Submitted:
04 November 2024
Posted:
06 November 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Epidemiology
1.2. Etiology
1.3. Treatment
1.4. Prognosis
2. Diagnosis of Bladder Cancer
Classic Approach
Epigenetic Biomarkers in the Diagnosis of BC
2.1. Available Tests Based on Analysis of Epigenetic Changes for BC Diagnosis
2.2. Epigenetic Biomarkers in BC – Cohort Studies
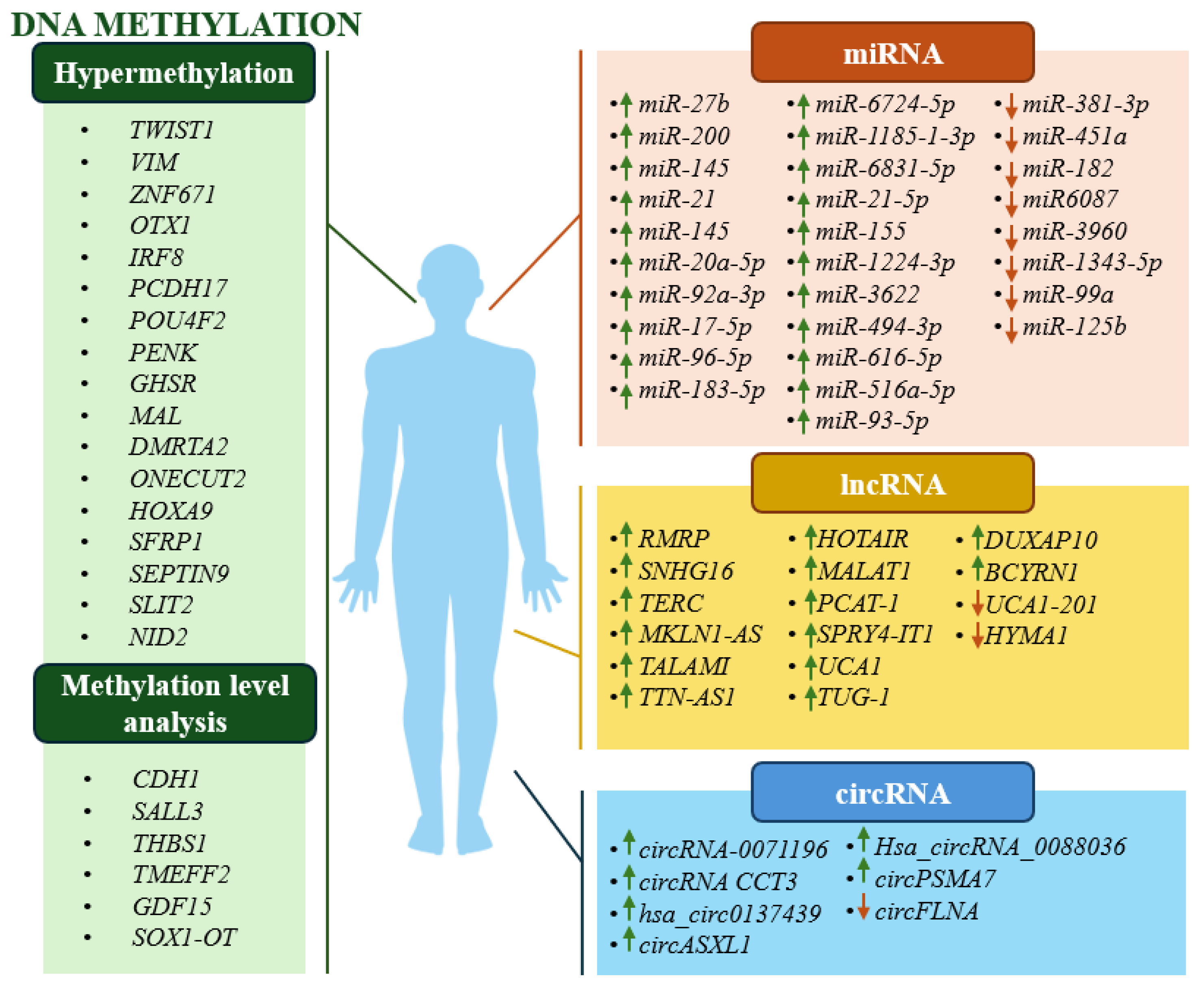
2.3. Potential Epigenetic Biomarkers in BC (Bioinformatics Analyses and Cell Line Studies)
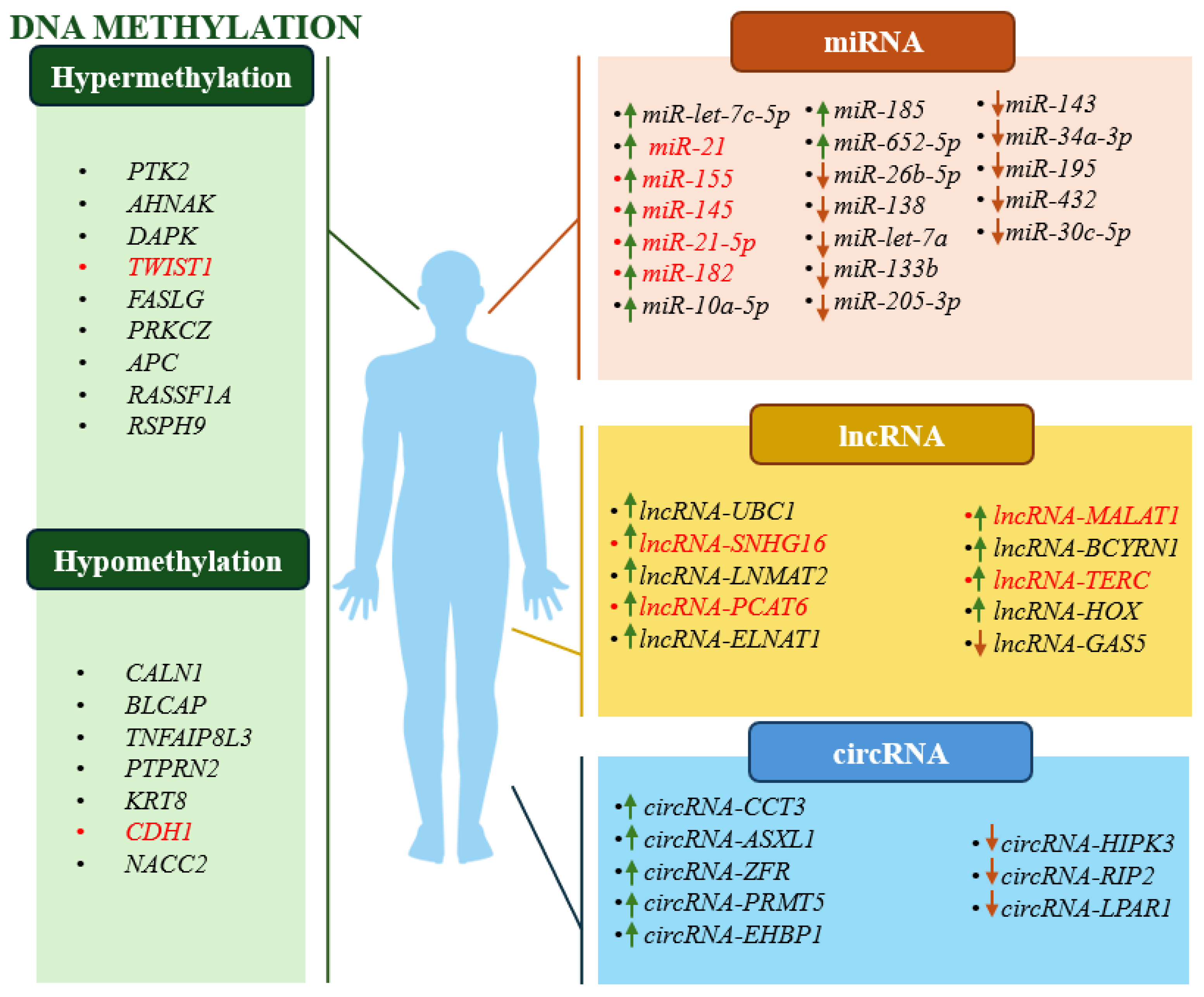
3. Epigenetic Biomarkers in Clinical Prognosis of BC
3.1. DNA Methylation Biomarkers – Cohort Studies
3.2. ncRNAs Biomarkers – Cohort Studies
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Freedman, N.D.; Silverman, D.T.; Hollenbeck, A.R.; Schatzkin, A.; Abnet, C.C. Association between smoking and risk of bladder cancer among men and women. Jama 2011, 306, 737–745. [Google Scholar] [CrossRef] [PubMed]
- International Agency for Research on Cancer. List of classifications by cancer sites with sufficient or limited evidence in humans. IARC monographs volumes 1–132. July 1, 2022. Available online: https://monographs. iarc.who.int/wp-content/uploads/2019/07/Classifications_by_cancer_ site.pdf.
- Dobruch, J.; Oszczudłowski, M. Bladder cancer: current challenges and future directions. Medicina 2021, 57, 749. [Google Scholar] [CrossRef] [PubMed]
- Henning, G.M.; Barashi, N.S.; Smith, Z.L. Advances in biomarkers for detection, surveillance, and prognosis of bladder cancer. Clinical Genitourinary Cancer 2021, 19, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Kluth, L.A.; Black, P.C.; Bochner, B.H.; Catto, J.; Lerner, S.P.; Stenzl, A.; Shariat, S.F. Prognostic and prediction tools in bladder cancer: a comprehensive review of the literature. European urology 2015, 68, 238–253. [Google Scholar] [CrossRef]
- Gilyazova, I.; Enikeeva, K.; Rafikova, G.; Kagirova, E.; Sharifyanova, Y.; Asadullina, D.; Pavlov, V. Epigenetic and Immunological Features of Bladder Cancer. International Journal of Molecular Sciences 2023, 24, 9854. [Google Scholar] [CrossRef]
- Lobo, N.; Afferi, L.; Moschini, M.; Mostafid, H.; Porten, S.; Psutka, S.P.; Lotan, Y. Epidemiology, screening, and prevention of bladder cancer. European urology oncology 2022, 5, 628–639. [Google Scholar] [CrossRef]
- Dyrskjøt, L.; Hansel, D.E.; Efstathiou, J.A.; Knowles, M.A.; Galsky, M.D.; Teoh, J.; Theodorescu, D. Bladder cancer. Nature Reviews Disease Primers 2023, 9, 58. [Google Scholar] [CrossRef]
- Hurst, C.D.; Alder, O.; Platt, F.M.; Droop, A.; Stead, L.F.; Burns, J.E.; Knowles, M.A. Genomic subtypes of non-invasive bladder cancer with distinct metabolic profile and female gender bias in KDM6A mutation frequency. Cancer cell 2017, 32, 701–715. [Google Scholar] [CrossRef]
- Ségal-Bendirdjian, E.; Geli, V. Non-canonical roles of telomerase: unraveling the imbroglio. Frontiers in Cell and Developmental Biology 2019, 7, 332. [Google Scholar] [CrossRef]
- Agarwal, N.; Rinaldetti, S.; Cheikh, B.B.; Zhou, Q.; Hass, E.P.; Jones, R.T.; Theodorescu, D. TRIM28 is a transcriptional activator of the mutant TERT promoter in human bladder cancer. Proceedings of the National Academy of Sciences 2021, 118, e2102423118. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, E.; L’hôte, C.G.; Kennedy, W.; Tomlinson, D.C.; Knowles, M.A. Mutant fibroblast growth factor receptor 3 induces intracellular signaling and cellular transformation in a cell type-and mutation-specific manner. Oncogene 2009, 28, 4306–4316. [Google Scholar] [CrossRef] [PubMed]
- Hurst, C.D.; Cheng, G.; Platt, F.M.; Castro, M.A.; Nour-Al-Dain, S.M.; Eriksson, P.; Knowles, M.A. Stage-stratified molecular profiling of non-muscle-invasive bladder cancer enhances biological, clinical, and therapeutic insight. Cell Reports Medicine 2021, 2. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Stratton, M.R. ALEXANDROV, Ludmil, B.; et al. Erratum: Signatures of mutational processes in human cancer (Nature (2013) 500 (415-421. Nature 2013, 502, 258. [Google Scholar] [CrossRef]
- Robertson, A.G.; Kim, J.; Al-Ahmadie, H.; Bellmunt, J.; Guo, G.; Cherniack, A.D.; Tam, A. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 2017, 171, 540–556. [Google Scholar] [CrossRef]
- Li, Q.; Damish, A.W.; Frazier, Z.; Liu, D.; Reznichenko, E.; Kamburov, A.; Mouw, K.W. ERCC2 helicase domain mutations confer nucleotide excision repair deficiency and drive cisplatin sensitivity in muscle-invasive bladder cancer. Clinical Cancer Research 2019, 25, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.V.; Hurst, C.D.; Knowles, M.A. Oncogenic FGFR3 gene fusions in bladder cancer. Human molecular genetics 2013, 22, 795–803. [Google Scholar] [CrossRef]
- Huan, J.; Grivas, P.; Birch, J.; Hansel, D.E. Emerging roles for mammalian target of rapamycin (mTOR) complexes in bladder cancer progression and therapy. Cancers 2022, 14, 1555. [Google Scholar] [CrossRef] [PubMed]
- Goriki, A.; Seiler, R.; Wyatt, A.W.; Contreras-Sanz, A.; Bhat, A.; Matsubara, A.; Black, P.C. Unravelling disparate roles of NOTCH in bladder cancer. Nature Reviews Urology 2018, 15, 345–357. [Google Scholar] [CrossRef]
- Li, S.; Xin, K.; Pan, S.; Wang, Y.; Zheng, J.; Li, Z.; Chen, X. Blood-based liquid biopsy: Insights into early detection, prediction, and treatment monitoring of bladder cancer. Cellular & Molecular Biology Letters 2023, 28, 28. [Google Scholar]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. verview of microRNA biogenesis, mechanisms of actions, and circulation. Frontiers in endocrinology 2018, 9, 402. [Google Scholar] [CrossRef]
- Robinson, E.K.; Covarrubias, S.; Carpenter, S. The how and why of lncRNA function: an innate immune perspective. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 2020, 1863, 194419. [Google Scholar] [CrossRef]
- Chen, L.L.; Yang, L. Regulation of circRNA biogenesis. RNA biology 2015, 12, 381–388. [Google Scholar] [CrossRef]
- Suzuki, H.; Tsukahara, T. A view of pre-mRNA splicing from RNase R resistant RNAs. International journal of molecular sciences 2014, 15, 9331–9342. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Jiang, J.; Wu, C. Function and clinical significance of circRNAs in solid tumors. Journal of hematology & oncology 2018, 11, 1–20. [Google Scholar]
- Poletajew, S.; Krajewski, W.; Kaczmarek, K.; Kopczyński, B.; Stamirowski, R.; Tukiendorf, A.; Radziszewski, P. The learning curve for transurethral resection of bladder tumour: how many is enough to be independent, safe and effective surgeon? Journal of Surgical Education 2020, 77, 978–985. [Google Scholar] [CrossRef]
- Morales, A.; Eidinger, D.; Bruce, A.W. Intracavitary Bacillus Calmette-Guerin in the treatment of superficial bladder tumors. The Journal of urology 1976, 116, 180–182. [Google Scholar] [CrossRef]
- Babjuk, M.; Burger, M.; Capoun, O.; Cohen, D.; Compérat, E.M.; Escrig, J.L.D.; Sylvester, R.J. European Association of Urology guidelines on non–muscle-invasive bladder cancer (Ta, T1, and carcinoma in situ). European urology 2022, 81, 75–94. [Google Scholar] [CrossRef]
- Huncharek, M.; Geschwind, J.F.; Witherspoon, B.; McGarry, R.; Adcock, D. Intravesical chemotherapy prophylaxis in primary superficial bladder cancer: a meta-analysis of 3703 patients from 11 randomized trials. Journal of clinical epidemiology 2000, 53, 676–680. [Google Scholar] [CrossRef] [PubMed]
- Witjes, J.A.; Bruins, H.M.; Cathomas, R.; Compérat, E.M.; Cowan, N.C.; Gakis, G.; van Der Heijden, A.G. European Association of Urology guidelines on muscle-invasive and metastatic bladder cancer: summary of the 2020 guidelines. European urology 2021, 79, 82–104. [Google Scholar] [CrossRef] [PubMed]
- Compérat, E.; Amin, M.B.; Cathomas, R.; Choudhury, A.; De Santis, M.; Kamat, A.; Witjes, J.A. Current best practice for bladder cancer: a narrative review of diagnostics and treatments. The Lancet 2022, 400, 1712–1721. [Google Scholar] [CrossRef]
- De Santis, M.; Bellmunt, J.; Mead, G.; Kerst, J. M.; Leahy, M.; Maroto, P.; Gil, T.; Marreaud, S.; Daugaard, G.; Skoneczna, I.; Collette, S.; Lorent, J.; de Wit, R.; Sylvester, R. Randomized phase II/III trial assessing gemcitabine/carboplatin and methotrexate/carboplatin/vinblastine in patients with advanced urothelial cancer who are unfit for cisplatin-based chemotherapy: EORTC study 30986. J Clin Oncol. 2012, 30, 191–9. [Google Scholar] [CrossRef]
- Loriot, Y.; Necchi, A.; Park, S.H.; Garcia-Donas, J.; Huddart, R.; Burgess, E.; Siefker-Radtke, A.O. Erdafitinib in locally advanced or metastatic urothelial carcinoma. New England Journal of Medicine 2019, 381, 338–348. [Google Scholar] [CrossRef]
- Powles, T.; Rosenberg, J.E.; Sonpavde, G.P.; Loriot, Y.; Durán, I.; Lee, J.L.; Petrylak, D.P. Enfortumab vedotin in previously treated advanced urothelial carcinoma. New England Journal of Medicine 2021, 384, 1125–1135. [Google Scholar] [CrossRef]
- Hoimes, C.J.; Flaig, T.W.; Milowsky, M.I.; Friedlander, T.W.; Bilen, M.A.; Gupta, S.; Rosenberg, J.E. Enfortumab vedotin plus pembrolizumab in previously untreated advanced urothelial cancer. Journal of clinical oncology 2023, 41, 22–31. [Google Scholar] [CrossRef]
- Tagawa, S.T.; Balar, A.V.; Petrylak, D.P.; Kalebasty, A.R.; Loriot, Y.; Fléchon, A.; Grivas, P. TROPHY-U-01: a phase II open-label study of sacituzumab govitecan in patients with metastatic urothelial carcinoma progressing after platinum-based chemotherapy and checkpoint inhibitors. Journal of Clinical Oncology 2021, 39, 2474–2485. [Google Scholar] [CrossRef]
- Pan, S.; Zhan, Y.; Chen, X.; Wu, B.; Liu, B. Bladder cancer exhibiting high immune infiltration shows the lowest response rate to immune checkpoint inhibitors. Frontiers in Oncology 2019, 9, 1101. [Google Scholar] [CrossRef]
- Zhou, B.; Guo, R. Integrative analysis of genomic and clinical data reveals intrinsic characteristics of bladder urothelial carcinoma progression. Genes 2019, 10, 464. [Google Scholar] [CrossRef]
- Matuszczak, M.; Kiljańczyk, A.; Salagierski, M. A liquid biopsy in bladder Cancer—the current Landscape in urinary biomarkers. International Journal of Molecular Sciences 2022, 23, 8597. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, D.; Mou, S.I.; Sultana, T.; Hosen, M.I.; Faruk, M.O. Identification and validation of prognostic signature genes of bladder cancer by integrating methylation and transcriptomic analysis. Scientific Reports 2024, 14, 368. [Google Scholar] [CrossRef]
- Cancer.gov. Available online: https://www.cancer.gov/types/bladder/survival.
- Lucca, I.; de Martino, M.; Klatte, T.; Shariat, S.F. Novel biomarkers to predict response and prognosis in localized bladder cancer. Urologic Clinics 2015, 42, 225–233. [Google Scholar] [CrossRef]
- Martinez, V.G.; Munera-Maravilla, E.; Bernardini, A.; Rubio, C.; Suarez-Cabrera, C.; Segovia, C.; Paramio, J.M. Epigenetics of bladder cancer: where biomarkers and therapeutic targets meet. Frontiers in Genetics 2019, 10, 1125. [Google Scholar] [CrossRef]
- Hu, X.; Li, G.; Wu, S. Advances in diagnosis and therapy for bladder cancer. Cancers 2022, 14, 3181. [Google Scholar] [CrossRef]
- Veeratterapillay, R.; Gravestock, P.; Nambiar, A.; Gupta, A.; Aboumarzouk, O.; Rai, B.; Heer, R. Time to turn on the blue lights: a systematic review and meta-analysis of photodynamic diagnosis for bladder cancer. European urology open science 2021, 31, 17–27. [Google Scholar] [CrossRef]
- Yafi, F.A.; Brimo, F.; Steinberg, J.; Aprikian, A.G.; Tanguay, S.; Kassouf, W. Prospective analysis of sensitivity and specificity of urinary cytology and other urinary biomarkers for bladder cancer. In: Urologic oncology: seminars and original investigations. Elsevier, 2015. p. 66. e25-66. e31.
- Tomiyama, E.; Fujita, K.; Hashimoto, M.; Uemura, H.; Nonomura, N. Urinary markers for bladder cancer diagnosis: A review of current status and future challenges. International Journal of Urology 2024, 31, 208–219. [Google Scholar] [CrossRef]
- Schmidbauer, J.; Remzi, M.; Klatte, T.; Waldert, M.; Mauermann, J.; Susani, M.; Marberger, M. Fluorescence cystoscopy with high-resolution optical coherence tomography imaging as an adjunct reduces false-positive findings in the diagnosis of urothelial carcinoma of the bladder. European urology 2009, 56, 914–919. [Google Scholar] [CrossRef]
- Li, Q.; Tang, J.; He, E.; Li, Y.; Zhou, Y.; Wang, B. Differentiation between high-and low-grade urothelial carcinomas using contrast enhanced ultrasound. Oncotarget 2017, 8, 70883. [Google Scholar] [CrossRef]
- Stav, K.; Leibovici, D.; Goren, E.; Livshitz, A.; Siegel, Y.I.; Lindner, A.; Zisman, A. Adverse effects of cystoscopy and its impact on patients’ quality of life and sexual performance. Isr Med Assoc J 2004, 6, 474–8. [Google Scholar]
- Witjes, J.A.; Morote, J.; Cornel, E.B.; Gakis, G.; van Valenberg, F.J.P.; Lozano, F.; Leibovitch, I. Performance of the bladder EpiCheck™ methylation test for patients under surveillance for non–muscle-invasive bladder cancer: results of a multicenter, prospective, blinded clinical trial. European urology oncology 2018, 1, 307–313. [Google Scholar] [CrossRef]
- Feber, A.; Dhami, P.; Dong, L.; de Winter, P.; Tan, W.S.; Martínez-Fernández, M.; Kelly, J.D. UroMark—a urinary biomarker assay for the detection of bladder cancer. Clinical epigenetics 2017, 9, 1–10. [Google Scholar] [CrossRef]
- Pharo, H.D.; Jeanmougin, M.; Ager-Wick, E.; Vedeld, H.M.; Sørbø, A.K.; Dahl, C.; Lind, G.E. BladMetrix: a novel urine DNA methylation test with high accuracy for detection of bladder cancer in hematuria patients. Clinical Epigenetics 2022, 14, 115. [Google Scholar] [CrossRef]
- Piatti, P.; Chew, Y.C.; Suwoto, M.; Yamada, T.; Jara, B.; Jia, X.Y.; Liang, G. Clinical evaluation of Bladder CARE, a new epigenetic test for bladder cancer detection in urine samples. Clinical epigenetics 2021, 13, 1–13. [Google Scholar] [CrossRef]
- Bang, B.R.; Zhong, J.; Oh, T.J.; Lee, J.Y.; Seo, Y.; Woo, M.A.; An, S. EarlyTect BCD, a Streamlined PENK Methylation Test in Urine DNA, Effectively Detects Bladder Cancer in Patients with Hematuria. The Journal of Molecular Diagnostics, 2024. [Google Scholar]
- Steinbach, D.; Kaufmann, M.; Hippe, J.; Gajda, M.; Grimm, M.O. High Detection Rate for Non–Muscle-Invasive Bladder Cancer Using an Approved DNA Methylation Signature Test. Clinical Genitourinary Cancer 2020, 18, 210–221. [Google Scholar] [CrossRef]
- Fang, Q.; Zhang, X.; Nie, Q.; Hu, J.; Zhou, S.; Wang, C. Improved urine DNA methylation panel for early bladder cancer detection. BMC cancer 2022, 22, 237. [Google Scholar] [CrossRef]
- Harsanyi, S.; Novakova, Z.V.; Bevizova, K.; Danisovic, L.; Ziaran, S. Biomarkers of bladder cancer: cell-free DNA, epigenetic modifications and non-coding RNAs. International Journal of Molecular Sciences 2022, 23, 13206. [Google Scholar] [CrossRef]
- Hentschel, A.E.; Beijert, I.J.; Bosschieter, J.; Kauer, P.C.; Vis, A.N.; Lissenberg-Witte, B.I.; Nieuwenhuijzen, J.A. Bladder cancer detection in urine using DNA methylation markers: a technical and prospective preclinical validation. Clinical epigenetics 2022, 14, 19. [Google Scholar] [CrossRef]
- Jiang, Y.H.; Liu, Y.S.; Wei, Y.C.; Jhang, J.F.; Kuo, H.C.; Huang, H.H.; Wang, H.J. Hypermethylation Loci of ZNF671, IRF8, and OTX1 as Potential Urine-Based Predictive Biomarkers for Bladder Cancer. Diagnostics 2024, 14, 468. [Google Scholar] [CrossRef]
- Wu, Y.; Jiang, G.; Zhang, N.; Liu, S.; Lin, X.; Perschon, C.; Xu, J. HOXA9, PCDH17, POU4F2, and ONECUT2 as a urinary biomarker combination for the detection of bladder cancer in Chinese patients with hematuria. European urology focus 2020, 6, 284–291. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, J.; Ruan, W.; Huang, M.; Wang, C.; Wang, H.; Lin, T. Urine DNA methylation assay enables early detection and recurrence monitoring for bladder cancer. The Journal of clinical investigation 2020, 130, 6278–6289. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, X.; Wang, T.; Lu, Y.; Lu, Z.; Wang, T.; Pan, Z. Clinical performance and utility of a noninvasive urine-based methylation biomarker: TWIST1/Vimentin to detect urothelial carcinoma of the bladder. Scientific Reports 2024, 14, 7941. [Google Scholar] [CrossRef]
- Deng, L.; Chao, H.; Deng, H.; Yu, Z.; Zhao, R.; Huang, L.; Zou, H. A novel and sensitive DNA methylation marker for the urine-based liquid biopsies to detect bladder cancer. BMC cancer 2022, 22, 510. [Google Scholar] [CrossRef]
- Ruan, W.; Chen, X.; Huang, M.; Wang, H.; Chen, J.; Liang, Z.; Fan, J.B. A urine-based DNA methylation assay to facilitate early detection and risk stratification of bladder cancer. Clinical Epigenetics 2021, 13, 91. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.Q.; Xiong, G.Y.; Yang, K.W.; Zhang, L.; He, S.M.; Gong, Y.Q.; Zhou, L.Q. Detection of urothelial carcinoma, upper tract urothelial carcinoma, bladder carcinoma, and urothelial carcinoma with gross hematuria using selected urine-DNA methylation biomarkers: A prospective, single-center study. In: Urologic Oncology: Seminars and Original Investigations. Elsevier, 2018. p. 342. e15-342. e23.
- Roperch, J.P.; Grandchamp, B.; Desgrandchamps, F.; Mongiat-Artus, P.; Ravery, V.; Ouzaid, I.; Incitti, R. Promoter hypermethylation of HS3ST2, SEPTIN9 and SLIT2 combined with FGFR3 mutations as a sensitive/specific urinary assay for diagnosis and surveillance in patients with low or high-risk non-muscle-invasive bladder cancer. BMC cancer 2016, 16, 1–9. [Google Scholar] [CrossRef]
- Yeh, C.M.; Chen, P.C.; Hsieh, H.Y.; Jou, Y.C.; Lin, C.T.; Tsai, M.H.; Chan, M.W. Methylomics analysis identifies ZNF671 as an epigenetically repressed novel tumor suppressor and a potential non-invasive biomarker for the detection of urothelial carcinoma. Oncotarget 2015, 6, 29555. [Google Scholar] [CrossRef]
- Yegin, Z.; Gunes, S.; Buyukalpelli, R. Hypermethylation of TWIST1 and NID2 in tumor tissues and voided urine in urinary bladder cancer patients. DNA and Cell Biology 2013, 32, 386–392. [Google Scholar] [CrossRef]
- Wang, J.; Peng, X.; Li, R.; Liu, K.; Zhang, C.; Chen, X.; Lai, Y. Evaluation of serum miR-17-92 cluster as noninvasive biomarkers for bladder cancer diagnosis. Frontiers in Oncology 2021, 11, 795837. [Google Scholar] [CrossRef]
- Wang, P.; Wei, X.; Qu, X.; Zhu, Y. Potential clinical application of microRNAs in bladder cancer. The Journal of Biomedical Research, 2024. [Google Scholar]
- Yu, Z.; Lu, C.; Lai, Y. A serum miRNAs signature for early diagnosis of bladder cancer. Annals of Medicine 2023, 55, 736–745. [Google Scholar] [CrossRef]
- Suarez-Cabrera, C.; Estudillo, L.; Ramón-Gil, E.; Martínez-Fernández, M.; Peral, J.; Rubio, C.; Dueñas, M. BlaDimiR: a urine-based miRNA score for accurate bladder cancer diagnosis and follow-up. European urology 2022, 82, 663–667. [Google Scholar] [CrossRef]
- El-Shal, A.S.; Shalaby, S.M.; Abouhashem, S.E.; Elbary, E.H.A.; Azazy, S.; Rashad, N.M.; Sarhan, W. Urinary exosomal microRNA-96-5p and microRNA-183-5p expression as potential biomarkers of bladder cancer. Molecular biology reports 2021, 48, 4361–4371. [Google Scholar] [CrossRef]
- Yu, S.; Lu, Z.; Liu, C.; Meng, Y.; Ma, Y.; Zhao, W.; Chen, J. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer research 2010, 70, 6015–6025. [Google Scholar] [CrossRef]
- Li, J.; Fu, H.; Xu, C.; Tie, Y.; Xing, R.; Zhu, J.; Zheng, X. miR-183 inhibits TGF-β1-induced apoptosis by downregulation of PDCD4 expression in human hepatocellular carcinoma cells. BMC cancer 2010, 10, 1–10. [Google Scholar] [CrossRef]
- Myatt, S.S.; Wang, J.; Monteiro, L.J.; Christian, M.; Ho, K.K.; Fusi, L.; Lam, E.W. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer research 2010, 70, 367–377. [Google Scholar] [CrossRef]
- Lin, H.; Shi, X.; Li, H.; Hui, J.; Liu, R.; Chen, Z.; Tan, W. Urinary Exosomal miRNAs as biomarkers of bladder Cancer and experimental verification of mechanism of miR-93-5p in bladder Cancer. Bmc Cancer 2021, 21, 1–17. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Fujita, K.; Jingushi, K.; Kawashima, A.; Ujike, T.; Nagahara, A.; Nonomura, N. MiR-21-5p in urinary extracellular vesicles is a novel biomarker of urothelial carcinoma. Oncotarget 2017, 8, 24668. [Google Scholar] [CrossRef]
- Zhang, D.Z.; Lau, K.M.; Chan, E.S.; Wang, G.; Szeto, C.C.; Wong, K.; Ng, C.F. Cell-free urinary microRNA-99a and microRNA-125b are diagnostic markers for the non-invasive screening of bladder cancer. PloS one 2014, 9, e100793. [Google Scholar] [CrossRef]
- Lu, C.; Lin, S.; Wen, Z.; Sun, C.; Ge, Z.; Chen, W.; Lai, Y. Testing the accuracy of a four serum microRNA panel for the detection of primary bladder cancer: a discovery and validation study. Biomarkers just-accepted: 1-21. 2024. [Google Scholar] [CrossRef]
- Grimaldi, A.M.; Lapucci, C.; Salvatore, M.; Incoronato, M.; Ferrari, M. Urinary miRNAs as a diagnostic tool for bladder cancer: a systematic review. Biomedicines 2022, 10, 2766. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, X.; Zhang, D.; Yu, Y.; Cai, L.; Zhang, C. Long non-coding RNA urothelial carcinoma–associated 1 as a tumor biomarker for the diagnosis of urinary bladder cancer. Tumor Biology 2017, 39, 1010428317709990. [Google Scholar] [CrossRef]
- Zhan, Y.; Du, L.; Wang, L.; Jiang, X.; Zhang, S.; Li, J.; Wang, C. Expression signatures of exosomal long non-coding RNAs in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Molecular Cancer 2018, 17, 1–5. [Google Scholar] [CrossRef]
- Yu, X.; Wang, R.; Han, C.; Wang, Z.; Jin, X. A panel of urinary long non-coding RNAs differentiate bladder cancer from urocystitis. Journal of Cancer 2020, 11, 781. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, X.; Wang, X.; Sun, R.; Li, Y.; Li, J.; Sun, Z. The clinical value of rapidly detecting urinary exosomal lncRNA RMRP in bladder cancer with an RT-RAA-CRISPR/Cas12a method. Clinica Chimica Acta 2024, 562, 119855. [Google Scholar] [CrossRef]
- Chen, C.; Shang, A.; Sun, Z.; Gao, Y.; Huang, J.; Ping, Y. ; Li, D Urinary exosomal long noncoding RNA TERC as a noninvasive diagnostic and prognostic biomarker for bladder urothelial carcinoma. Journal of Immunology Research 2022, 2022, 9038808. [Google Scholar] [CrossRef]
- Yang, Y.; Li, J.; Yao, W.; Zou, G.; Ye, X.; Mo, Q. Diagnostic value of urine cyclic RNA-0071196 for bladder urothelial carcinoma. BMC urology 2024, 24, 1–6. [Google Scholar] [CrossRef]
- Song, Z.; Zhang, Q.; Zhu, J.; Yin, G.; Lin, L.; Liang, C. Identification of urinary hsa_circ_0137439 as a potential biomarker and tumor regulator of bladder cancer. Neoplasma 2020, 67. [Google Scholar] [CrossRef] [PubMed]
- Mamdouh, S.; Sherif, H.; Romeih, M.; Elesaily, K. Urine micro-RNA signature as a potential non-invasive diagnostic biomarker in bladder cancer. Asian Pacific Journal of Cancer Prevention: APJCP 2023, 24, 121. [Google Scholar] [PubMed]
- Zhang, X.F.; Zhang, X.Q.; Chang, Z.X.; Wu, C.C.; Guo, H. microRNA-145 modulates migration and invasion of bladder cancer cells by targeting N-cadherin. Molecular Medicine Reports 2018, 17, 8450–8456. [Google Scholar] [CrossRef]
- Zhang, H.H.; Qi, F.; Cao, Y.H.; Zu, X.B.; Chen, M.F. Expression and clinical significance of microRNA-21, maspin and vascular endothelial growth factor-C in bladder cancer. Oncology letters 2015, 10, 2610–2616. [Google Scholar] [CrossRef]
- Piao, X.M.; Jeong, P.; Kim, Y.H.; Byun, Y.J.; Xu, Y.; Kang, H.W.; Kim, W.J. Urinary cell-free microRNA biomarker could discriminate bladder cancer from benign hematuria. International journal of cancer 2019, 144, 380–388. [Google Scholar] [CrossRef]
- Usuba, W.; Urabe, F.; Yamamoto, Y.; Matsuzaki, J.; Sasaki, H.; Ichikawa, M.; Ochiya, T. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer science 2019, 110, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhao, H.; Qian, X.; Qiu, J. MiR-20a in cell-free urine as a potential diagnostic biomarker for non-muscle invasive bladder cancer: a Chinese population-based study. Int J Clin Exp Med 2018, 11, 209–216. [Google Scholar]
- Urquidi, V.; Netherton, M.; Gomes-Giacoia, E.; Serie, D.J.; Eckel-Passow, J.; Rosser, C.J.; Goodison, S. A microRNA biomarker panel for the non-invasive detection of bladder cancer. Oncotarget 2016, 7, 86290. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Liu, X.; Fang, A.; Wang, J.; Yang, Y.; Wang, C. Direct quantitative detection for cell-free miR-155 in urine: a potential role in diagnosis and prognosis for non-muscle invasive bladder cancer. Oncotarget 2016, 7, 3255. [Google Scholar] [CrossRef] [PubMed]
- Miah, S.; Dudziec, E.; Drayton, R.M.; Zlotta, A.R.; Morgan, S.L.; Rosario, D.J.; Catto, J.W.F. An evaluation of urinary microRNA reveals a high sensitivity for bladder cancer. British journal of cancer 2012, 107, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xu, P.; Shao, S.; Wang, F.; Zheng, Z.; Li, S.; Li, G. The value of urinary exosomal lncRNA SNHG16 as a diagnostic biomarker for bladder cancer. Molecular Biology Reports 2023, 50, 8297–8304. [Google Scholar] [CrossRef]
- Bian, B.; Li, L.; Ke, X.; Chen, H.; Liu, Y.; Zheng, N.; Shen, L. Urinary exosomal long non-coding RNAs as noninvasive biomarkers for diagnosis of bladder cancer by RNA sequencing. Frontiers in Oncology 2022, 12, 976329. [Google Scholar] [CrossRef]
- Sarfi, M.; Abbastabar, M.; Khalili, E. Increased expression of urinary exosomal LnCRNA TUG-1 in early bladder cancer. Gene Reports 2021, 22, 101010. [Google Scholar] [CrossRef]
- Luo, L.; Xie, Q.; Wu, Y.; Li, P.; Qin, F.; Liao, D.; Wang, K. Circular RNA CCT3 is a unique molecular marker in bladder cancer. BMC cancer 2023, 23, 977. [Google Scholar] [CrossRef]
- Tang, G.; Xie, W.; Qin, C.; Zhen, Y.; Wang, Y.; Chen, F.; Hu, H. Expression of circular RNA circASXL1 correlates with TNM classification and predicts overall survival in bladder cancer. International Journal of Clinical and Experimental Pathology 2017, 10, 8495. [Google Scholar] [PubMed]
- Cheng, H.; Liu, Y.; Chen, G. Identification of potential DNA methylation biomarkers related to diagnosis in patients with bladder cancer through integrated bioinformatic analysis. BMC urology 2023, 23, 135. [Google Scholar] [CrossRef]
- Li, T.; Liu, X.; Yang, A.; Fu, W.; Yin, F.; Zeng, X. Associations of tumor suppressor SPARCL1 with cancer progression and prognosis. Oncology Letters 2017, 14, 2603–2610. [Google Scholar] [CrossRef]
- Hu, H.; Zhang, H.; Ge, W.; Liu, X.; Loera, S.; Chu, P.; Zheng, S. Secreted protein acidic and rich in cysteines-like 1 suppresses aggressiveness and predicts better survival in colorectal cancers. Clinical Cancer Research 2012, 18, 5438–5448. [Google Scholar] [CrossRef] [PubMed]
- Esposito, I.; Kayed, H.; Keleg, S.; Giese, T.; Sage, E.H.; Schirmacher, P.; Kleeff, J. Tumor-suppressor function of SPARC-like protein 1/Hevin in pancreatic cancer. Neoplasia 2007, 9, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Turtoi, A.; Musmeci, D.; Naccarato, A.G.; Scatena, C.; Ortenzi, V.; Kiss, R.; Castronovo, V. Sparc-like protein 1 is a new marker of human glioma progression. Journal of proteome research 2012, 11, 5011–5021. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Y.; Wang, Y.X.; Shen, A.; Yang, X.Q.; Liang, C.C.; Huang, R.J.; Yuan, S.Q. Construction of a gene model related to the prognosis of patients with gastric cancer receiving immunotherapy and exploration of COX7A1 gene function. European Journal of Medical Research 2024, 29, 180. [Google Scholar] [CrossRef]
- Shen, H.; Zhang, L.; Zhou, J.; Chen, Z.; Yang, G.; Liao, Y.; Zhu, M. Epidermal growth factor-containing fibulin-like extracellular matrix protein 1 (EFEMP1) acts as a potential diagnostic biomarker for prostate cancer. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research 2017, 23, 216. [Google Scholar] [CrossRef]
- Fu, S.; Luan, T.; Jiang, C.; Huang, Y.; Li, N.; Wang, H.; Wang, J. miR-3622a promotes proliferation and invasion of bladder cancer cells by downregulating LASS2. Gene 2019, 701, 23–31. [Google Scholar] [CrossRef]
- Xu, X.H.; Sun, J.M.; Chen, X.F.; Zeng, X.Y.; Zhou, H.Z. MicroRNA-494-3p facilitates the progression of bladder cancer by mediating the KLF9/RGS2 axis. The Kaohsiung Journal of Medical Sciences 2022, 38, 1070–1079. [Google Scholar] [CrossRef]
- Ren, W.; Hu, J.; Li, H.; Chen, J.; Ding, J.; Zu, X.; Fan, B. miR-616-5p promotes invasion and migration of bladder cancer via downregulating NR2C2 expression. Frontiers in Oncology 2021, 11, 762946. [Google Scholar] [CrossRef]
- Lv, X.Y.; Ma, L.; Chen, J.F.; Yu, R.; Li, Y.; Yan, Z.J.; Ma, Q. Knockdown of DUXAP10 inhibits proliferation and promotes apoptosis in bladder cancer cells via PI3K/Akt/mTOR signaling pathway. International journal of oncology 2017, 52, 288–294. [Google Scholar] [CrossRef]
- Guo, H.; German, P.; Bai, S.; Barnes, S.; Guo, W.; Qi, X.; Ding, Z. The PI3K/AKT pathway and renal cell carcinoma. Journal of genetics and genomics 2015, 42, 343–353. [Google Scholar] [CrossRef]
- Arima, J.; Yoshino, H.; Fukumoto, W.; Kawahara, I.; Saito, S.; Li, G.; Enokida, H. LncRNA BCYRN1 as a Potential Therapeutic Target and Diagnostic Marker in Serum Exosomes in Bladder Cancer. International Journal of Molecular Sciences 2024, 25, 5955. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Shi, G.; Li, Q.; Li, W.; Zhou, H. Long noncoding RNA XIST participates in bladder cancer by downregulating p53 via binding to TET1. Journal of cellular biochemistry 2019, 120, 6330–6338. [Google Scholar] [CrossRef] [PubMed]
- Li, H.J.; Gong, X.; Li, Z.K.; Qin, W.; He, C.X.; Xing, L.; Cao, H.L. Role of long non-coding RNAs on bladder cancer. Frontiers in cell and developmental biology 2021, 9, 672679. [Google Scholar] [CrossRef]
- Zhou, X.; Xiao, L.; Meng, F.; Zuo, F.; Wu, W.; Li, G.; Shen, H. GAS6-AS1 drives bladder cancer progression by increasing MMP7 expression in a ceRNA-and RBP-dependent manner. Translational Oncology 2024, 48, 102065. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Qi, M.; Fei, X.; Wang, X.; Wang, K. Hsa_circRNA_0088036 acts as a ceRNA to promote bladder cancer progression by sponging miR-140-3p. Cell Death & Disease 2022, 13, 322. [Google Scholar]
- Lin, S.; Wang, L.; Shi, Z.; Zhu, A.; Zhang, G.; Hong, Z.; Cheng, C. Circular RNA circFLNA inhibits the development of bladder carcinoma through microRNA miR-216a-3p/BTG2 axis. Bioengineered 2021, 12, 11376–11389. [Google Scholar] [CrossRef]
- Yi, J.; Ma, X.; Ying, Y.; Liu, Z.; Tang, Y.; Shu, X.; Xie, L. N6-methyladenosine-modified CircPSMA7 enhances bladder cancer malignancy through the miR-128–3p/MAPK1 axis. Cancer Letters 2024, 585, 216613. [Google Scholar] [CrossRef]
- Oliver, J.; Garcia-Aranda, M.; Chaves, P.; Alba, E.; Cobo-Dols, M.; Onieva, J.L.; Barragan, I. Emerging noninvasive methylation biomarkers of cancer prognosis and drug response prediction. In: Seminars in cancer biology. Academic Press, 2022. p. 584-595.
- Yoon, H.Y.; Kim, Y.J.; Kim, J.S.; Kim, Y.W.; Kang, H.W.; Kim, W.T.; Kim, W.J. RSPH9 methylation pattern as a prognostic indicator in patients with non-muscle invasive bladder cancer. Oncology Reports 2016, 35, 1195–1203. [Google Scholar] [CrossRef]
- Zhan, L.; Zhang, B.; Tan, Y.; Yang, C.; Huang, C.; Wu, Q.; Shu, A. Quantitative assessment of the relationship between RASSF1A gene promoter methylation and bladder cancer (PRISMA). Medicine 2017, 96, e6097. [Google Scholar] [CrossRef]
- Shivakumar, M.; Lee, Y.; Bang, L.; Garg, T.; Sohn, K.A.; Kim, D. Identification of epigenetic interactions between miRNA and DNA methylation associated with gene expression as potential prognostic markers in bladder cancer. BMC medical genomics 2017, 10, 65–75. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, L.; Zang, Y.; Xu, Z. Identification of core genes and key pathways via integrated analysis of gene expression and DNA methylation profiles in bladder cancer. Medical science monitor: international medical journal of experimental and clinical research 2018, 24, 3024. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, J.; Xu, M.; Zhu, T. Exploring prognostic DNA methylation genes in bladder cancer: a comprehensive analysis. Discover Oncology 2024, 15, 331. [Google Scholar] [CrossRef]
- Takagi, K.; Naruse, A.; Akita, K.; Muramatsu-Maekawa, Y.; Kawase, K.; Koie, T.; Kikuchi, A. CALN1 hypomethylation as a biomarker for high-risk bladder cancer. BMC urology 2022, 22, 176. [Google Scholar] [CrossRef]
- Chen, J.Q.; Salas, L.A.; Wiencke, J.K.; Koestler, D.C.; Molinaro, A.M.; Andrew, A.S.; Christensen, B.C. Immune profiles and DNA methylation alterations related with non-muscle-invasive bladder cancer outcomes. Clinical Epigenetics 2022, 14, 14. [Google Scholar] [CrossRef]
- Zhang, C.; Shen, K.; Zheng, Y.; Qi, F.; Luo, J. Genome-wide screening of abberant methylated drivers combined with relative risk loci in bladder cancer. Cancer Medicine 2020, 9, 768–782. [Google Scholar] [CrossRef]
- Kim, C.; Oh, S.; Im, H.; Gim, J. Unveiling Bladder Cancer Prognostic Insights by Integrating Patient-Matched Sample and CpG Methylation Analysis. Medicina 2024, 60. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, J.; Zhang, Q.; Liang, Y.; Du, Y.; Wang, G. Identification of prognostic biomarkers for bladder cancer based on DNA methylation profile. Frontiers in cell and developmental biology 2022, 9, 817086. [Google Scholar] [CrossRef]
- Koukourikis, P.; Papaioannou, M.; Georgopoulos, P.; Apostolidis, I.; Pervana, S.; Apostolidis, A. A Study of DNA Methylation of Bladder Cancer Biomarkers in the Urine of Patients with Neurogenic Lower Urinary Tract Dysfunction. Biology 2023, 12, 1126. [Google Scholar] [CrossRef]
- Azzouzi, M.; El Ahanidi, H.; Alaoui, C.H.; Chaoui, I.; Benbacer, L.; Tetou, M.; Attaleb, M. Evaluation of DNA methylation in promoter regions of hTERT, TWIST1, VIM and NID2 genes in Moroccan bladder cancer patients. Cancer Genetics 2022, 260, 41–45. [Google Scholar] [CrossRef]
- Guo, Y.; Yin, J.; Dai, Y.; Guan, Y.; Chen, P.; Chen, Y.; Song, D. A novel CpG methylation risk indicator for predicting prognosis in bladder cancer. Frontiers in Cell and Developmental Biology 2021, 9, 642650. [Google Scholar] [CrossRef]
- Zhou, Q.; Chen, Q.; Chen, X.; Hao, L. Bioinformatics analysis to screen DNA methylation-driven genes for prognosis of patients with bladder cancer. Translational Andrology and Urology 2021, 10, 3604. [Google Scholar] [CrossRef] [PubMed]
- Mitash, N.; Agnihotri, S.; Tiwari, S.; Agrawal, V.; Mandhani, A. MicroRNA-21 could be a molecular marker to predict the recurrence of nonmuscle invasive bladder cancer. Indian Journal of Urology 2017, 33, 283–290. [Google Scholar] [PubMed]
- Chen, X.; Wu, B.; Xu, Z.; Li, S.; Tan, S.; Liu, X.; Wang, K. Downregulation of miR-133b predict progression and poor prognosis in patients with urothelial carcinoma of bladder. Cancer Medicine 2016, 5, 1856–1862. [Google Scholar] [CrossRef]
- Zeng, W.; Zhu, J.F.; Liu, J.Y.; Li, Y.L.; Dong, X.; Huang, H.; Shan, L. miR-133b inhibits cell proliferation, migration and invasion of esophageal squamous cell carcinoma by targeting EGFR. Biomedicine & Pharmacotherapy 2019, 111, 476–484. [Google Scholar]
- Avgeris, M.; Tsilimantou, A.; Levis, P.K.; Tokas, T.; Sideris, D.C.; Stravodimos, K.; Scorilas, A. Loss of GAS5 tumour suppressor lncRNA: an independent molecular cancer biomarker for short-term relapse and progression in bladder cancer patients. British journal of cancer 2018, 119, 1477–1486. [Google Scholar] [CrossRef]
- Jiao, R.; Jiang, W.; Wei, X.; Zhang, M.; Zhao, S.; Huang, G. Clinicopathological significance and prognosis of long noncoding RNA SNHG16 expression in human cancers: a meta-analysis. BMC cancer 2020, 20, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Luo, Y.; He, W.; Zhao, Y.; Kong, Y.; Liu, H.; Lin, T. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. The Journal of clinical investigation 2020, 130, 404–421. [Google Scholar] [CrossRef]
- Lin, G.; Sheng, H.; Xie, H.; Zheng, Q.; Shen, Y.; Shi, G.; Ye, D. circLPAR1 is a novel biomarker of prognosis for muscle-invasive bladder cancer with invasion and metastasis by miR-762. Oncology Letters 2019, 17, 3537–3547. [Google Scholar] [CrossRef]
- Liu, J.; Liu, T.; Wang, X.; He, A. Circles reshaping the RNA world: from waste to treasure. Molecular cancer 2017, 16, 1–12. [Google Scholar] [CrossRef]
- Andrew, A.S.; Karagas, M.R.; Schroeck, F.R.; Marsit, C.J.; Schned, A.R.; Pettus, J.R.; Seigne, J.D. MicroRNA Dysregulation and Non-Muscle–Invasive Bladder Cancer Prognosis. Cancer Epidemiology, Biomarkers & Prevention 2019, 28, 782–788. [Google Scholar]
- Miyamoto, K.; Seki, N.; Matsushita, R.; Yonemori, M.; Yoshino, H.; Nakagawa, M.; Enokida, H. Tumour-suppressive miRNA-26a-5p and miR-26b-5p inhibit cell aggressiveness by regulating PLOD2 in bladder cancer. British journal of cancer 2016, 115, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Borkowska, E.M.; Konecki, T.; Pietrusiński, M.; Borowiec, M.; Jabłonowski, Z. MicroRNAs which can prognosticate aggressiveness of bladder cancer. Cancers 2019, 11, 1551. [Google Scholar] [CrossRef] [PubMed]
- Juracek, J.; Stanik, M.; Vesela, P.; Radova, L.; Dolezel, J.; Svoboda, M.; Slaby, O. Tumor expression of miR-34a-3p is an independent predictor of recurrence in non–muscle-invasive bladder cancer and promising additional factor to improve predictive value of EORTC nomogram. In: Urologic Oncology: Seminars and Original Investigations. Elsevier, 2019. p. 184. e1-184. e7.
- Ding, Z.S.; He, Y.H.; Deng, Y.S.; Peng, P.X.; Wang, J.F.; Chen, X.; Zhou, X.F. MicroRNA-34a inhibits bladder cancer cell migration and invasion and upregulates PTEN expression. Oncology letters 2019, 18, 5549–5554. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Sun, H.F.; Guo, L.Q.; Cao, H.B. MiR-10a-5p: A promising biomarker for early diagnosis and prognosis evaluation of bladder cancer. Cancer Management and Research 2021, 7841–7850. [Google Scholar] [CrossRef] [PubMed]
- Bogale, D. E The roles of FGFR3 and c-MYC in urothelial bladder cancer. Discover Oncology 2024, 15, 295. [Google Scholar] [CrossRef]
- Yin, X.H.; Jin, Y.H.; Cao, Y.; Wong, Y.; Weng, H.; Sun, C.; Zeng, X.T. Development of a 21-miRNA signature associated with the prognosis of patients with bladder cancer. Frontiers in oncology 2019, 9, 729. [Google Scholar] [CrossRef]
- Yerukala Sathipati, S.; Tsai, M.J.; Shukla, S.K.; Ho, S.Y.; Liu, Y.; Beheshti, A. MicroRNA signature for estimating the survival time in patients with bladder urothelial carcinoma. Scientific reports 2022, 12, 4141. [Google Scholar] [CrossRef]
- Zhu, Q.L.; Zhan, D.M.; Chong, Y.K.; Ding, L.; Yang, Y.G. MiR-652-3p promotes bladder cancer migration and invasion by targeting KCNN3. European Review for Medical & Pharmacological Sciences 2019, 23. [Google Scholar]
- Chen, C.; Zheng, H.; Luo, Y.; Kong, Y.; An, M.; Li, Y.; Lin, T. UMOylation promotes extracellular vesicle–mediated transmission of lncRNA ELNAT1 and lymph node metastasis in bladder cancer. The Journal of Clinical Investigation 2021, 131. [Google Scholar]
- Liang, T.; Xu, F.; Wan, P.; Zhang, L.; Huang, S.; Yang, N.; Wang, Y. Malat-1 expression in bladder carcinoma tissues and its clinical significance. American Journal of Translational Research 2021, 13, 3555. [Google Scholar]
- Zheng, H.; Chen, C.; Luo, Y.; Yu, M.; He, W.; An, M.; Lin, T. Tumor-derived exosomal BCYRN1 activates WNT5A/VEGF-C/VEGFR3 feedforward loop to drive lymphatic metastasis of bladder cancer. Clinical and translational medicine 2021, 11, e497. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Fujita, K.; Netto, G.J.; Nonomura, N. Clinical application of TERT promoter mutations in urothelial carcinoma. Frontiers in oncology 2021, 11, 705440. [Google Scholar] [CrossRef]
- Xie, F.; Zhao, N.; Zhang, H.; Xie, D. Circular RNA CircHIPK3 promotes gemcitabine sensitivity in bladder cancer. Journal of Cancer 2020, 11, 1907. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.X.; Lu, J.; Xie, H.; Wang, D.P.; Ni, H.E.; Zhu, Y.; Wang, R.L. circHIPK3 regulates lung fibroblast-to-myofibroblast transition by functioning as a competing endogenous RNA. Cell death & disease 2019, 10, 182. [Google Scholar]
- Su, Y.; Feng, W.; Shi, J.; Chen, L.; Huang, J.; Lin, T. circRIP2 accelerates bladder cancer progression via miR-1305/Tgf-β2/smad3 pathway. Molecular cancer 2020, 19, 1–13. [Google Scholar] [CrossRef]
- Zhu, J.; Luo, Y.; Zhao, Y.; Kong, Y.; Zheng, H.; Li, Y.; Chen, C. circEHBP1 promotes lymphangiogenesis and lymphatic metastasis of bladder cancer via miR-130a-3p/TGFβR1/VEGF-D signaling. Molecular Therapy 2021, 29, 1838–1852. [Google Scholar] [CrossRef] [PubMed]
- Zhenhai, Z.; Qi, C.; Shuchao, Z.; Zhongqi, W.; Xue, S.; Zhijun, G.; Yuanyuan, G. MiR-205-3p suppresses bladder cancer progression via GLO1 mediated P38/ERK activation. BMC cancer 2023, 23, 956. [Google Scholar] [CrossRef]
- Hao, Y.; Zhu, Y.; Sun, F.; Xu, D.; Wang, C. MicroRNA-30c-5p arrests bladder cancer G2/M phase and suppresses its progression by targeting PRC1-mediated blocking of CDK1/Cyclin B1 axis. Cellular Signalling 2023, 110, 110836. [Google Scholar] [CrossRef]
- Awadalla, A.; Abol-Enein, H.; Hamam, E.T.; Ahmed, A.E.; Khirallah, S.M.; El-Assmy, A.; Harraz, A.M. Identification of epigenetic interactions between miRNA and gene expression as potential prognostic markers in bladder cancer. Genes 2022, 13, 1629. [Google Scholar] [CrossRef]
- Zhang, Z.; Sang, Y.; Liu, Z.; Shao, J. Negative correlation between circular RNA SMARC5 and MicroRNA 432, and their clinical implications in bladder cancer patients. Technology in Cancer Research & Treatment 2021, 20, 15330338211039110. [Google Scholar]
- Spagnuolo, M.; Costantini, M.; Ferriero, M.; Varmi, M.; Sperduti, I.; Regazzo, G.; Rizzo, M.G. Urinary expression of let-7c cluster as non-invasive tool to assess the risk of disease progression in patients with high grade non-muscle invasive bladder Cancer: A pilot study. Journal of Experimental & Clinical Cancer Research 2020, 39, 1–11. [Google Scholar]
- Braicu, C.; Buiga, R.; Cojocneanu, R.; Buse, M.; Raduly, L.; Pop, L.A.; Berindan-Neagoe, I. Connecting the dots between different networks: miRNAs associated with bladder cancer risk and progression. Journal of Experimental & Clinical Cancer Research 2019, 38, 1–17. [Google Scholar]
- Chen, H.; Xu, L.; Wang, L. Expression of miR-182 and Foxo3a in patients with bladder cancer correlate with prognosis. International Journal of Clinical and Experimental Pathology 2019, 12, 4193. [Google Scholar]
- Yang, K.; Tang, H.; Ding, M.; Guo, Y.; Kai, K.; Xiao, J.; Zhou, R. Expression of miR-195 and MEK1 in patients with bladder cancer and their relationship to prognosis. International Journal of Clinical and Experimental Pathology 2019, 12, 843. [Google Scholar] [PubMed]
- Li, D.; Hao, X.; Song, Y. An integrated analysis of key microRNAs, regulatory pathways and clinical relevance in bladder cancer. OncoTargets and therapy 2018, 3075–3085. [Google Scholar] [CrossRef]
- Martins, E.P.; Vieira de Castro, J.; Fontes, R.; Monteiro-Reis, S.; Henrique, R.; Jerónimo, C.; Costa, B.M. Relevance of HOTAIR rs920778 and rs12826786 Genetic Variants in Bladder Cancer Risk and Survival. Cancers 2024, 16, 434. [Google Scholar] [CrossRef]
- Novikova, E.L.; Kulakova, M.A. There and back again: hox clusters use both DNA strands. Journal of developmental biology 2021, 9, 28. [Google Scholar] [CrossRef]
- Xia, W.; Chen, C.; Zhang, M.R.; Zhu, L.N. LncRNA PCAT6 aggravates the progression of bladder cancer cells by targeting miR-513a-5p. Eur Rev Med Pharmacol Sci 2020, 24, 9908–9914. [Google Scholar]
- Zhang, S.; Du, L.; Wang, L.; Jiang, X.; Zhan, Y.; Li, J.; Wang, C. Evaluation of serum exosomal Lnc RNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. Journal of cellular and molecular medicine 2019, 23, 1396–1405. [Google Scholar] [CrossRef]
- Luo, L.; Miao, P.; Ming, Y.; Tao, J.; Shen, H. Circ-ZFR promotes progression of bladder cancer by upregulating WNT5A via sponging miR-545 and miR-1270. Frontiers in oncology 2021, 10, 596623. [Google Scholar] [CrossRef]
- Chen, X.; Chen, R.X.; Wei, W.S.; Li, Y.H.; Feng, Z.H.; Tan, L.; Xie, D. PRMT5 Circular RNA Promotes Metastasis of Urothelial Carcinoma of the Bladder through Sponging miR-30c to Induce Epithelial–Mesenchymal Transition. Clinical Cancer Research 2021, 27, 2664–2664. [Google Scholar] [CrossRef] [PubMed]
| Authors | Test Name | Target biomarker |
Method | Study material |
Diagnostic performance |
Study group (n) | Race/ Nationality |
|---|---|---|---|---|---|---|---|
| Bang et al. (2024) [56] | EarlyTest BCD | PENK | qPCR | Voided urine | SN=81% SP=91,5% |
210 (21 BC, 189 non-BC based on cystoscopy) | Korean, American |
| Pharo et al. (2022) [54] | BladMetrix | 8 methylated gene regions | ddPCR | Urinary exfoliated cell DNA | SN=92,1% SP=93,3% | 273 (with gross hematuria) (93 BC) | European |
| Piatti et al. (2021) [55] | Bladder CARE | DNA methylation in TRNA-Cys, SIM2 and NKX1 | Methylation-sensitive restriction enzymes with qPCR | Urine | SN=93,5% SP=92,6% | 213 (77 BC, 136 non-BC) | Caucasian, Asian, African American, Hispanic |
| Steinbach et al. (2020) [57] | GynTect | DNA methylation in ASTN1, DLX1, ITGA4, RXFP3, SOX17 and ZNF671 | GynTect Assay | Urine | SN=60% SP=96,7% |
70 (40 BC, 15 BPH, 15 Urolithiasis) | German |
| Witjes et al. (2018) [52] | Bladder EpiCheck | 15 methylated gene regions | qPCR | Urinary exfoliated cell DNA | SN=68,2% SP=88% |
353 (UC) | Caucasian |
| Feber et al. (2017) [53] | UroMark | 150 CpG loci methylation | Targeted bisulfite sequencing | Urinary exfoliated cell DNA | SN=98% SP=97% |
274 (107 BC, 167 non-BC) | English |
| Authors | Study biomarker |
Methylation Status |
Study material |
Diagnostic performance |
Study group (n) | Race/ Nationality |
|---|---|---|---|---|---|---|
| Zhang et al. (2024) [64] | TWIST1, VIM | Methylation level analysis | Urine cells sediment | SN=78% SP=83% | 231 (77 BC, 81 other urological malignancies, 19 benign disease, 26 UTUC, 28 healthy) | China |
| Jiang et al. (2024) [61] |
ZNF671, OTX1, IRF8 | Hypermethylation | Voided urine | SN=75% SP=90,9% | 114 (61 BC, 53 non-BC) | Taiwan |
| Fang et al. (2022) [58] |
PCDH17, POU4F2, PENK | Hypermethylation | Urine cells sediment | SN=87% SP=97% | 207 (107 BC, 100 non-BC) | China |
| Hentschel et al. (2022) [60] |
GHSR, MAL | Hypermethylation | Urine pellet | SN=80% SP=93% | 208 (108 BC, 34 benign hematuria, 43 other benign urological conditions, 23 healthy) | Dutch |
| Deng et al. (2022) [65] | DMRTA2 | Hypermethylation | Urine | SN=82,9% SP=92,5% | 520 (79 BC, 107 other malignancies, 22 benign tumors of bladder8 recurring cancers, 304 healthy | China |
| Ruan et al. (2021) [66] | ONECUT2, VIM | Hypermethylation | Urine | Cohort 1: SN=88,1% SP=89,7% Cohort 2: SN=91,2% SP=85.7% |
Cohort 1: 98 (patients suspected of BC) (59 BC, 39 Non-Bc); Cohort 2: 174(hematuria patients) (34BC, 140 non-BC) |
China |
| Wu et al. (2020) [62] | ONECUT2, HOXA9, PCDH17, POU4F2 | Hypermethylation | Urine | SN=90,5% SP=73,2% | 111 (53 BC, 58 non-BC) | China |
| Chen et al. (2020) [63] |
OTX1, SOX1-OT | Methylation level analysis | Urine | SN=91,7% SP=77,3% | 175 (109 BC, 66 benign diseases) | China |
| Guo et al. (2018) [67] | VIM, CDH1, SALL3, THBS1, TMEFF2, GDF15 | Methylation level analysis | Voided urine | SN=89% SP=74% | 473 (217 UC, 256 controls) | China |
| Roperch et al. (2016) [68] |
SEPTIN9+SLIT2 | Hypermethylation | Urine | SN=91% SP=71.4% | 272 (167 NMIBC, 105 controls) | France |
| Yen et al. (2015) [69] |
ZNF671, SFRP1, IRF8 | Hypermethylation | Urine | SN=96.2% SP=84.2% | 45 (26 UC, 19 non-cancerous) | Taiwan |
| Yegin et al. (2013) [70] | TWIST1, NID2 | Hypermethylation | Urine |
TWIST1: SN=87.5% SP=93.3% NID2: SN=95.8% SP=100% |
39 (24 BC, 15 non-cancerous) | Turkey |
| Authors | Study biomarker |
Biomarker change |
Biomarker targets | Study Material | Diagnostic performance |
Study group (n) | Race/ Nationality |
|---|---|---|---|---|---|---|---|
| miRNA | |||||||
| Lu et al. (2024) [82] | miR-221-5p, miR-181a-5p, miR-15a-5p, miR-222-3p | Aberrantly expressed |
Not specified | Serum | SN=82.1% SP=85.7% | 224 (112 BC, 112 controls) | China |
| Yu et al. (2023) [73] |
miR-27b, miR-381-3p, miR-451a |
Overexpression: miR-27b Underexpression: miR-381-3p, miR-451a |
SMAD4, FOXO1 | Serum | SN=86.7% SP=77.4% | 224 (112 BC, 112 healthy) | China |
| Mamdouh et al. (2023) [91] *Zhang et al. (2018) [92] **Zhang et al. (2015) [93] |
miR-200 | Overexpression | Not specified | Urine and tissue |
Tissue: SN=93,3% SP=100% Urine: SN=62,2% SP=100% |
136 (111 BC, 25 healthy) |
Egyptian |
| miR-145 | N-cadherin* |
Tissue: SN=80% SP=100% Urine: SN=78,4% SP=91,7% |
|||||
| miR-21 | maspin, VEGF-C** |
Tissue: SN=73.3% SP=80% Urine: SN=83.3% SP=100% |
|||||
| SurezCabrera et al. (2022) [74] |
miR-145, miR-182 |
Overexpression (miR-145), Underexpression (miR-182) |
FCS1 | Urine | SN=93% SP=86% |
82 (40 BC, 42 controls) |
European |
| Wang et al. (2021) [71] | miR-20a-5p, miR-92a-3p, miR-17-5p | Overexpression | p21, PTEN | Serum | SN=90.4% SP=94.4% | 164 (74BC, 90 healthy) |
China |
| El-Shal et al. (2021) [75] | miR-96-5p, miR-183-5p | Overexpression | FOXO, KRAS PDCD4 | Voided urine |
miR-96 alone: SN=80.4% SP=91.8% miR-183 alone: SN=78.4% SP=81.6% both combined: SN=88.2% SP=87.8% |
100 (51 BC, 21 benign bladder lesions, 28 healthy) |
Egyptian |
| Lin et al. (2021) [79] | miR-516a-5p, miR-93-5p | Overexpression | miR-516a-5p (not specified) miR-93-5p: PEDF, EGFR, FoxO pathway, PI3K-Akt pathway, BTG2 | Midstream urine |
miR-93-5p alone: SN=74.1% SP=90.2% miR-516a-5p alone: SN=72.9% SP=89.9% both combined: SN=85.2% SP=82.4% |
104 (53 BC, 51 healthy) |
China |
| Piao et al. (2019) [94] | miR-6124, miR-4511 | Aberrantly expressed |
Not specified | urine | SN=91.5% SP=76.2% (ratio miR-6124 to miR-4511) |
543 (326 BC, 174 hematuria, non-BC pyuria) | South Korea |
| Usuba et al. (2018) [95] |
miR-6087, miR-6724-5p, miR-3960, miR-1343-5p, miR-1185-1-3p, miR-6831-5p, miR-4695-5p combined |
miR-4695-5p: no significant change Underexpression: miR6087, miR-3960, miR-1343-5p Overexpression: miR-6724-5p, miR-1185-1-3p, miR-6831-5p |
Not specified | serum | SN=95% SP=87% |
972 (392 BC, 100 non-BC, 480 other cancers) |
Japan |
| Huang et al. (2018) [96] |
miR-20a | Overexpression | Not specified | Urine | SN=72.1% SP=87.5% | 166 (80 NMIBC, 86 healthy) | China |
| Matsuzaki et al.e (2017) [80] |
miR-21-5p | Overexpression | Not specified | Urine | SN=75% SP=98% |
60 (24 controls, 36 UC) |
Japan |
| Urquidi et al. (2016) [97] | 25 mi-RNAs combined | Aberrantly expressed |
Not specified | Midstream urine |
SN=87% SP=100% |
121 (61 cases, 60 controls) |
USA |
| Zhang et al. (2016) [98] | miR-155 | Overexpression | APC, VHL, PIK3R1, MLH1 | Voided urine | SN=80.2% SP=84.6% |
324 (162 NIMBC, 86 cystitis, 76 healthy) |
China |
| Zhang et al. (2014) [81] | miR-99a, miR-125b | Underexpression |
SMARCA5, SMARCD1, mTOR kinase, STAT3, E2F3 |
Urine supernatant |
SN=86.7% SP=81.1% | 71 (50 UCB, 21 controls) | China |
| Miah et al. (2012) [99] | miR-1224-3p | Overexpression | Not specified | Urine sediment |
SN=75.9% SP=82.4% | 121 (68 BC, 53 controls) | UK |
| lncRNA | |||||||
| Gao et al. (2024) [87] | RMRP | Overexpression | Not specified | Urine | SN=83% SP=70% (RT-qPCR) SN=95% SP=92.5% (RT-RAA- CRISPR/Cas12a) |
339 (229 BC, 110 benign lesions) | China |
| Liu et al. (2023) [100] | SNHG16 | Overexpression |
possibly Wnt/β-catenin pathway |
Urine | SN=61.9% SP=83.3% | 84 (42 BC, 42 Healthy) | China |
| Chen et al. (2022) [88] | TERC | Overexpression | Not specified | Urine | SN=78.7% SP=77.8% | 152 (89 BLCA, 63 Healthy) | China |
| Bian et al. (2022) [101] |
MKLN1-AS | Overexpression |
Not specified | Urine |
SN=79.1% SP=67.4% | 92 (46 BC, 46 Controls) | China |
| TALAM1 | Not specified |
SN=90.1% SP=55.8% |
|||||
| TTN-AS1 | Not specified |
SN=76.7% SP=76.7% |
|||||
| UCA1 |
PI3K-Akt-mTOR pathway, GLS2, HMGB1 p21 |
SN=90.7% SP=90.7% |
|||||
| Sarfi et al. (2021) [102] | TUG-1 | Overexpression |
Not specified |
Voided urine | SN=76.7% SP=77.8% | 40 (30 NMIBC, 10 Controls) | Iran |
| Yu et al. (2020) [86] | UCA-1-201, HOTAIR, HYMA1, MALAT1 | Overexpression: HOTAIR, MALAT1 Underexpression: UCA-1-201, HYMA1 |
Not specified | Urine |
SN=93.3% SP=96.7% |
120 (60 Urocystitis, 60 NMIBC) |
China |
| Zhan et al. (2018) [85] | MALAT1, PCAT-1, SPRY4-IT1 | Overexpression | Not specified | Urine | SN=62.5% SP=85.6% | 208 (104 BC, 104 healthy) | China |
| other ncRNA | |||||||
| Yang et al. (2024) [89] | circRNA-0071196 | Overexpression | CIT, miR-19b-3p, | Urine | SN=87.5% SP=85% |
70 (40 BUC, 30 non-BUC) | China |
| Luo et al. (2023) [103] | circRNA CCT3 | Overexpression | PP2A, miR-135a-5p | Plasma | SN=86.1% SP=60% |
125 (85 BC, 40 healthy) | China |
|
Song et al. (2020) [90] |
hsa_circ0137439 |
Overexpression | miR-142-5p | Urine | SN=87.9 SP=80.1% (BC vs. Controls) SN=88.6% SP=73.5% (NMIBC vs. MIBC) |
146 (62 NMIBC, 54 MIBC, 30 controls) |
China |
| Tang et al. (2017) [104] | circASXL1 | Overexpression | Not specified | Tumor tissue | SN=68.6% SP=76.9% | 61 pairs of tumor tissue and adjacent normal mucosa | China |
| Authors | Study biomarker |
Methylation status |
Study material |
Assessed Study Endpoints (Associated Change) |
Study group (n) |
Race/ Nationality |
|---|---|---|---|---|---|---|
| Kim et al. (2024) [133] | PTK2 | hypermethylation | Tissue | OS (↓) | BC patients (n=275) healthy donors (n=10) |
Republic of Korea |
| Zhang et al. (2024) [129] |
AHNAK | hypermethylation | Tissue | OS (↓) | BC patients (n=812) | China |
| Koukourikis et al. (2023) [135] |
DAPK | hypermethylation | Urine | OS (↓) | BC patients (n=414) healthy donors (n=10) |
Greece |
| El Azzouzi et al. (2022) [136] |
TWIST1 | hypermethylation | Tissue | PFS (↓) | BC patients (n=70) | Morocco |
| Takagi et al. (2022) [130] |
CALN1 | hypomethylation | Tissue | PFS (↓) | BC patients (n=82) | Japan |
| Zhang et al. (2022) [134] |
FASLG, PRKCZ | hypermethylation | Tissue | PFS (↓) | BC patients (n=408) healthy donors (n=14) |
China |
| Chen et al. (2022) [131] | BLCAP | hypomethylation | Peripheral blood | PFS (↓) OS (↓) | BC patients (n=603) |
USA |
| Guo et al. (2021) [137] | TNFAIP8L3 | hypomethylation | Tissue | PFS (↓) OS (↓) | BC patients (n=357) |
China |
| Zhou et al. (2021) [138] | PTPRN2 | hypomethylation | Tissue | OS (↓) | BC patients (n=399) |
China |
| Guo et al. (2021) [137] |
APC | hypermethylation | Tissue | PFS (↓) | BC patients (n=357) |
China |
| Zhang et al. (2020) [128] |
KRT8 | hypomethylation | Tissue | OS (↓) | BC patients (n=41) healthy donors (n=35) |
China |
| Zhang et al. (2018) [128] |
CDH1 | hypomethylation | Tissue | OS (↓) | BC patients (n=167) healthy donors (n=13) |
China |
| Zhan et al. (2017) [126] | RASSF1A | hypermethylation | Tissue | PFS (↓) OS (↓) | BC patients (n=389) |
China |
| Shivakumar et al. (2017) [127] |
NACC2 | hypomethylation | Tissue | OS (↓) | BC patients (n=403) | USA |
| Yoon et al. (2016) [125] | RSPH9 | hypermethylation | Tissue | PFS (↓) | BC patients (n=128) healthy donors (n=8) |
Republic of Korea |
| Authors | Study biomarker |
Biomarker change |
Biomarker targets | Study Material | Assessed Study Endpoints (Associated Change) |
Study group (n) | Race/ Nationality |
|---|---|---|---|---|---|---|---|
| miRNA | |||||||
| Zhenhai et al. (2023) [165] | miR-205-3p | Downregulated | GLO1 | Tissue | PFS (↓) OS (↓) | BC patients (n=35) |
China |
| Hao et al. (2023) [166] | miR-30c-5p | Downregulated | PRC1 | Tissue | OS (↓) | BC patients (n=445) |
China |
| Awadalla et al. (2022) [167] |
miR-138 | Downregulated | HIF1α | Tissue | CSS (↓) | BC patients (n=157) |
Egypt |
| Awadalla et al. (2022) [167] |
miR-let-7a | Downregulated | WNT7A | Tissue | CSS (↓) | BC patients (n=157) |
Egypt |
| Yerukala et al. (2022) [155] |
miR-652-5p | Upregulated | KCNN3 | Tissue | OS (↓) | BC patients (n=106) |
USA |
| Zhang et al. (2021) [168] |
miR-432 | Downregulated | SMARCA5 | Tissue | OS (↓) | BC patients (n=156) | China |
| Yang et al. (2021) [152] Borkowska et al. (2019) [149] |
miR-10a-5p | Upregulated | FGFR3 | Plasma | OS (↓) | BC patients (n=88) healthy donors (n=36) |
China |
| Andrew et al. (2019) [147] | miR-26b-5p | Downregulated | PLOD2 | Tissue | PFS (↓) | BC patients (n=231) | Lebanon |
| Spagnuolo et al. (2020) [169] |
miR-let-7c-5p | Upregulated | HRAS | Urine | PFS (↓) | BC patients (n=57) healthy donors (n=20) |
Italy |
| Borkowska et al. (2019) [149] | miR-21-5p | Upregulated | TP53 | Tissue | OS (↓) | BC patients (n=55) healthy donors (n=30) |
Poland |
| Braicu et al. (2019) [170] | miR-143 | Downregulated | TP53 | Tissue | OS (↓) | BC patients (n=409) healthy donors (n=19) |
Romania |
| Chen et al. (2019) [171] | miR-182 | Upregulated | FOXO3a | Tissue | OS (↓) | BC patients (n=60) healthy donors (n=20) |
China |
| Juracek et al. (2019) [150] Ding et al. (2019) [151] |
miR-34a-3p | Downregulated | PTEN | Tissue | OS (↓) | BC patients (n=78) | Czech Republic |
| Yin et al. (2019) [154] | miR-185 | Upregulated | ITGB5 | Tissue | OS (↓) | BC patients (n=408) healthy donors (n=19) |
China |
| Yang et al. (2019) [172] | miR-195 | Downregulated | MEK1 | Tissue | OS (↓) | BC patients (n=80) healthy donors (n=30) |
China |
| Li et al. (2018) [173] | miR-145 | Upregulated | CDK4 | Tissue | OS (↓) | BC patients (n=127) |
China |
| Mitash et al. (2017) [139] | miR-21 | Upregulated | PTEN | Tissue | RFS (↓) | BC patients (n=31) |
India |
| Zhang et al. (2016) [98] | miR-155 | Upregulated | APC | Urine | RFS (↓) PFS (↓) | BC patients (n=162) healthy donors (n=86) |
China |
| Chen et al. (2016) [140] | miR-133b | Downregulated | EGFR | Tissue | PFS (↓) OS (↓) | BC patients (n=146) | China |
| lncRNA | |||||||
| Martins et al. (2024) [174] Novikova et al. (2021) [175] | lncRNA-HOX | Upregulated | HOXD | Peripheral blood | OS (↓) | BC patients (n=106) healthy donors (n=199) |
Portugal |
| Chen et al. (2022) [88] | lncRNA-TERC | Upregulated | TERT | Tissue | OS (↓) | BC patients (n=89) healthy donors (n=63) |
China |
| Chen et al. (2021) [157] | lncRNA-ELNAT1 | Upregulated | UBC9 | Urine | OS (↓) | BC patients (n=242) |
China |
| Liang et al. (2021) [158] | lncRNA-MALAT1 | Upregulated | MDM2 | Tissue | OS (↓) | BC patients (n=90) |
China |
| Zheng et al. (2021) [159] | lncRNA-BCYRN1 | Upregulated | WNT5A | Tissue | OS (↓) | BC patients (n=210) |
China |
| Jiao et al. (2020) [143] | lncRNA-SNHG16 | Upregulated | CCL5 | Tissue | OS (↓) | BC patients (n=1148) |
China |
| Chen et al. (2020) [144] | lncRNA-LNMAT2 | Upregulated | PROX1 | Tissue | OS (↓) | BC patients (n=266) |
China |
| Xia et al. (2020) [176] | lncRNA-PCAT6 | Upregulated | MIR513A1 | Tissue | OS (↓) | BC patients (n=21) healthy donors (n=21) |
China |
| Zhang et al. (2019) [177] | lncRNA-UBC1 | Upregulated | PRC2 | Serum | RFS (↓) | BC patients (n=260) healthy donors (n=260) |
China |
| Avgeris et al. (2018) [142] | lncRNA-GAS5 | Downregulated | CDK6 | Tissue | PFS (↓) | BC patients (n=176) | Greece |
| other ncRNA | |||||||
| Luo et al. (2023) [103] | circRNA-CCT3 | Upregulated | PP2A | Urine | RFS (↓) OS (↓) | BC patients (n=85) healthy donors (n=40) |
China |
| Luo et al. (2021) [178] | circRNA-ZFR | Upregulated | WNT5A | Tissue | OS (↓) | BC patients (n=60) |
China |
| Chen et al. (2021) [179] | circRNA-PRMT5 | Upregulated | SNAIL1 | Tissue | OS (↓) | BC patients (n=119) |
China |
| Zhu et al. (2021) [164] | circRNA-EHBP1 | Upregulated | TGFβR1 | Tissue | OS (↓) | BC patients (n=186) |
China |
| Xie et al. (2020) [161] Zhang et al. (2019) [162] | circRNA-HIPK3 | Downregulated | SOX4 | Tissue | OS (↓) | BC patients (n=68) |
China |
| Su et al. (2020) [163] | circRNA-RIP2 | Downregulated | SMAD3 | Tissue | OS (↓) | BC patients (n=58) |
China |
| Lin et al. (2019) [145] | circRNA-LPAR1 | Downregulated | WNT5A | Tissue | DSS (↓) | BC patients (n=125) |
China |
| Tang et al. (2017) [104] | circRNA-ASXL1 | Upregulated | TP53 | Tissue | OS (↓) | BC patients (n=61) | China |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





