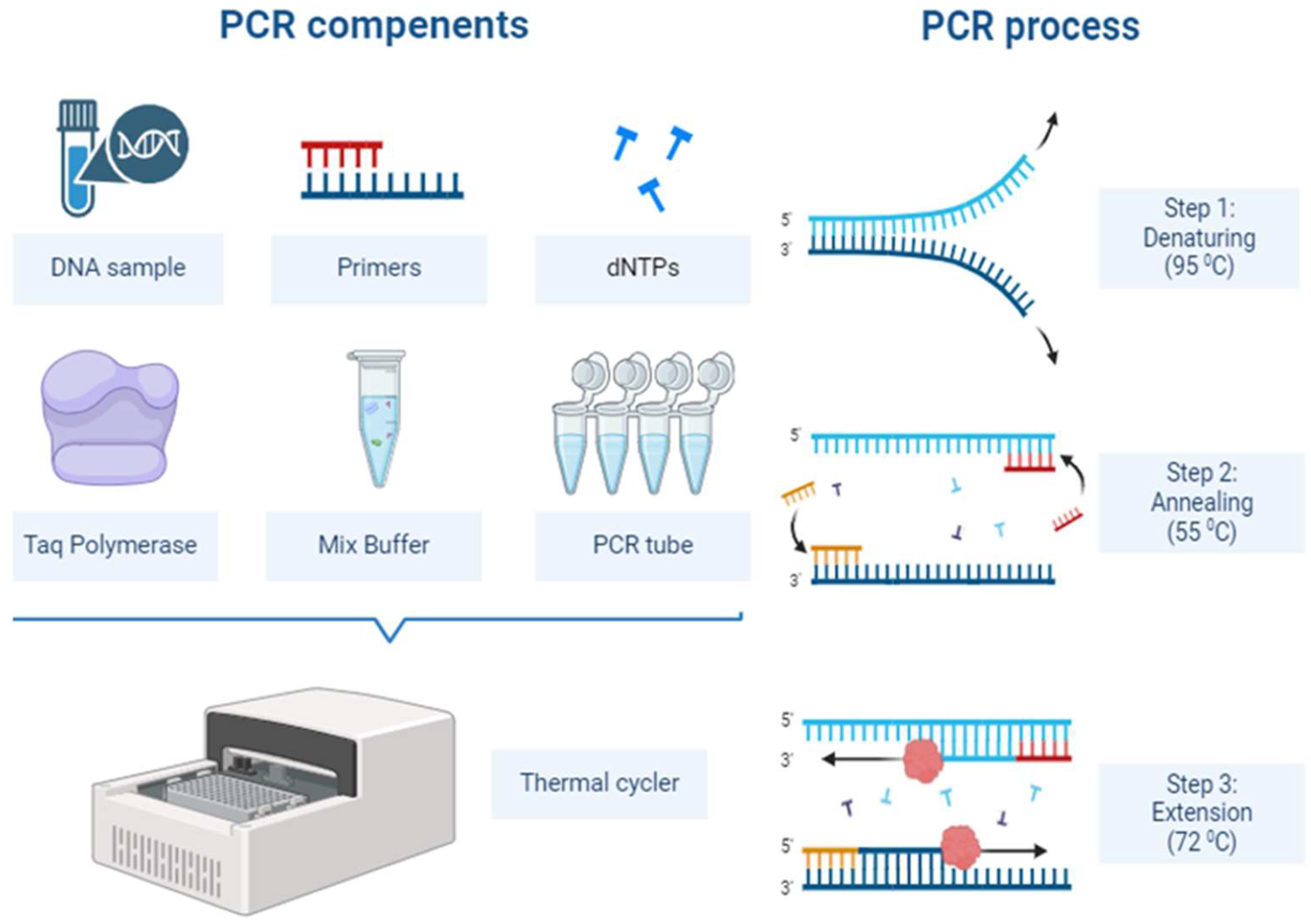
The principle of PCR involves targeting a specific region of DNA or RNA and amplifying this region using primers, allowing for the detection of the pathogen in a laboratory setting (
Figure 1). One of the main reasons for the widespread use of this method in infectious diseases is its ability to detect even very small amounts of pathogenic DNA or RNA [
17]. PCR provides highly sensitive and specific results, especially in the early stages of viral infections such as HIV, Hepatitis B (HBV), and Hepatitis C (HCV) [
18,
19]. Similarly, for bacterial infections like tuberculosis, Group B streptococcus, and Methicillin-resistant
Staphylococcus aureus, PCR offers faster and more reliable results compared to traditional culture methods [
20].
Over time, advanced variants of PCR have made significant progress in the diagnosis of infectious diseases. For example, Real-Time PCR (qPCR) performs DNA amplification while simultaneously measuring the amount of amplified DNA in real time. By using a fluorescent signal to monitor the amplification process, qPCR indicates how much target DNA is being replicated, allowing for both qualitative and quantitative analysis. This technology plays a crucial role in determining disease burden and monitoring response to treatment. qPCR offers high sensitivity and specificity, making it a powerful tool for pathogen detection [
21]. During the COVID-19 pandemic, it was the most widely used method for detecting SARS-CoV-2 [
22]. Additionally, because qPCR can detect multiple pathogens simultaneously, it plays an important role in identifying co-infections [
23].
Multiplex PCR is a PCR technique that allows the simultaneous amplification of multiple target DNA regions. This method enables the detection of more than one pathogen in the same sample. By using multiple pairs of primers, the genetic material of various pathogens can be amplified in the same reaction tube [
24]. This approach offers a significant advantage, especially in respiratory infections where multiple viruses or bacteria may be present simultaneously [
25]. Additionally, multiplex PCR can detect antimicrobial resistance genes at the same time, playing a crucial role in managing patient treatment [
26].
Digital PCR (dPCR) is a newer and more advanced form of PCR, in which the sample is partitioned into thousands of micro-reactions to enhance quantitative accuracy. dPCR allows for the more sensitive and precise detection of nucleic acids present in low copy numbers. As a result, it is used for detecting rare mutations or low levels of pathogens [
27].
Despite the widespread use of PCR in infectious diseases, it has some limitations. For instance, this method only indicates the presence of DNA or RNA, which does not always signify an active infection. Additionally, PCR testing is prone to technical errors that can result in false positive or negative results. Therefore, it is important to support PCR results with clinical findings.
2.1.1. Applications of PCR in the Diagnosis of Infectious Diseases
PCR has become an indispensable tool in modern clinical microbiology laboratories due to its wide range of applications in diagnosing infectious diseases [
28]. One of the major advantages of PCR is its ability to produce results quickly, with a typical PCR process completed within just a few hours, which is crucial for the rapid diagnosis of infections. Additionally, its high sensitivity allows for the detection of even very small amounts of pathogenic DNA, setting it apart from other diagnostic methods. It is widely used in the diagnosis of viral, bacterial, fungal, and parasitic infections, providing significant advantages in the clinical management of infections [
17,
29].
2.1.1.1. Detection of Viral Infections
PCR has been a groundbreaking method in the diagnosis of viral infections. Traditional serological methods can only yield positive results at specific stages of an infection, whereas PCR technology can detect the virus's genetic material at much earlier stages, thereby speeding up the diagnostic process [
30].
PCR technology is indispensable for the detection of HBV and HCV. The detection of HCV RNA using PCR can be performed in the early stages of infection, allowing for the rapid identification of these viruses, which have the potential to become chronic. Moreover, PCR enables the measurement of viral load, which is useful for monitoring the progression of the disease and the response to antiviral treatment [
31,
32].
PCR also plays a crucial role in the diagnosis of Human Immunodeficiency Virus (HIV) infection. It can detect the virus's RNA even during the pre-seroconversion period, providing an opportunity to initiate antiretroviral therapy (ART) early. The quantitative applications of PCR are essential for monitoring viral load in HIV patients, allowing for rapid intervention in cases of treatment failure or disease progression [
33,
34].
During the COVID-19 pandemic, PCR became one of the most widely used methods for detecting SARS-CoV-2. The amplification of RNA from nasopharyngeal swab samples using reverse transcription PCR (RT-PCR) emerged as the most common approach for diagnosing the disease. The high sensitivity of RT-PCR allowed for the detection of not only symptomatic cases but also asymptomatic carriers, thus playing a significant role in controlling the spread of the pandemic [
5,
35,
36].
2.1.1.2. Detection of Bacterial Infections
One of the main advantages of PCR in the diagnosis of bacterial infections is its ability to provide results much faster than traditional culture methods. The speed and accuracy offered by PCR are crucial for clinical outcomes, especially in serious bacterial infections that require rapid treatment [
37].
Sepsis is one of the most serious examples of bacterial infections that require rapid intervention. PCR allows for the quick identification of pathogens causing sepsis, enabling the prompt initiation of appropriate antibiotic treatment. While diagnosis using traditional blood culture methods can take several days, PCR can complete the process within hours. This has a direct impact on the course of the disease and the patient's chances of survival [
38].
In the diagnosis of chronic bacterial infections such as tuberculosis (TB), the speed and sensitivity provided by PCR make a significant difference. While culturing Mycobacterium tuberculosis, the causative agent of tuberculosis, using traditional methods can take weeks, PCR can diagnose tuberculosis within a few hours [
39,
40]. Moreover, PCR has the ability to detect antibiotic resistance genes, making it an important guide in treatment planning. For instance, the rapid identification of strains resistant to drugs such as rifampicin and isoniazid allows for the swift and effective management of the treatment process [
41,
42].
2.1.1.3. Detection of Fungal Infections
PCR also makes a significant contribution to the diagnosis of fungal infections. In particular, the ability of PCR to provide rapid and sensitive results in diagnosing invasive fungal infections allows for timely initiation of antifungal treatments. The detection of clinically important fungi such as
Candida and
Aspergillus species can take weeks with traditional methods, whereas PCR enables diagnosis within a few hours. This is especially crucial for immunocompromised patients, as early detection of invasive fungal infections in this group significantly reduces mortality rates [
43].
Moreover, one of the broad applications of PCR is the identification of antifungal resistance genes. In cases where pathogens such as
Candida albicans and
Aspergillus fumigatus have developed resistance to antifungal treatments, PCR-based tests enable the rapid detection of resistant isolates, guiding the clinical treatment process [
44,
45].
2.1.1.4. Detection of Parasitic Infections
The effectiveness of PCR in diagnosing parasitic infections is particularly evident in cases where traditional microscopic diagnostic methods are inadequate. Since PCR can directly detect parasite DNA, it is especially effective in identifying asymptomatic or low-intensity infections.
For example, in the diagnosis of malaria caused by
Plasmodium species, PCR can provide much more sensitive results compared to traditional microscopy methods. The specific detection of malaria species such as
Plasmodium falciparum is important for clinical management, and PCR can expedite this process [
46]. Additionally, the identification of drug-resistant parasites is also possible with PCR [
47].
In parasitic diseases such as leishmaniasis, PCR greatly facilitates the diagnostic process. In cases where microscopic diagnosis is challenging, such as with cutaneous and visceral leishmaniasis, PCR can amplify the pathogen's genomic material to provide a rapid and accurate diagnosis [
48,
49].
2.2. Loop-mediated Isothermal Amplification (LAMP)
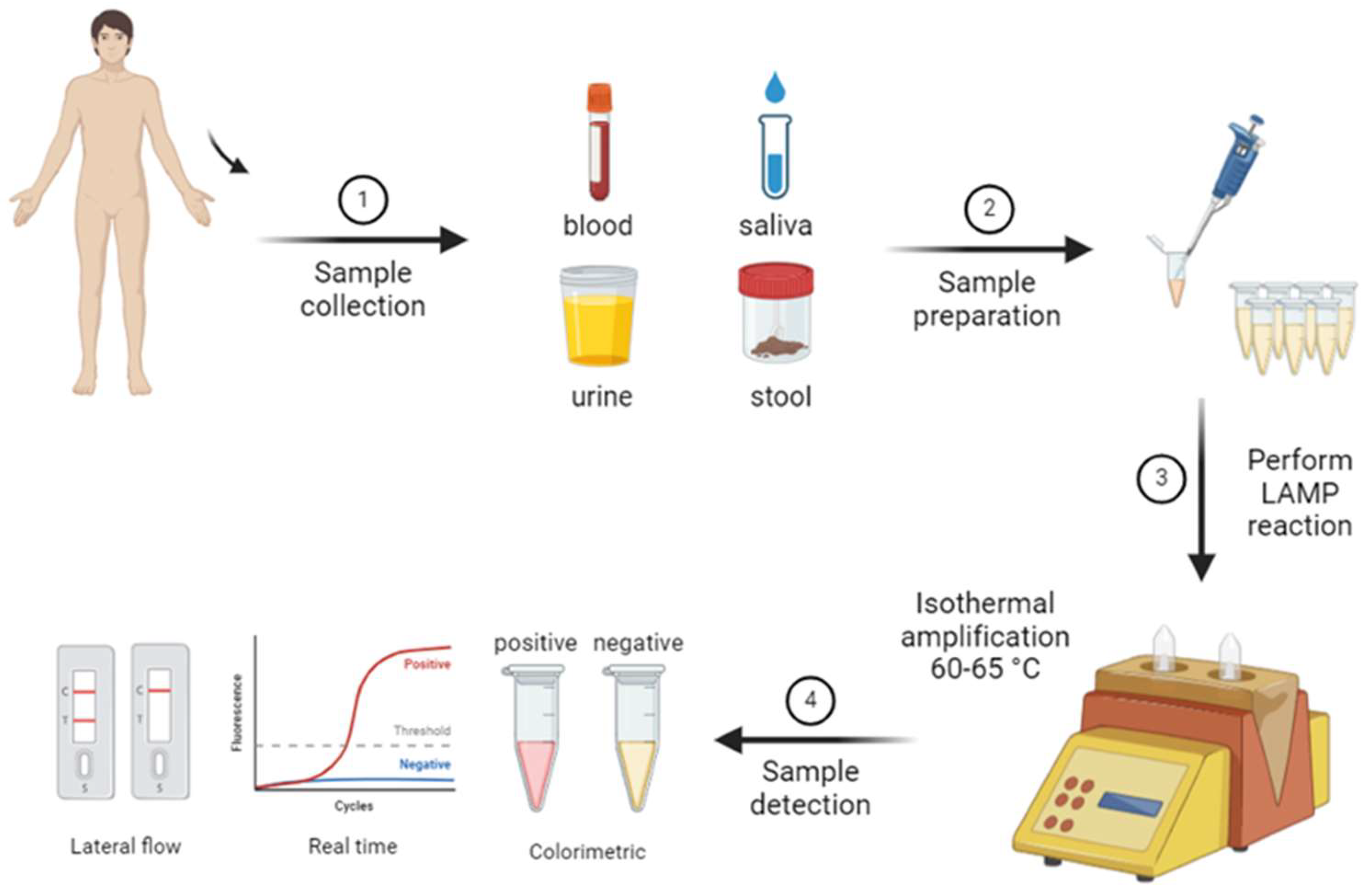
In recent years, alternative molecular diagnostic methods have been developed in addition to the advantages offered by PCR. These methods overcome some of the limitations of PCR, providing faster, more cost-effective, and portable solutions. One such innovative method is LAMP. LAMP is a rapid and sensitive molecular diagnostic technique that can amplify DNA or RNA at a constant temperature. Unlike PCR, the LAMP method does not require a thermal cycler and is carried out under isothermal conditions, meaning at a single constant temperature. Typically operating between 60-65°C, LAMP delivers quick results and is notable for its ease of use in field settings (
Figure 2) [
50].
2.2.1. Principles of the LAMP Method
Since LAMP performs the amplification process at a constant temperature, it does not require complex and expensive thermal cyclers. This feature provides a significant advantage for use in developing regions and fieldwork. The principle of LAMP involves the binding of four to six different primers to the target DNA sequence to initiate amplification, followed by the cyclic replication of DNA using DNA polymerase [
50]. Primer design is a critical step in ensuring the specificity of LAMP; therefore, primers must be carefully selected and specific to the target DNA.
During the LAMP reaction, the amplification of the target DNA results in the accumulation of magnesium pyrophosphate, a distinct byproduct. This accumulation creates a turbidity in the reaction tube that can be observed with the naked eye, allowing for visual evaluation of the results. Additionally, LAMP reactions can be monitored using fluorescent dyes or color changes, enabling users to easily interpret the outcomes [
51].
2.2.2. Advantages and Applications of LAMP
One of the main advantages of LAMP is its ability to deliver rapid results. While PCR typically requires a process lasting 1-2 hours, LAMP results can usually be obtained within 30 minutes to 1 hour. Moreover, since LAMP is an isothermal method, it does not require the cyclic temperature changes needed in PCR. This makes LAMP suitable for use in field conditions and easily applicable with portable devices [
52].
LAMP is more advantageous than PCR in terms of cost. The technique does not require thermal cyclers and can be performed with simple equipment, making it particularly appealing in regions with limited resources. Additionally, the ability to visually observe the byproducts produced during the LAMP reaction eliminates the need for expensive detection devices. These characteristics make LAMP an ideal option for low-cost, portable, and rapid diagnostic tests, especially for detecting outbreaks and tropical infections [
53,
54].
LAMP can be used for the detection of a wide range of pathogens. It has been effectively applied in diagnosing infections that are common in tropical regions, such as malaria, dengue, and Zika virus. In the detection of
Plasmodium species, the causative agent of malaria, LAMP provides much faster and more reliable results compared to traditional microscopic methods. RNA viruses like dengue and Zika can also be easily detected with LAMP following a reverse transcription step [
55,
56,
57].
During the COVID-19 pandemic, LAMP attracted significant attention. It was used as an alternative to PCR for detecting the SARS-CoV-2 virus, and its ability to deliver results in a shorter time contributed to the widespread adoption of rapid diagnostic tests. LAMP could be used to detect the virus in both symptomatic and asymptomatic individuals, and its low cost made it a preferred choice for mass screening tests. These characteristics have made LAMP an important diagnostic tool for managing pandemics, especially in regions with limited resources [
58,
59,
60].
2.2.3. Comparison of LAMP with Other Molecular Methods
Compared to PCR, LAMP offers significant advantages due to its simpler technology and operation under isothermal conditions. LAMP allows for rapid results while providing a similar level of sensitivity and specificity to PCR. Moreover, unlike PCR, it does not require complex laboratory equipment, making it easier to implement in the field. The low cost and speed of LAMP make it particularly valuable in resource-limited regions and during outbreaks requiring urgent diagnosis [
51,
61].
However, LAMP does have some limitations. The primer design for LAMP is quite complex, and the selection of appropriate primers plays a critical role in the success of the method. Additionally, while LAMP is typically optimized for the detection of a single pathogen, PCR has the capability to detect multiple targets simultaneously. Therefore, LAMP is generally used for the detection of single-pathogen infections, and in clinical samples containing multiple pathogens, PCR may be more advantageous than LAMP [
51,
62].
2.3. Next-Generation Sequencing (NGS) and Metagenomic Approaches
Following the success of PCR in diagnosing infectious diseases, another significant advancement in molecular diagnostics came with the introduction of NGS technologies in clinical applications. NGS revolutionized the field of genome sequencing by enabling the simultaneous sequencing of numerous DNA and RNA molecules. This technology allows for the high-capacity, sensitive, and rapid analysis of genetic material, breaking new ground in the molecular diagnosis of infectious diseases. In addition to providing comprehensive genome sequencing, NGS has also facilitated transcriptomic, epigenomic, metagenomic, and other omics studies [
63,
64].
The fundamental principle of NGS is its ability to read billions of base sequences in parallel. This technology allows for the analysis of the genetic material of specific pathogens as well as the detection of various infectious agents such as viruses, bacteria, and parasites. It also offers high efficiency in identifying genetic variations and mutations, and even discovering new pathogen species [
65]. One of the key features that sets NGS apart from other molecular methods is its ability to analyze the entire microbiome with a single test. However, the implementation of NGS is a costly process that requires technical infrastructure, which limits its widespread use in clinical laboratories. Nonetheless, the development of portable NGS devices and the reduction in costs are expected to make this technology more widely accessible in the near future [
66].
2.3.1. The Use of NGS in Diagnosis and Treatment
NGS technology plays a significant role not only in detecting pathogens but also in identifying mutations, antimicrobial resistance genes, and virulence factors. For example, the detection and monitoring of resistance genes in pathogens such as carbapenem-resistant
Klebsiella pneumoniae, a bacterium carrying antibiotic resistance, is possible with NGS technology [
67]. This is crucial for controlling hospital-acquired infections and tracking the spread of antibiotic resistance. Additionally, NGS enables the epidemiological monitoring of pathogens and understanding their spread dynamics, which plays a critical role in planning public health measures.
Especially during outbreaks, the genetic information provided by NGS offers rapid and comprehensive analysis, contributing to the shaping of public health policies. The interest in using this technology during the COVID-19 pandemic was particularly notable for tracking genetic mutations and different variants of the virus. NGS became an essential tool for understanding the evolution of SARS-CoV-2 and the spread of variants in different regions. Additionally, this information played a critical role in vaccine development efforts and public health measures [
68]. The emergence of variants such as Delta, Beta, and Omicron highlighted the vital importance of NGS in monitoring mutations.
2.3.2. Metagenomic Approaches and Clinical Applications
One of the innovations brought by NGS technologies is the use of metagenomic approaches in clinical applications. Metagenomics allows for the sequencing of all microbial genomes present in environmental samples or disease tissues, enabling the analysis of complex microbial communities without the need to detect individual pathogens. This technique is particularly useful for identifying pathogens that cannot be cultured by traditional methods or are previously unknown. Clinical metagenomics serves as an important diagnostic tool for rare pathogens or newly emerging disease agents [
69].
Metagenomic approaches offer a non-hypothesis-driven screening method. This allows for the sequencing and analysis of all microbial genomes present in a sample without prior knowledge of the infection's cause. Such an approach provides a significant advantage in resolving complex or unknown infections. For example, metagenomic analysis is one of the most effective methods for diagnosing conditions like meningitis or sepsis when the cause cannot be determined. Additionally, it plays a crucial role in outbreak management and individual patient care by providing information on the origins and transmission pathways of diseases [
69,
70,
71].
Metagenomic applications provide important insights not only into infectious diseases but also into immune responses and pathogen-host interactions by studying the human microbiome. For example, metagenomic analyses in infectious diseases can reveal how pathogens and host defense mechanisms interact, thereby aiding in the development of more personalized treatment approaches [
72].
2.3.3. The Future of NGS and Metagenomics
The widespread use of NGS in clinical applications is currently limited due to high costs and technical infrastructure requirements. However, this situation is beginning to change with the rapid development of portable NGS devices. The advancement of portable NGS technology and the reduction in sequencing costs are significant developments that will enable more widespread use of this technology in clinical laboratories [
66].
In the future, as NGS and metagenomic approaches become more cost-effective and faster, they are expected to become standard methods for diagnosing infectious diseases. The role of these technologies will especially grow in the fields of personalized medicine and pathogen monitoring, providing effective results even in more complex clinical samples.
In addition to its advantages in diagnosing infectious diseases, NGS has important applications in areas such as monitoring viral genomic variations, identifying antimicrobial resistance genes, and tracking gene transfer between pathogens. This plays a crucial role in controlling outbreaks and determining treatment strategies [
65].
2.4. CRISPR-Based Diagnostic Methods
Another innovative approach is diagnostic methods based on CRISPR-Cas technology. Although CRISPR-Cas technology is primarily known as a gene-editing tool, it has also been successfully used for the specific detection of genetic material. This method stands out for its ability to deliver rapid, sensitive, and specific results [
73]. CRISPR-based diagnostic platforms such as SHERLOCK, DETECTR, FELUDA, CONAN, and VaNGuard have been successfully applied in diagnosing viral infections like COVID-19 [
74].
In the future, CRISPR-based diagnostic systems are expected to develop further and become more widespread. Additionally, the impact of CRISPR technology is increasing in personalized medicine and the diagnosis of genetic diseases. The flexibility offered by CRISPR-Cas systems will allow this technology to be integrated into new diagnostic platforms and find a broader range of applications.
3. Future Perspectives and Challenges
3.1. The Future Role of Molecular Diagnostic Methods
In the future, the integration of artificial intelligence (AI) and machine learning (ML) technologies into the analysis of molecular diagnostic data will make diagnostic processes faster and more accurate. Especially in the analysis of NGS and metagenomic data, where large amounts of data are generated, AI and ML algorithms can contribute in the following ways:
Automation of Data Analysis: By using artificial intelligence in the analysis of large-scale genetic data, it will be possible to rapidly detect and classify pathogens. This significantly reduces diagnostic time and minimizes human error.
Detection of Resistance Genes and Mutations: AI-assisted analyses increase the accuracy in identifying antibiotic resistance and genetic variations of pathogens, enabling more precise results. This allows for the rapid determination of appropriate treatment options.
3.2. The Role of Artificial Intelligence and Machine Learning
The use of molecular diagnostic methods in clinical laboratories enables diagnostic processes to be carried out more rapidly and accurately. In the future, advanced technologies such as PCR, NGS, and CRISPR are expected to become more widespread and accessible. As these technologies become standard practices in healthcare, significant advancements are anticipated in the following areas:
Widespread Use of Rapid Diagnostic Tests: The development of portable and low-cost molecular diagnostic devices will enable the expansion of rapid diagnostic tests in the field and rural areas. This provides a critical advantage, particularly for the early detection of outbreaks and controlling the spread of infectious diseases.
Personalized Medicine Applications: Molecular diagnostic technologies will support personalized medicine approaches, such as genetic profiling and the identification of individual disease risks, allowing for more effective treatment planning.
3.3. Clinical and Ethical Challenges
The widespread use of molecular diagnostic methods also brings certain clinical and ethical challenges.
Cost and Infrastructure Requirements: The need for high-cost equipment and technical infrastructure limits the use of these technologies in low- and middle-income countries. In developing countries, innovative solutions should be developed to reduce costs and promote the widespread use of portable devices.
Detection of Resistance Genes and Mutations: Comprehensive genetic analyses such as NGS can result in the acquisition of sensitive information about individuals' genetic data. This raises ethical concerns and issues related to data security. Therefore, legal regulations should be developed to ensure the protection and ethical use of genetic data.
3.4. Development of Portable and Low-Cost Diagnostic Devices
In recent years, the development of low-cost molecular diagnostic systems, such as portable PCR and NGS devices, has made it possible to conduct diagnostics in field conditions. These innovations will play a significant role, especially in monitoring and controlling infectious disease outbreaks. In the future, the widespread adoption of such devices and further reductions in costs are expected.
3.5. The Future of CRISPR-Based Diagnostic Systems
The use of CRISPR technology for diagnostic purposes holds great potential, particularly for the rapid and sensitive detection of DNA and RNA targets. In the future, CRISPR-based diagnostic systems are expected to further develop and become more widespread. These innovative diagnostic methods could have a significant impact in the following areas:
Personalized Medicine: Diagnostic and treatment approaches tailored to patients' genetic profiles could be offered.
New Diagnostic Platforms: CRISPR-based systems can enable the development of portable and low-cost diagnostic devices, providing ideal solutions for rapid diagnostics in field conditions.
4. Conclusions
Molecular diagnostic methods have significantly advanced the diagnosis and management of infectious diseases, offering rapid, sensitive, and specific detection capabilities that surpass traditional techniques. The emergence of technologies such as PCR, NGS, and CRISPR has transformed clinical microbiology by enabling the early identification of pathogens, detection of antimicrobial resistance, and monitoring of genetic variations. As these methods continue to evolve, they are becoming more accessible and practical, with the development of portable, low-cost diagnostic devices expanding their use in resource-limited settings. However, the high costs and infrastructure requirements associated with these technologies, along with the need for skilled personnel, still pose challenges, particularly in low- and middle-income countries. Innovative solutions to reduce costs and simplify the technology are essential to overcoming these barriers and promoting global health equity.
Future directions in molecular diagnostics will likely involve the integration of artificial intelligence and machine learning for the automated analysis of complex datasets generated by technologies such as NGS and metagenomics. This approach could significantly enhance the accuracy and speed of diagnostic processes, while also aiding in the identification of resistance genes and novel pathogens. Moreover, ethical considerations related to genetic data privacy and the use of sensitive information must be addressed through the establishment of appropriate regulatory frameworks. As CRISPR-based diagnostics mature, they could further revolutionize the field, providing personalized medicine solutions and rapid, field-deployable testing for emerging infectious diseases. Together, these advancements hold the promise of more effective disease control, improved patient outcomes, and the potential to transform global health practices.
Author Contributions
Conceptualization, A.N.S. and S.S.; validation, S.S., Y.Y. and Z.Z.; resources, S.S.; writing—original draft preparation, A.N.S., T.G.T. and A.M.; writing—review and editing, S.S. and T.G.T.; visualization, T.G.T.; supervision, A.M.; funding acquisition, A.N.S. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was funded by Libyan Academy Attaché.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study.
Acknowledgments
Figures were created with BioRender.com. We thank BioRender for providing the platform to produce scientific illustrations.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Global Tuberculosis Report 2018. Available online: https://www.who.int/tb/publications/global_report/en/ (accessed on 30 October 2018).
- Schubert, A.M.; Kostrzynska, M. Advances in the molecular-based techniques for the detection of bacterial pathogens. J. Microbiol. Methods 2001, 47, 235–252. [Google Scholar]
- Lamb, L.E.; Bartolone, S.N.; Ward, E.; Chancellor, M.B. Rapid detection of novel coronavirus (COVID-19) by reverse transcription-loop-mediated isothermal amplification. PLoS ONE 2020, 15, e0234682. [Google Scholar] [CrossRef] [PubMed]
- Mullis, K.; Faloona, F.; Scharf, S.; Saiki, R.; Horn, G.; Erlich, H. Specific enzymatic amplification of DNA in vitro: The polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 1986, 51, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef]
- Dramé, M.; Tabue Teguo, M.; Proye, E.; Hequet, F.; Hentzien, M.; Kanagaratnam, L.; Godaert, L. Should RT-PCR be considered a gold standard in the diagnosis of COVID-19? J. Med. Virol. 2020, 92, 2312–2313. [Google Scholar] [CrossRef]
- Metzker, M.L. Sequencing technologies—the next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef]
- Didelot, X.; Bowden, R.; Wilson, D.J.; Peto, T.E.A.; Crook, D.W. Transforming clinical microbiology with bacterial genome sequencing. Nat. Rev. Genet. 2012, 13, 601–612. [Google Scholar] [CrossRef]
- Binnie, A.; Fernandes, E.; Almeida-Lousada, H.; de Mello, R.A.; Castelo-Branco, P. CRISPR-based strategies in infectious disease diagnosis and therapy. Infection 2021, 49, 377–385. [Google Scholar] [CrossRef]
- Kostyusheva, A.; Brezgin, S.; Babin, Y.; Vasilyeva, I.; Glebe, D.; Kostyushev, D.; Chulanov, V. CRISPR-Cas systems for diagnosing infectious diseases. Methods 2022, 203, 431–446. [Google Scholar] [CrossRef]
- Petti, C.A.; Polage, C.R.; Quinn, T.C.; Ronald, A.R.; Sande, M.A. Laboratory Medicine in Africa: A Barrier to Effective Health Care. Clin. Infect. Dis. 2006, 42, 377–382. [Google Scholar] [CrossRef]
- Kadja, T.; Liu, C.; Sun, Y.; Chodavarapu, V.P. Low-Cost, Real-Time Polymerase Chain Reaction System for Point-of-Care Medical Diagnosis. Sensors 2022, 22, 2320. [Google Scholar] [CrossRef] [PubMed]
- Sebastião, C.S.; Pingarilho, M.; Bathy, J.; Bonfim, E.; Toancha, K.; Miranda, M.N.; Martins, M.R.O.; Gomes, P.; Lázaro, L.; Pina-Araujo, I.; et al. MARVEL-minimising the emergence and dissemination of HIV-1 drug resistance in Portuguese-speaking African Countries (PALOP): low-cost portable NGS platform for HIV-1 surveillance in Africa. BMC Infect. Dis. 2024, 24, 1–8. [Google Scholar] [CrossRef]
- Schmitz, J.E.; Stratton, C.W.; Persing, D.H.; Tang, Y.-W. Forty Years of Molecular Diagnostics for Infectious Diseases. J. Clin. Microbiol. 2022, 60, e0244621. [Google Scholar] [CrossRef] [PubMed]
- Krishna, N.K.; Cunnion, K.M. Role of Molecular Diagnostics in the Management of Infectious Disease Emergencies. Med Clin. North Am. 2012, 96, 1067–1078. [Google Scholar] [CrossRef]
- Yang, S.; E Rothman, R. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 2004, 4, 337–348. [Google Scholar] [CrossRef]
- Espy, M.J.; Uhl, J.R.; Sloan, L.M.; Buckwalter, S.P.; Jones, M.F.; Vetter, E.A.; Yao, J.D.C.; Wengenack, N.L.; Rosenblatt, J.E.; Cockerill, F.R.; et al. Real-Time PCR in Clinical Microbiology: Applications for Routine Laboratory Testing. Clin. Microbiol. Rev. 2006, 19, 165–256. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Bennett, B.; Sullivan, T.; Parker, M.M.; Heffelfinger, J.D.; Sullivan, P.S. Rapid HIV screening: Missed opportunities for HIV diagnosis and prevention. J. Clin. Virol. 2012, 54, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Zauli, D.A.G.; de Menezes, C.L.P.; de Oliveira, C.L.; Mateo, E.C.C.; Ferreira, A.C.d.S. In-house quantitative real-time PCR for the diagnosis of hepatitis B virus and hepatitis C virus infections. Braz. J. Microbiol. 2016, 47, 987–992. [Google Scholar] [CrossRef]
- Niemz, A.; Ferguson, T.M.; Boyle, D.S. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011, 29, 240–250. [Google Scholar] [CrossRef]
- Jung, R.; Soondrum, K.; Neumaier, M. Quantitative PCR. Clin. Chem. Lab. Med. 2000, 38, 833–836. [Google Scholar] [CrossRef]
- Ahmadzadeh, M.; Vahidi, H.; Mahboubi, A.; Hajifathaliha, F.; Nematollahi, L.; Mohit, E. Different Respiratory Samples for COVID-19 Detection by Standard and Direct Quantitative RT-PCR: A Literature Review. Iran J. Pharm. Res. 2021, 20, 285–299. [Google Scholar] [PubMed]
- de Freitas, V.L.T.; Novaes, C.T.G.; Sartori, A.M.C.; Carvalho, N.B.; da Silva, S.C.V.; Nakanishi. S.; Salvador, F.; de Castro, C.N.; Bezerra, R.C.; Westphalen, E.V.N.; et al. Quantitative PCR as a marker for preemptive therapy and its role in therapeutic control in Trypanosoma cruzi/HIV coinfection. PLOS Neglected Trop. Dis. 2024, 18, e0011961. [Google Scholar] [CrossRef] [PubMed]
- Markoulatos, P.; Siafakas, N.; Moncany, M. Multiplex polymerase chain reaction: A practical approach. J. Clin. Lab. Anal. 2002, 16, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, C.; Hirsch, H.H. Comparing Luminex NxTAG-Respiratory Pathogen Panel and RespiFinder-22 for multiplex detection of respiratory pathogens. J. Med. Virol. 2016, 88, 1319–1324. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Alvarez, S.; Pérez-Roth, E. La PCR múltiple en microbiología clínica [Multiplex PCR in clinical microbiology]. Enferm. Infecc. Microbiol. Clin. 2004, 22, 183–191. [Google Scholar] [CrossRef]
- Kuypers, J.; Jerome, K.R. Applications of Digital PCR for Clinical Microbiology. J. Clin. Microbiol. 2017, 55, 1621–1628. [Google Scholar] [CrossRef]
- Bartlett, J.M.; Stirling, D. A short history of the polymerase chain reaction. Methods Mol. Biol. 2003, 226, 3–6. [Google Scholar]
- Mackay, I. Real-time PCR in the microbiology laboratory. Clin. Microbiol. Infect. 2004, 10, 190–212. [Google Scholar] [CrossRef]
- Harsh; Tripathi, P. Medical viruses: diagnostic techniques. Virol. J. 2023, 20, 1–13. [Google Scholar] [CrossRef]
- Villar, L.M.; Cruz, H.M.; Barbosa, J.R.; Bezerra, C.S.; Portilho, M.M.; Scalioni, L.d.P. Update on hepatitis B and C virus diagnosis. World J. Virol. 2015, 4, 323–342. [Google Scholar] [CrossRef]
- Chakravarty, R. Diagnosis and monitoring of chronic viral hepatitis serologic and molecular markers. Front. Biosci. 2011, S3, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Constantine, N.; Zhao, R. Molecular-based laboratory testing and monitoring for human immunodeficiency virus infections. . 2005, 18, 263–70. [Google Scholar]
- Tsang, H.-F.; Chan, L.W.-C.; Tong, J.C.-H.; Wong, H.-T.; Lai, C.K.-C.; Au, T.C.-C.; Chan, A.K.-C.; Ng, L.P.-W.; Cho, W.C.-S.; Wong, S.-C.C. Implementation and new insights in molecular diagnostics for HIV infection. Expert Rev. Mol. Diagn. 2018, 18, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Sule, W.F.; Oluwayelu, D.O. Real-time RT-PCR for COVID-19 diagnosis: challenges and prospects. Pan Afr. Med. J. 2020, 35, 121. [Google Scholar] [CrossRef]
- van Kasteren, P.B.; van Der Veer, B.; van den Brink, S.; Wijsman, L.; de Jonge, J.; van den Brandt, A.; Molenkamp, R.; Reusken, C.B.E.M.; Meijer, A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020, 128, 104412. [Google Scholar] [CrossRef]
- Järvinen, A.-K.; Laakso, S.; Piiparinen, P.; Aittakorpi, A.; Lindfors, M.; Huopaniemi, L.; Piiparinen, H.; Mäki, M. Rapid identification of bacterial pathogens using a PCR- and microarray-based assay. BMC Microbiol. 2009, 9, 161–161. [Google Scholar] [CrossRef]
- Trung, N.T.; Thau, N.S.; Bang, M.H.; Song, L.H. PCR-based Sepsis@Quick test is superior in comparison with blood culture for identification of sepsis-causative pathogens. Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Broccolo, F.; Scarpellini, P.; Locatelli, G.; Zingale, A.; Brambilla, A.M.; Cichero, P.; Sechi, L.A.; Lazzarin, A.; Lusso, P.; Malnati, M.S. Rapid Diagnosis of Mycobacterial Infections and Quantitation of Mycobacterium tuberculosis Load by Two Real-Time Calibrated PCR Assays. J. Clin. Microbiol. 2003, 41, 4565–4572. [Google Scholar] [CrossRef]
- Kay, A.; Vasiliu, A.; Carratala-Castro, L.; Mtafya, B.; Reyes, J.E.M.; Maphalala, N.; Munguambe, S.; Mulengwa, D.; Ness, T.; Saavedra, B.; et al. Performance of a stool-based quantitative PCR assay for the diagnosis of tuberculosis in adolescents and adults: a multinational, prospective diagnostic accuracy study. Lancet Microbe 2024, 5, e433–e441. [Google Scholar] [CrossRef]
- Helb, D.; Jones, M.; Story, E.; Boehme, C.; Wallace, E.; Ho, K.; Kop, J.; Owens, M.R.; Rodgers, R.; Banada, P.; Safi, H.; Blakemore, R.; Lan, N.T.; Jones-López, E.C.; Levi, M.; Burday, M.; Ayakaka, I.; Mugerwa, R.D.; McMillan, B.; Winn-Deen, E.; Christel, L.; Dailey, P.; Perkins, M.D.; Persing, D.H.; Alland, D. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 2010, 48, 229–237. [Google Scholar] [CrossRef]
- Anupurba, S.; Sinha, P.; Banerjee, T.; Srivastava, G. Rapid detection of drug-resistant Mycobacterium tuberculosis directly from clinical specimens using allele-specific polymerase chain reaction assay. Indian J. Med Res. 2019, 150, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Klingspor, L.; Jalal, S. Molecular detection and identification of Candida and Aspergillus spp. from clinical samples using real-time PCR. Clin. Microbiol. Infect. 2006, 12, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Jahanshiri, Z.; Manifar, S.; Arastehnazar, F.; Hatami, F.; Lotfali, E. Azole Resistance in Candida albicans Isolates from Oropharyngeal Candidiasis is Associated with ERG11 Mutation and Efflux Overexpression. Jundishapur J. Microbiol. 2022, 15. [Google Scholar] [CrossRef]
- Scharmann, U.; Kirchhoff, L.; Hain, A.; Buer, J.; Koldehoff, M.; Steinmann, J.; Rath, P.-M. Evaluation of Three Commercial PCR Assays for the Detection of Azole-Resistant Aspergillus fumigatus from Respiratory Samples of Immunocompromised Patients. J. Fungi 2021, 7, 132. [Google Scholar] [CrossRef]
- Gadalla, A.A.H.; Siciliano, G.; Farid, R.; Alano, P.; Ranford-Cartwright, L.; McCarthy, J.S.; Thompson, J.; A Babiker, H. Real-time PCR assays for detection and quantification of early P. falciparum gametocyte stages. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhu, D.; Wu, K.; He, W.; Li, L.; Li, T.; Liu, L.; Liu, Z.; Song, X.; Cheng, W.; et al. Establishment and evaluation of a qPCR method for the detection of pfmdr1 mutations in Plasmodium falciparum, the causal agent of fatal malaria. Diagn. Microbiol. Infect. Dis. 2024, 110, 116400. [Google Scholar] [CrossRef]
- Sirekbasan, S.; Polat, E. Real-time PCR using high-resolution melting analysis technology for diagnosis of Leishmania and determination of types of clinical samples. Turk. J. Med Sci. 2018, 48, 1358–1363. [Google Scholar] [CrossRef]
- Wu, Y.; Tian, X.; Song, N.; Huang, M.; Wu, Z.; Li, S.; Waterfield, N.R.; Zhan, B.; Wang, L.; Yang, G. Application of Quantitative PCR in the Diagnosis and Evaluating Treatment Efficacy of Leishmaniasis. Front. Cell. Infect. Microbiol. 2020, 10, 581639. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Soroka, M.; Wasowicz, B.; Rymaszewska, A. Loop-Mediated Isothermal Amplification (LAMP): The Better Sibling of PCR? Cells 2021, 10, 1931. [Google Scholar] [CrossRef]
- Parida, M.; Sannarangaiah, S.; Dash, P.K.; Rao, P.V.L.; Morita, K. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med Virol. 2008, 18, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.D.R.; Balzan, L.D.R.; Inamine, E.; Pancotto, L.R.; Gaboardi, G.; Cantarelli, V.V. Performance of Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) Targeting the RNA Polymerase Gene for the Direct Detection of SARS-CoV2 in Nasopharyngeal Swabs. Int. J. Mol. Sci. 2023, 24, 13056. [Google Scholar] [CrossRef] [PubMed]
- Vanhomwegen, J.; Kwasiborski, A.; Diop, A.; Boizeau, L.; Hoinard, D.; Vray, M.; Bercion, R.; Ndiaye, B.; Dublineau, A.; Michiyuki, S.; et al. Development and clinical validation of loop-mediated isothermal amplification (LAMP) assay to diagnose high HBV DNA levels in resource-limited settings. Clin. Microbiol. Infect. 2021, 27, 1858–e9. [Google Scholar] [CrossRef]
- Puri, M.; Brar, H.K.; Madan, E.; Srinivasan, R.; Rawat, K.; Gorthi, S.S.; Kumari, G.; Sah, R.; Ojha, S.B.; Panigrahi, S.; et al. Rapid diagnosis of Plasmodium falciparum malaria using a point-of-care loop-mediated isothermal amplification device. Front. Cell. Infect. Microbiol. 2022, 12, 961832. [Google Scholar] [CrossRef]
- Kumar, S.; Sharma, S.; Bhardwaj, N.; Pande, V.; Savargaonkar, D.; Anvikar, A.R. Advanced Lyophilised Loop Mediated Isothermal Amplification (L-LAMP) based point of care technique for the detection of dengue virus. J. Virol. Methods 2021, 293, 114168. [Google Scholar] [CrossRef]
- da Silva, S.J.R.; Pardee, K.; Balasuriya, U.B.R.; Pena, L. Development and validation of a one-step reverse transcription loop-mediated isothermal amplification (RT-LAMP) for rapid detection of ZIKV in patient samples from Brazil. Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef]
- Werbajh, S.; Larocca, L.; Carrillo, C.; Stolowicz, F.; Ogas, L.; Pallotto, S.; Cassará, S.; Mammana, L.; Zapiola, I.; Bouzas, M.B.; Vojnov, A.A. Colorimetric RT-LAMP Detection of Multiple SARS-CoV-2 Variants and Lineages of Concern Direct from Nasopharyngeal Swab Samples without RNA Isolation. Viruses 2023, 15, 1910. [Google Scholar] [CrossRef] [PubMed]
- Alhamid, G.; Tombuloglu, H.; Al-Suhaimi, E. Development of loop-mediated isothermal amplification (LAMP) assays using five primers reduces the false-positive rate in COVID-19 diagnosis. Sci. Rep. 2023, 13, 1–13. [Google Scholar] [CrossRef]
- Borah Slater, K.; Ahmad, M.; Poirier, A.; Stott, A.; Siedler, B.S.; Brownsword, M.; Mehat, J.; Urbaniec, J.; Locker, N.; Zhao, Y.; La Ragione, R.; Silva, S.R.P.; McFadden, J. Development of a loop-mediated isothermal amplification (LAMP)-based electrochemical test for rapid detection of SARS-CoV-2. iScience 2023, 26, 107570. [Google Scholar] [CrossRef]
- Mori, Y.; Nagamine, K.; Tomita, N.; Notomi, T. Detection of Loop-Mediated Isothermal Amplification Reaction by Turbidity Derived from Magnesium Pyrophosphate Formation. Biochem. Biophys. Res. Commun. 2001, 289, 150–154. [Google Scholar] [CrossRef]
- Srivastava, P.; Prasad, D. Isothermal nucleic acid amplification and its uses in modern diagnostic technologies. 3 Biotech 2023, 13, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Rodino, K.G.; Simner, P.J. Status check: next-generation sequencing for infectious-disease diagnostics. J. Clin. Investig. 2023, 134. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.E.; Myers, R.M. Advancements in Next-Generation Sequencing. Annu. Rev. Genom. Hum. Genet. 2016, 17, 95–115. [Google Scholar] [CrossRef] [PubMed]
- Gwinn, M.; MacCannell, D.; Armstrong, G.L. Next-Generation Sequencing of Infectious Pathogens. JAMA 2019, 321, 893–894. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef]
- Zhou, X.; Yang, M.; Chen, F.; Wang, L.; Han, P.; Jiang, Z.; Shen, S.; Rao, G.; Yang, F. Prediction of antimicrobial resistance in Klebsiella pneumoniae using genomic and metagenomic next-generation sequencing data. J. Antimicrob. Chemother. 2024, 79, 2509–2517. [Google Scholar] [CrossRef]
- John, G.; Sahajpal, N.S.; Mondal, A.K.; Ananth, S.; Williams, C.; Chaubey, A.; Rojiani, A.M.; Kolhe, R. Next-Generation Sequencing (NGS) in COVID-19: A Tool for SARS-CoV-2 Diagnosis, Monitoring New Strains and Phylodynamic Modeling in Molecular Epidemiology. Curr. Issues Mol. Biol. 2021, 43, 845–867. [Google Scholar] [CrossRef]
- Gu, W.; Miller, S.; Chiu, C.Y. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 319–338. [Google Scholar] [CrossRef] [PubMed]
- Brenner, T.; Decker, S.O.; Grumaz, S.; Stevens, P.; Bruckner, T.; Schmoch, T.; Pletz, M.W.; Bracht, H.; Hofer, S.; Marx, G.; Weigand, M.A.; Sohn, K. ; TIFOnet Critical Care Trials Group. Next-generation sequencing diagnostics of bacteremia in sepsis (Next GeneSiS-Trial): Study protocol of a prospective, observational, noninterventional, multicenter, clinical trial. Medicine (Baltimore) 2018, 97, e9868. [Google Scholar]
- Wilson, M.R.; Sample, H.A.; Zorn, K.C.; Arevalo, S.; Yu, G.; Neuhaus, J.; Federman, S.; Stryke, D.; Briggs, B.; Langelier, C.; et al. Clinical Metagenomic Sequencing for Diagnosis of Meningitis and Encephalitis. New Engl. J. Med. 2019, 380, 2327–2340. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Miller, S.A. Clinical metagenomics. Nat. Rev. Genet. 2019, 20, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Ji, S.; Koh, H.R. CRISPR as a Diagnostic Tool. Biomolecules 2021, 11, 1162. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, S.; Khanbabaei, H.; Abbasi, S.; Fani, M.; Soltani, S.; Zandi, M.; Najafimemar, Z. CRISPR-Cas System: A Promising Diagnostic Tool for Covid-19. Avicenna J. Med Biotechnol. 2022, 14, 3–9. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).